Function and Molecular Mechanism of Circhomer1 in Myogenesis
Abstract
1. Introduction
2. Results
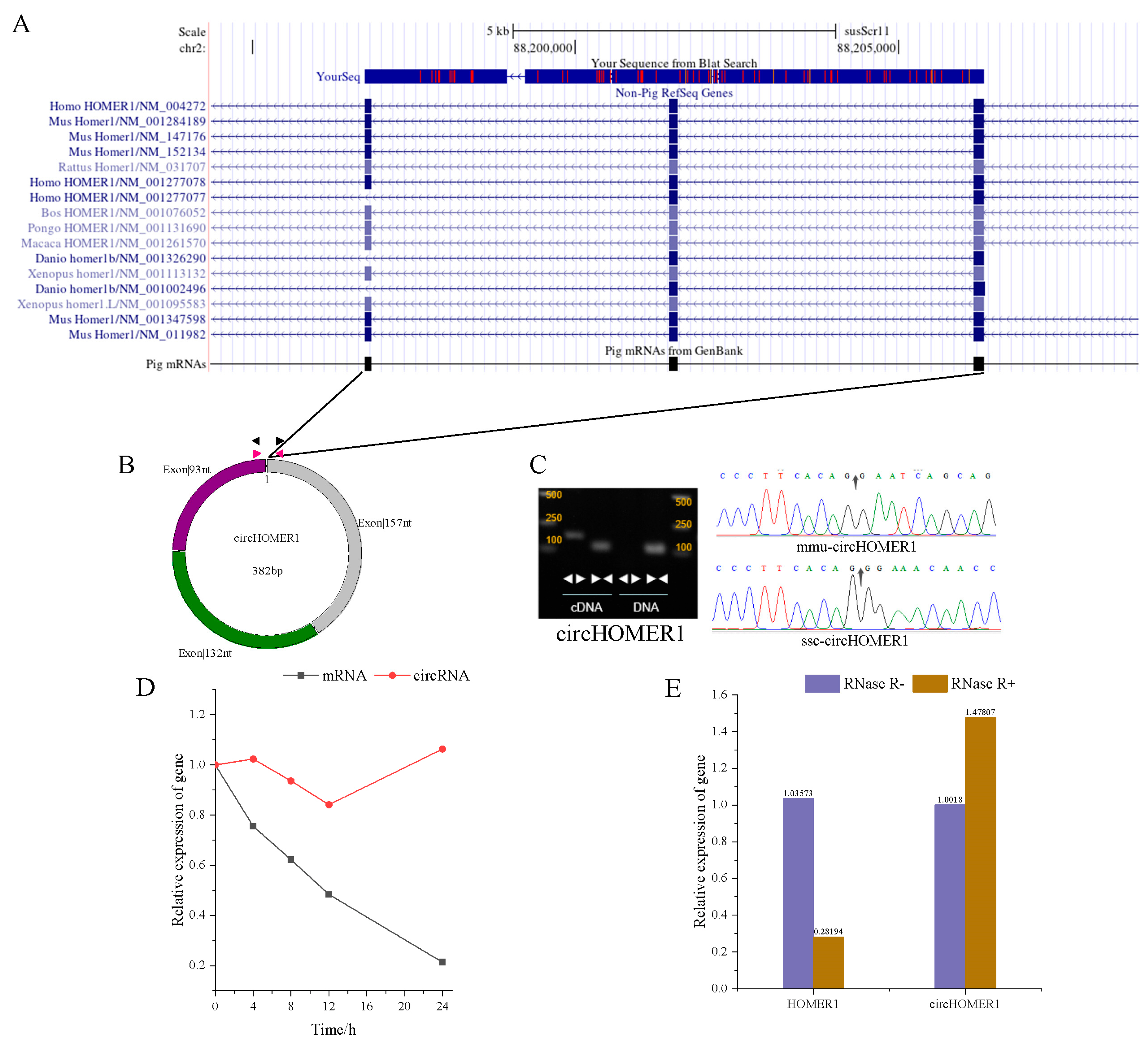
2.1. Characteristics Analysis of circHOMER1
2.2. Homologous Transformation of circHOMER1
2.3. Detection of Expression Pattern of circHOMER1
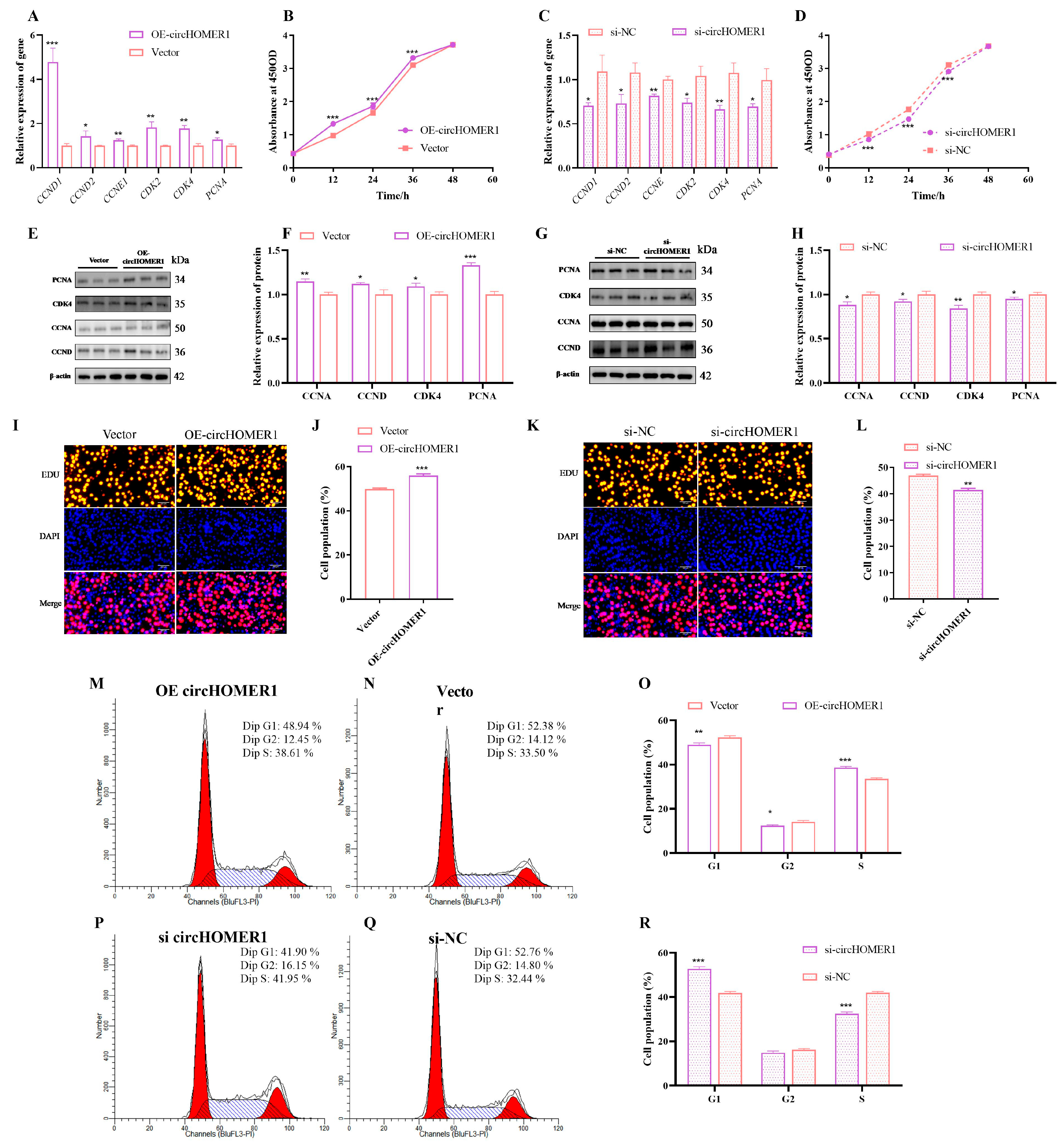
2.4. Detection Overexpression and Interference Efficiency for circHOMER1
2.5. circHOMER1 Promotes the Proliferation of Myoblasts
2.6. circHOMER1 Promotes Myoblast Apoptosis
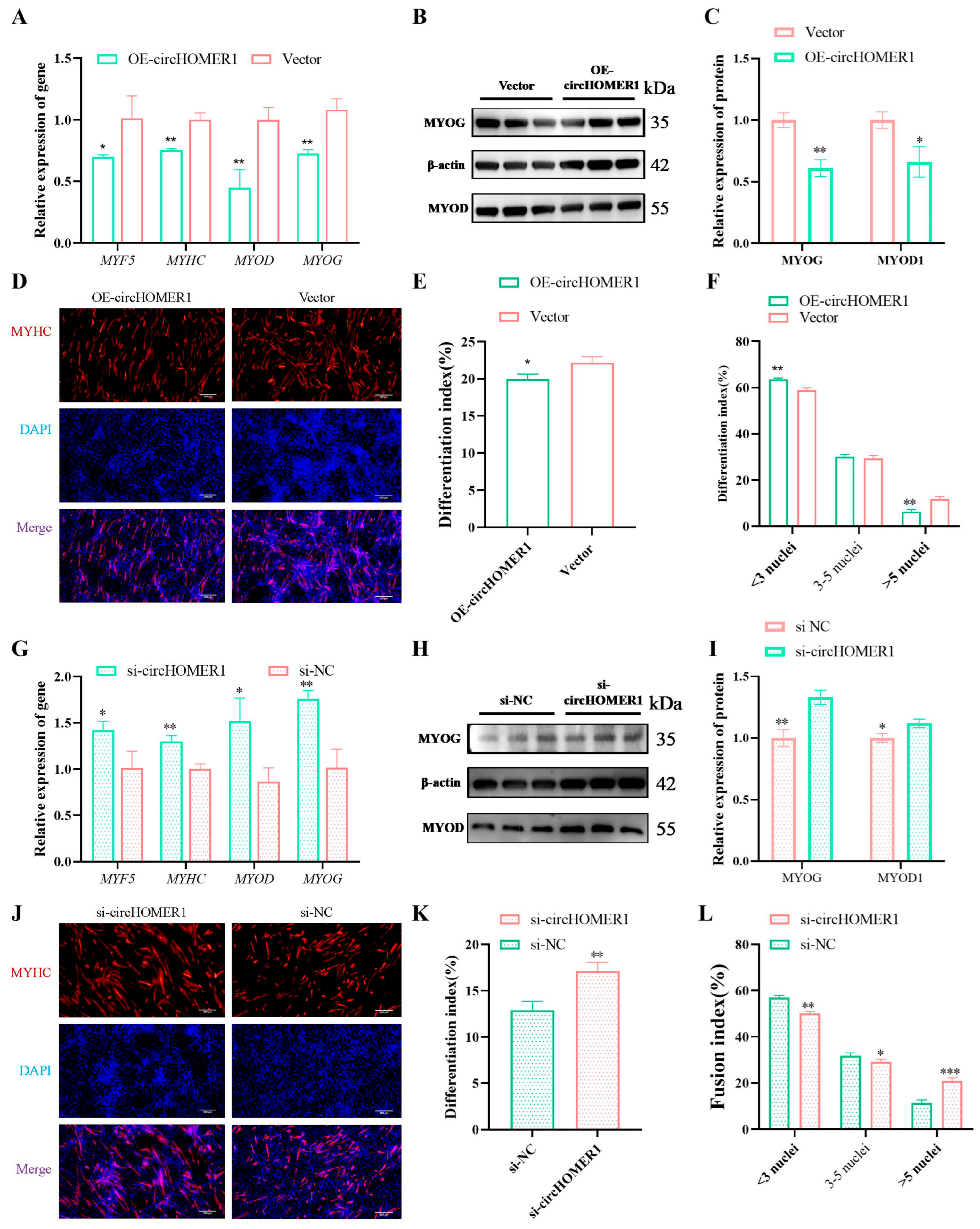
2.7. circHOMER1 Inhibits Myoblast Differentiation
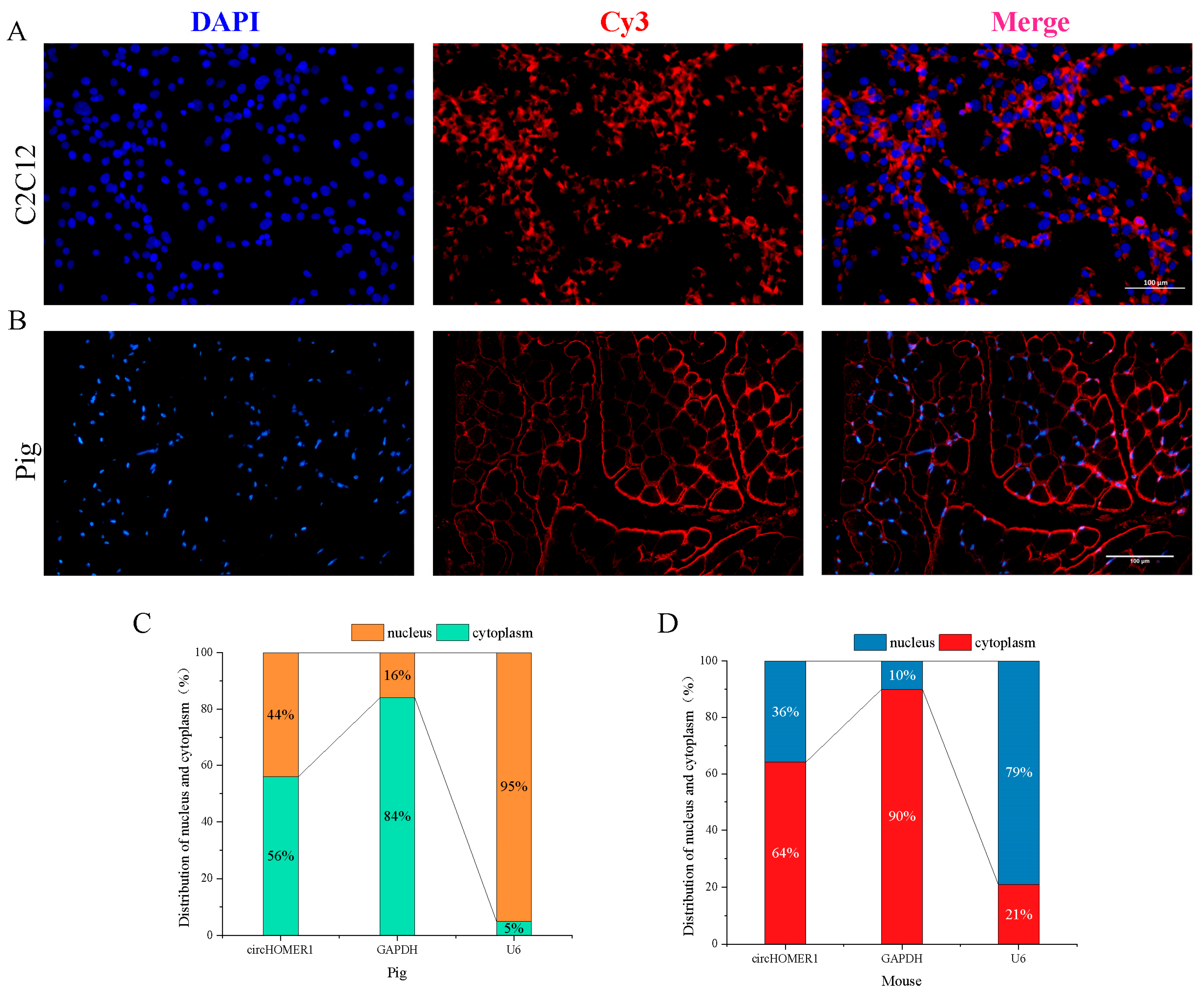
2.8. Subcellular Localization Analysis of circHOMER1
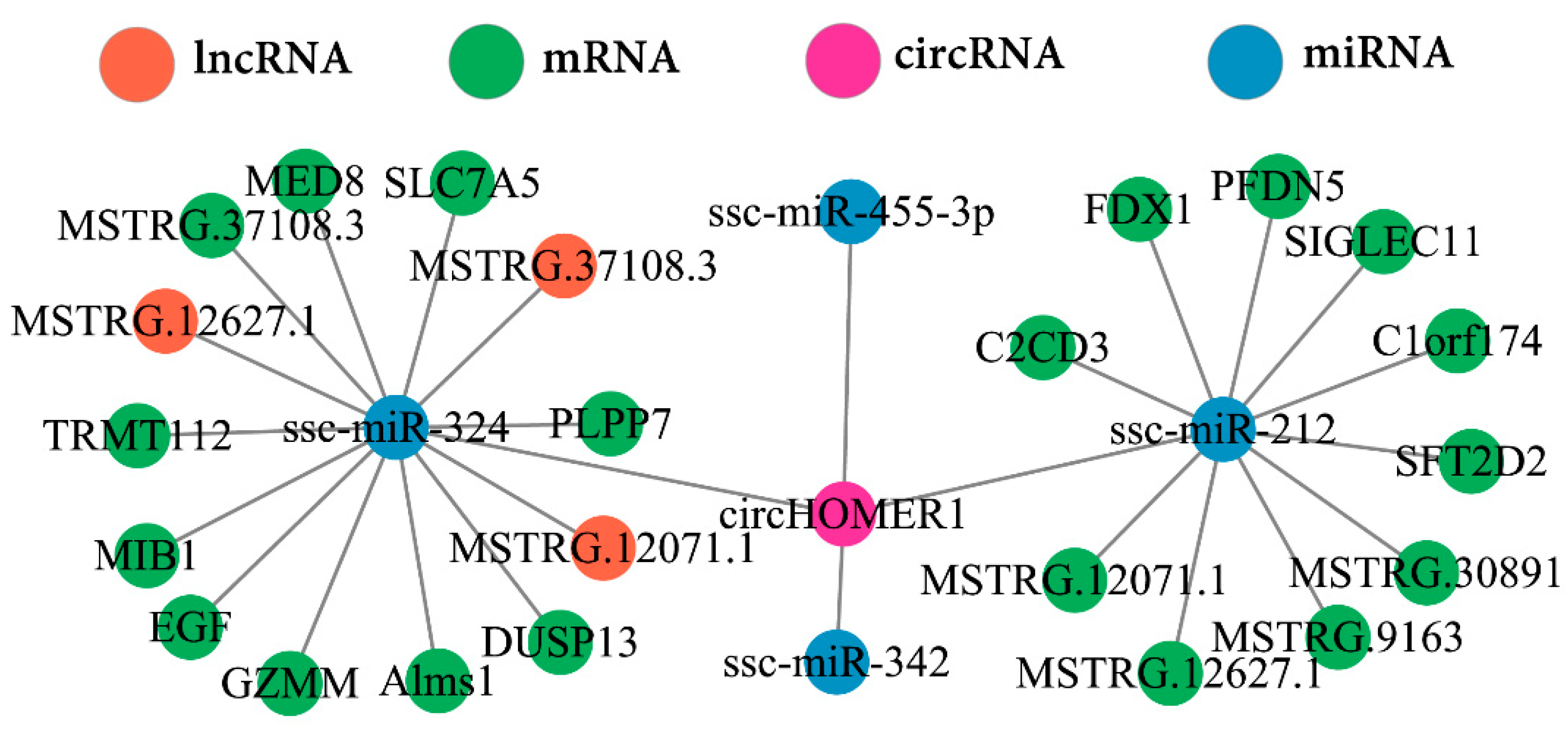
2.9. Construction and Verification of ceRNA Network Mediated by circHOMER1
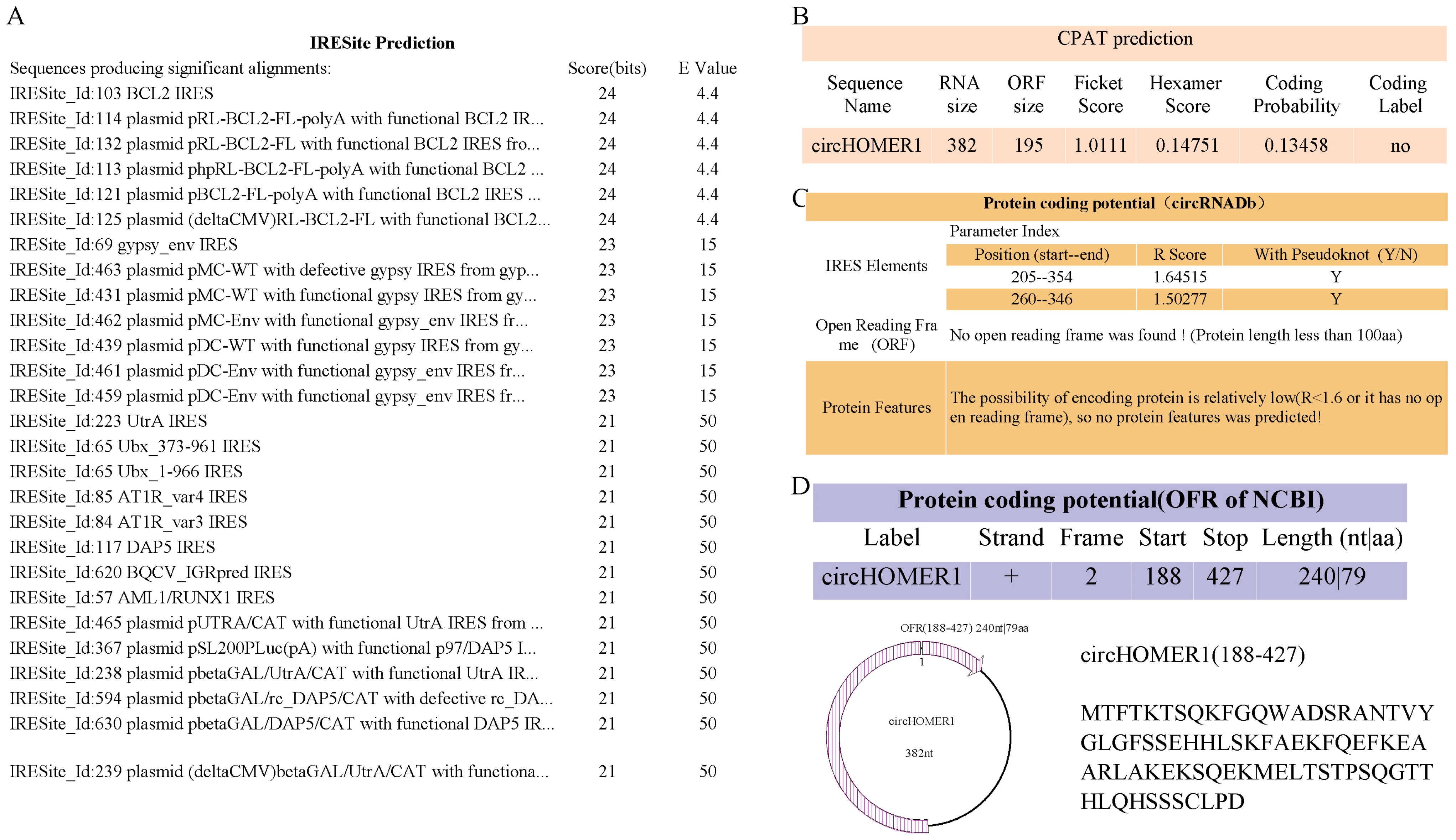
2.10. Prediction of Coding Ability of circHOMER1
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Samples
4.2. Plasmid Construction and RNA Interference
4.3. Cell Transfection
4.4. RNA Isolation, Reverse Transcription and qPCR
4.5. Western Blot Assay
4.6. Proliferation Experiment of CCK-8 and EdU
4.7. Immunofluorescence Staining
4.8. Flow Cytometry Assay
4.9. Subcellular Fractionation and RNA Extraction
4.10. Fluorescence In Situ Hybridization (FISH)
4.11. Prediction of Protein Coding Ability of circRNA
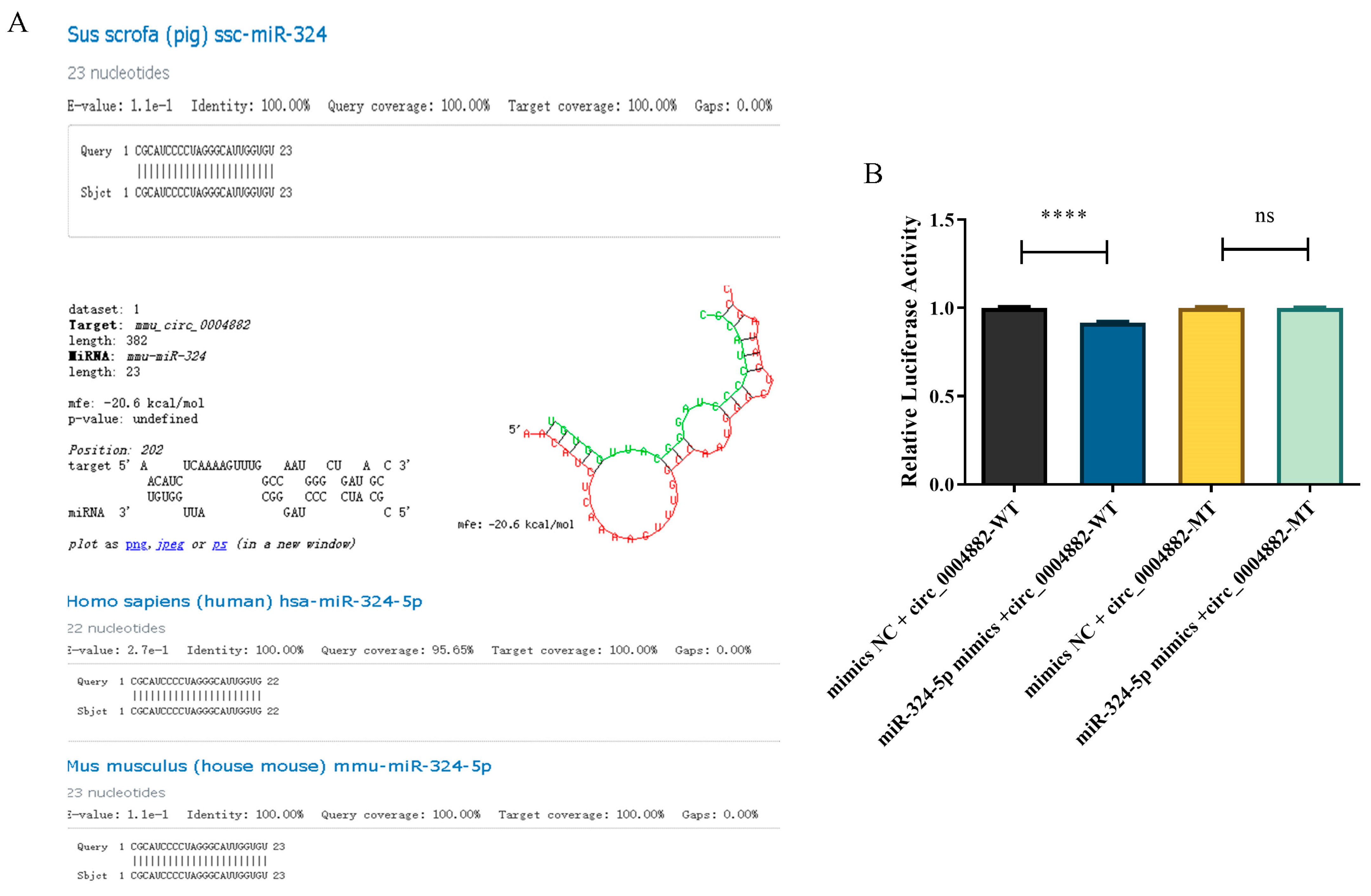
4.12. Homology Transformation of circRNA and Conservation Analysis of miRNA
4.13. Double Luciferase Report Assay
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, B.; Gao, H.; Yang, F.; Li, Y.; Yang, Q.; Liao, Y.; Guo, H.; Xu, K.; Tang, Z.; Gao, N.; et al. Comparative Characterization of Volatile Compounds of Ningxiang Pig, Duroc and Their Crosses (Duroc × Ningxiang) by Using SPME-GC-MS. Foods 2023, 12, 1059. [Google Scholar] [CrossRef] [PubMed]
- Lassar, A.B.; Buskin, J.N.; Lockshon, D.; Davis, R.L.; Apone, S.; Hauschka, S.D.; Weintraub, H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell 1989, 58, 823–831. [Google Scholar] [CrossRef]
- Vicente-García, C.; Hernández-Camacho, J.D.; Carvajal, J.J. Regulation of myogenic gene expression. Exp. Cell Res. 2022, 419, 113299. [Google Scholar] [CrossRef] [PubMed]
- Hasty, P.; Bradley, A.; Morris, J.H.; Edmondson, D.G.; Venuti, J.M.; Olson, E.N.; Klein, W.H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 1993, 364, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.N.; Tajbakhsh, S.; Mouly, V.; Cossu, G.; Buckingham, M.; Butler-Browne, G.S. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell Sci. 1999, 112 Pt 17, 2895–2901. [Google Scholar] [CrossRef]
- Morisaki, T.; Sermsuvitayawong, K.; Byun, S.H.; Matsuda, Y.; Hidaka, K.; Morisaki, H.; Mukai, T. Mouse Mef2b gene: Unique member of MEF2 gene family. J. Biochem. 1997, 122, 939–946. [Google Scholar] [CrossRef]
- Kim, M.S.; Fielitz, J.; McAnally, J.; Shelton, J.M.; Lemon, D.D.; McKinsey, T.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol. Cell. Biol. 2008, 28, 3600–3609. [Google Scholar] [CrossRef]
- Kaushal, S.; Schneider, J.W.; Nadal-Ginard, B.; Mahdavi, V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science 1994, 266, 1236–1240. [Google Scholar] [CrossRef]
- Rao, P.K.; Kumar, R.M.; Farkhondeh, M.; Baskerville, S.; Lodish, H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. 2006, 103, 8721–8726. [Google Scholar] [CrossRef]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Y.; Li, T.; Ma, Z.; Jia, H.; Chen, Q.; Zhao, Y.; Zhai, L.; Zhong, R.; Li, C.; et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017, 8, 14016. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Li, Z.; Ma, M.; Wang, Z.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. LncRNA-Six1 Encodes a Micropeptide to Activate Six1 in Cis and Is Involved in Cell Proliferation and Muscle Growth. Front. Physiol. 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, B.; Shen, M.; Cao, Y.; Zhang, X.; Li, W.; Tao, J.; Wu, W.; Liu, H. LncRNA 2310043L19Rik inhibits differentiation and promotes proliferation of myoblast by sponging miR-125a-5p. Aging 2020, 12, 5625–5639. [Google Scholar] [CrossRef]
- Xu, J.; Wen, Y.; Li, X.; Peng, W.; Zhang, Z.; Liu, X.; Yang, P.; Chen, N.; Lei, C.; Zhang, J.; et al. Bovine enhancer-regulated circSGCB acts as a ceRNA to regulate skeletal muscle development via enhancing KLF3 expression. Int. J. Biol. Macromol. 2024, 261, 129779. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Z.; Xie, T.; Wang, X.; Guo, D.; Lin, S.; An, L.; Lin, J.; Zhang, L. Novel Circular RNA CircSLC2A13 Regulates Chicken Muscle Development by Sponging MiR-34a-3p. J. Agric. Food Chem. 2024, 72, 15530–15540. [Google Scholar] [CrossRef]
- Lin, Z.; Xie, F.; He, X.; Wang, J.; Luo, J.; Chen, T.; Jiang, Q.; Xi, Q.; Zhang, Y.; Sun, J. A novel protein encoded by circKANSL1L regulates skeletal myogenesis via the Akt-FoxO3 signaling axis. Int. J. Biol. Macromol. 2024, 257, 128609. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Chen, C.; Fang, F. hsa_circ_0006916 Exerts Effect on Amyloid Beta-Induced Neuron Injury by Targeting miR-217/HOMER1. Ann. Clin. Lab. Sci. 2023, 53, 181–191. [Google Scholar]
- Shu, S.; Xu, Z.; Lu, H.; Li, Z.; Zhang, Y. CircHOMER1aggravates oxidative stress, inflammation and extracellular matrix deposition in high glucose-induced human mesangial cells. Nephrology 2022, 27, 983–993. [Google Scholar] [CrossRef]
- Papageorgiou, G.; Amoah, S.K.; Pierotti, C.; Otero, M.; Eckel, S.; Coffey, K.; Allan, A.M.; Caldwell, K.K.; Mellios, N. Prenatal alcohol exposure results in brain region- and sex-specific changes in circHomer1 expression in adult mouse brain. Front. Neurosci. 2023, 17, 1087950. [Google Scholar] [CrossRef]
- Hafez, A.K.; Zimmerman, A.J.; Papageorgiou, G.; Chandrasekaran, J.; Amoah, S.K.; Lin, R.; Lozano, E.; Pierotti, C.; Dell Orco, M.; Hartley, B.J.; et al. A bidirectional competitive interaction between circHomer1 and Homer1b within the orbitofrontal cortex regulates reversal learning. Cell Reports 2022, 38, 110282. [Google Scholar] [CrossRef] [PubMed]
- Urdánoz-Casado, A.; Sánchez-Ruiz De Gordoa, J.; Robles, M.; Acha, B.; Roldan, M.; Zelaya, M.V.; Blanco-Luquin, I.; Mendioroz, M. Gender-Dependent Deregulation of Linear and Circular RNA Variants of HOMER1 in the Entorhinal Cortex of Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 9205. [Google Scholar] [CrossRef]
- Zhao, M.; Dong, G.; Meng, Q.; Lin, S.; Li, X. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J. Cell. Biochem. 2020, 121, 4440–4449. [Google Scholar] [CrossRef]
- Cervera-Carles, L.; Dols-Icardo, O.; Molina-Porcel, L.; Alcolea, D.; Cervantes-Gonzalez, A.; Muñoz-Llahuna, L.; Clarimon, J. Assessing circular RNAs in Alzheimer’s disease and frontotemporal lobar degeneration. Neurobiol. Aging 2020, 92, 7–11. [Google Scholar] [CrossRef]
- Li, J.; Sun, Q.; Zhu, S.; Xi, K.; Shi, Q.; Pang, K.; Liu, X.; Li, M.; Zhang, Y.; Sun, J. Knockdown of circHomer1 ameliorates METH-induced neuronal injury through inhibiting Bbc3 expression. Neurosci. Lett. 2020, 732, 135050. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, L.; Ma, H.; Wang, Y.; Wang, P. Lidocaine Suppresses Cell Proliferation and Aerobic Glycolysis by Regulating circHOMER1/miR-138-5p/HEY1 Axis in Colorectal Cancer. Cancer Manag. Res. 2020, 12, 5009–5022. [Google Scholar] [CrossRef]
- Veno, M.T.; Hansen, T.B.; Veno, S.T.; Clausen, B.H.; Grebing, M.; Finsen, B.; Holm, I.E.; Kjems, J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015, 16, 245. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21. [Google Scholar] [CrossRef]
- Shenje, L.T.; Andersen, P.; Halushka, M.K.; Lui, C.; Fernandez, L.; Collin, G.B.; Amat-Alarcon, N.; Meschino, W.; Cutz, E.; Chang, K.; et al. Mutations in Alström protein impair terminal differentiation of cardiomyocytes. Nat. Commun. 2014, 5, 3416. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, P.; Sciancalepore, M.; Veltruska, K.; Lorenzon, P.; Bandiera, A. Epidermal Growth Factor–based adhesion substrates elicit myoblast scattering, proliferation, differentiation and promote satellite cell myogenic activation. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2019, 1866, 504–517. [Google Scholar] [CrossRef]
- Seo, J.; Kang, J.; Kim, Y.L.; Jo, Y.; Kim, J.; Hann, S.; Park, J.; Park, I.; Park, H.; Yoo, K.; et al. Maintenance of type 2 glycolytic myofibers with age by Mib1-Actn3 axis. Nat. Commun. 2021, 12, 1294. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Voza, F.; Ezzeddine, N.; Frasch, M. Drosophila mind bomb2 is required for maintaining muscle integrity and survival. J. Cell Biol. 2007, 179, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Collao, N.; Akohene-Mensah, P.; Nallabelli, J.; Binet, E.R.; Askarian, A.; Lloyd, J.; Niemiro, G.M.; Beals, J.W.; van Vliet, S.; Rajgara, R.; et al. The role of L-type amino acid transporter 1 (Slc7a5) during in vitro myogenesis. Am. J. Physiol.-Cell Physiol. 2022, 323, C595–C605. [Google Scholar] [CrossRef] [PubMed]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S.A. ORF-FINDER: A vector for high-throughput gene identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Chen, C.; Cheng, R.; Demeter, J.; Chen, J.; Weingarten-Gabbay, S.; Jiang, L.; Snyder, M.P.; Weissman, J.S.; Segal, E.; Jackson, P.K.; et al. Structured elements drive extensive circular RNA translation. Mol. Cell 2021, 81, 4300–4318. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]

| Source | ID |
|---|---|
| circRNAdb | hsa_circ_22696 |
| circbank | hsa_circHOMER1_003 |
| circAtlas3.0 | hsa-HOMER1_0008 |
| Basic information | |
| circBank ID: hsa_circHOMER1_003 | Host gene Symbol: HOMER1 |
| circBase ID: hsa_circ_0073132 | bestTranscript: NM_004272 |
| Position: chr5: 78742875-78752841 strand: - | Annotation: ANNOTATED, CDS, coding, INTERNAL, OVCODE, OVEXON |
| Length: 382 | |
| RNA sequence: | |
| GGAACAACCTATCTTCAGCACTCGAGCTCATGTCTTCCAAATTGACCCAAACACAAAGAAGAACTGGGTACCCACCAGCAAGCATGCAGTTACTGTGTCTTATTTCTATGACAGCACAAGAAATGTGTATAGGATAATCAGTTTAGATGGCTCAAAGGCAATAATAAATAGTACCATCACCCCAAACATGACATTTACTAAAACATCTCAGAAGTTTGGCCAGTGGGCTGATAGCCGGGCAAACACCGTTTATGGATTGGGATTCTCCTCTGAGCATCATCTTTCGAAATTTGCAGAAAAGTTTCAGGAATTTAAAGAAGCTGCTCGACTAGCAAAGGAAAAATCACAAGAGAAGATGGAACTTACCAGTACACCTTCACAG | |
| conserved mm9 circRNA: >mmu_circ_0004882 | |
| GGAGCAACCTATCTTCAGCACTCGAGCTCATGTCTTCCAGATTGACCCGAACACAAAGAAGAACTGGGTACCCACCAGCAAGCATGCAGTTACTGTATCTTATTTTTATGACAGCACAAGAAATGTGTATAGGATAATCAGTTTAGATGGCTCAAAGGCAATAATAAATAGCACCATCACACCAAACATGACATTTACTAAAACATCTCAAAAGTTTGGCCAATGGGCTGATAGCCGGGCAAACACTGTTTATGGACTGGGATTCTCCTCTGAGCATCATCTTTCAAAATTCGCAGAAAAGTTTCAGGAATTTAAGGAAGCTGCTCGGCTTGCAAAGGAGAAGTCGCAGGAGAAGATGGAGCTGACCAGTACCCCTTCACAG | |
| coding_potential_assessment | |
| circBank ID: hsa_circHOMER1_003 | Fickett_score: 0.8529 |
| circRNA_size: 1146 | Hexamer_score: 0.0431 |
| ORF_size: 240 | coding_prob: 0.1187 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Wang, K.; Chen, B.; Liu, J.; Chen, W.; Ma, H. Function and Molecular Mechanism of Circhomer1 in Myogenesis. Int. J. Mol. Sci. 2025, 26, 6264. https://doi.org/10.3390/ijms26136264
Yu Z, Wang K, Chen B, Liu J, Chen W, Ma H. Function and Molecular Mechanism of Circhomer1 in Myogenesis. International Journal of Molecular Sciences. 2025; 26(13):6264. https://doi.org/10.3390/ijms26136264
Chicago/Turabian StyleYu, Zonggang, Kaiming Wang, Bohe Chen, Jingwen Liu, Wenwu Chen, and Haiming Ma. 2025. "Function and Molecular Mechanism of Circhomer1 in Myogenesis" International Journal of Molecular Sciences 26, no. 13: 6264. https://doi.org/10.3390/ijms26136264
APA StyleYu, Z., Wang, K., Chen, B., Liu, J., Chen, W., & Ma, H. (2025). Function and Molecular Mechanism of Circhomer1 in Myogenesis. International Journal of Molecular Sciences, 26(13), 6264. https://doi.org/10.3390/ijms26136264




