DNA Methylation and Transcript Variant Analysis of CDKN2A Exon 2 Despite High Sequence Identity with CDKN2B Exon 2
Abstract
1. Introduction
2. Results and Discussion
2.1. DNA Methylation Analysis of CDKN2A Exon 2—Method Development and Validation
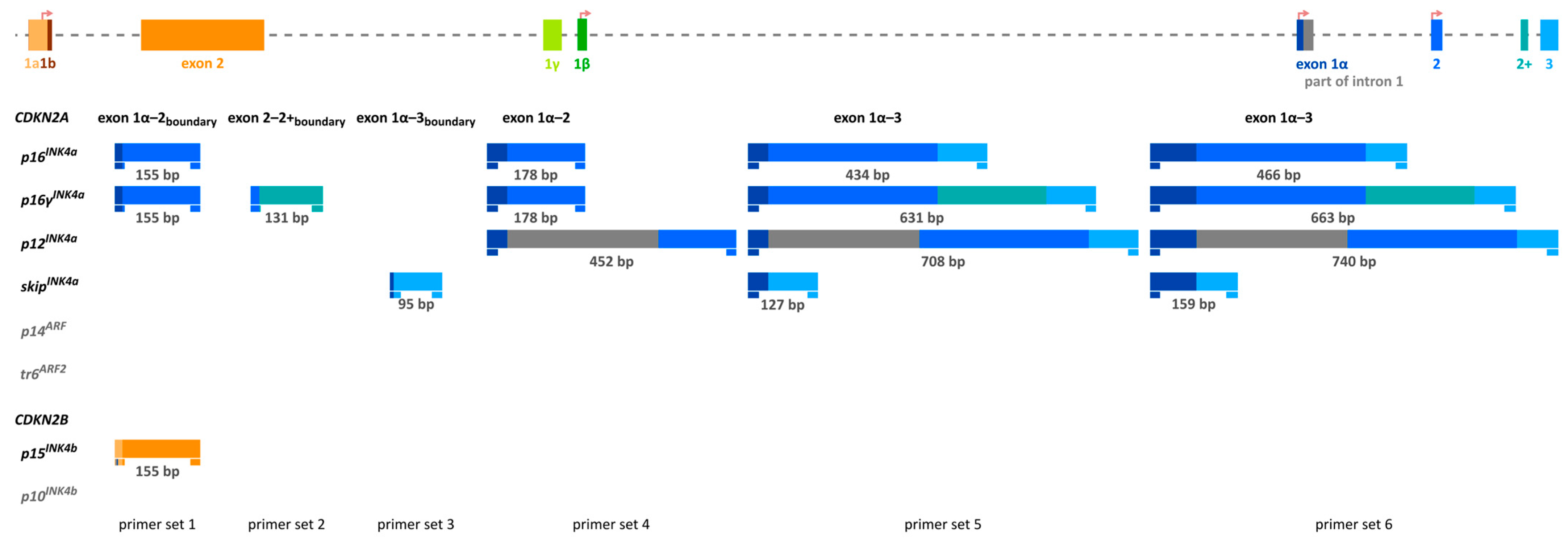
2.1.1. Primer Design and Characteristics of the Target Region
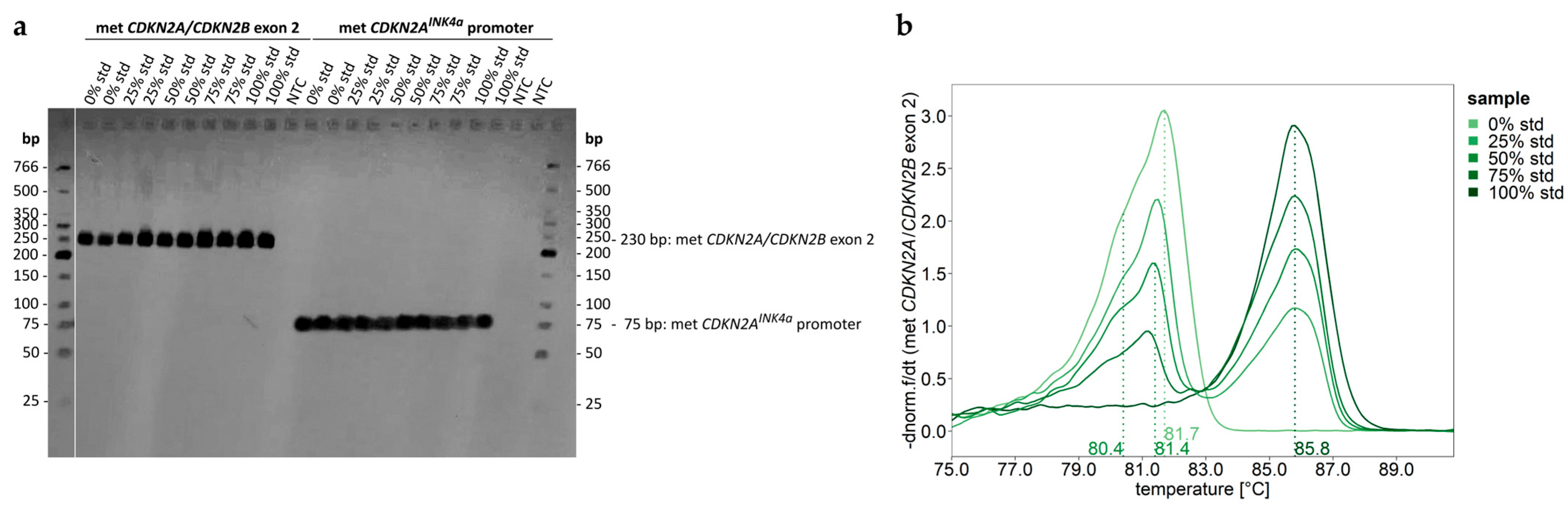
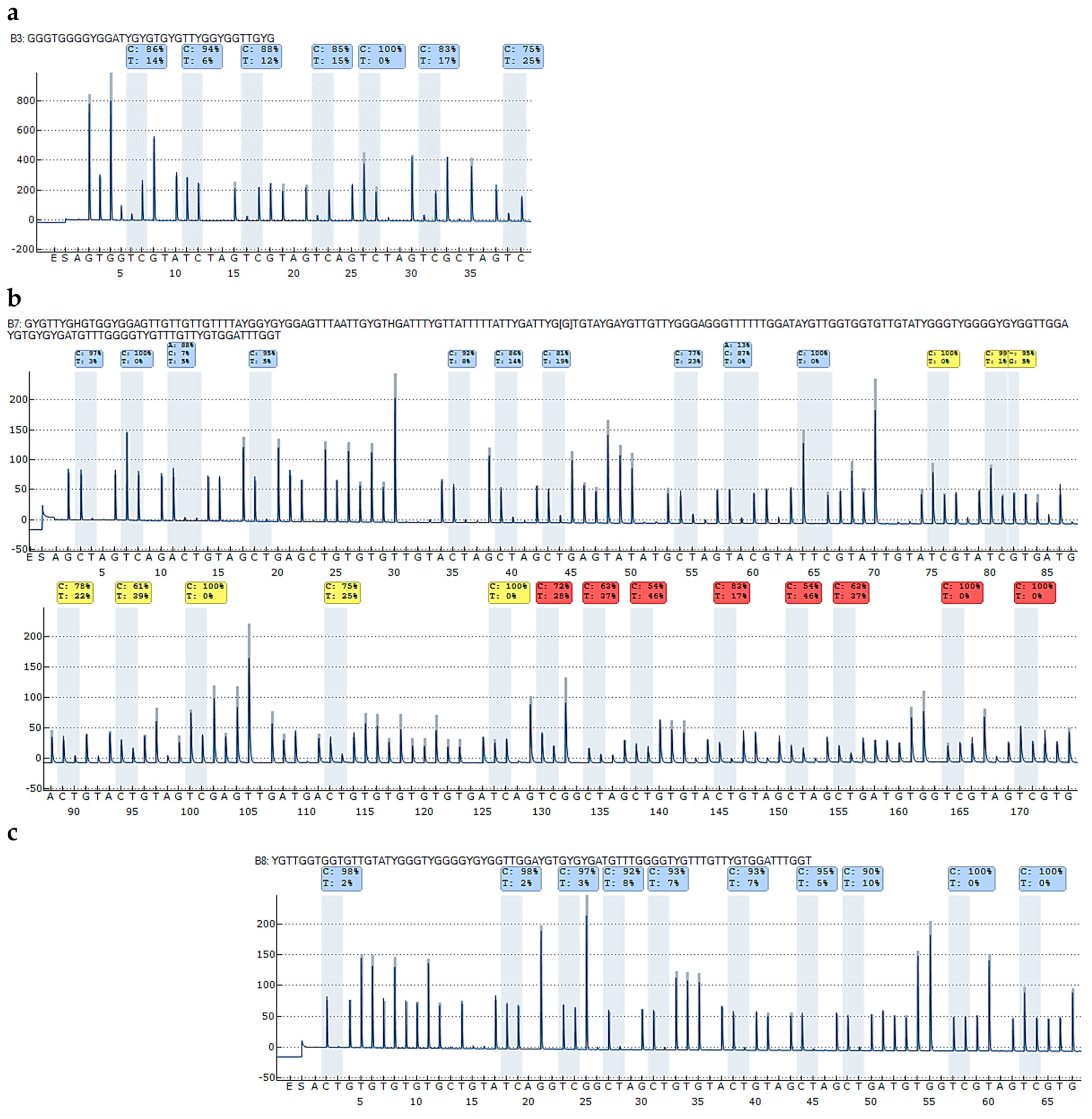
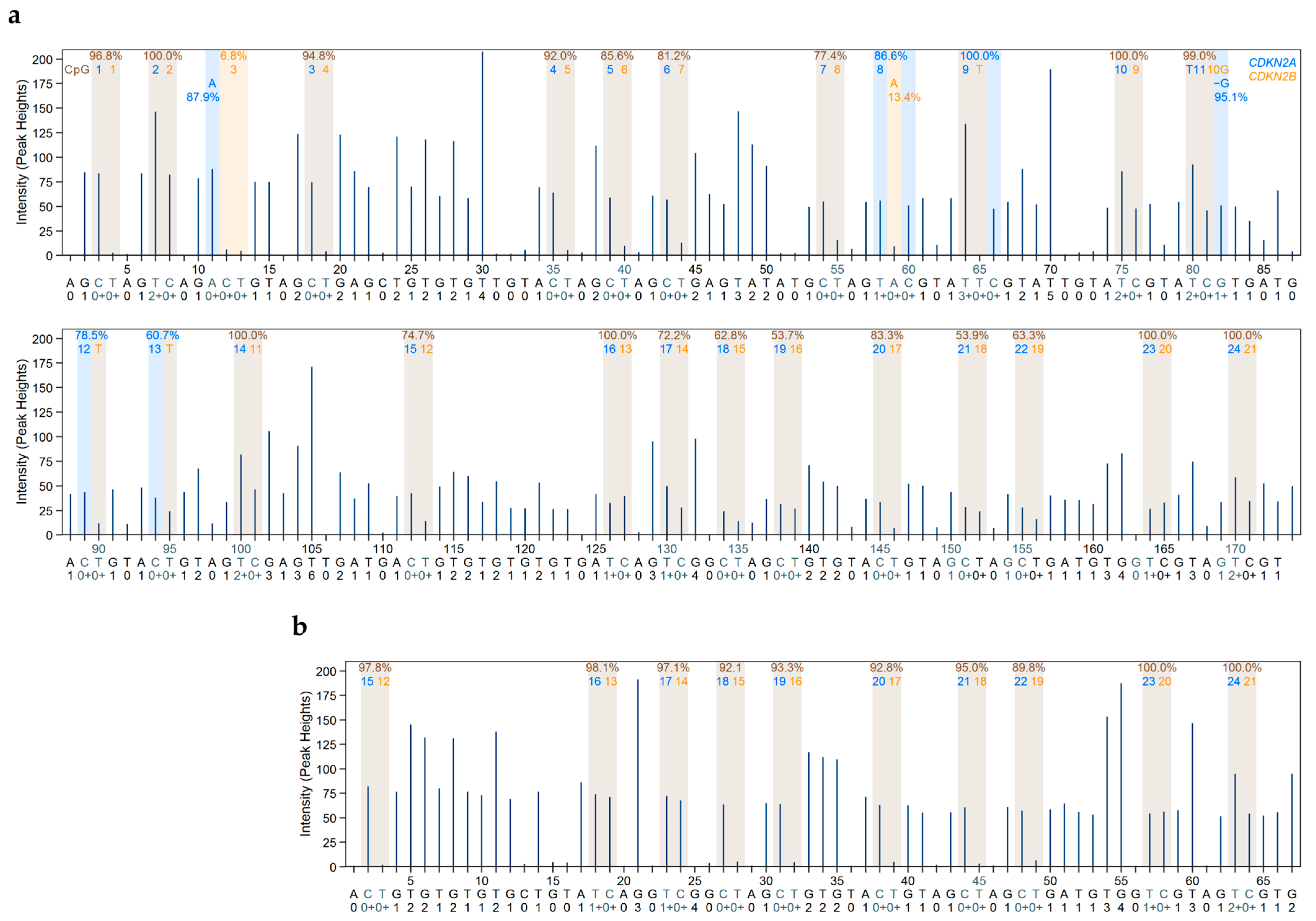
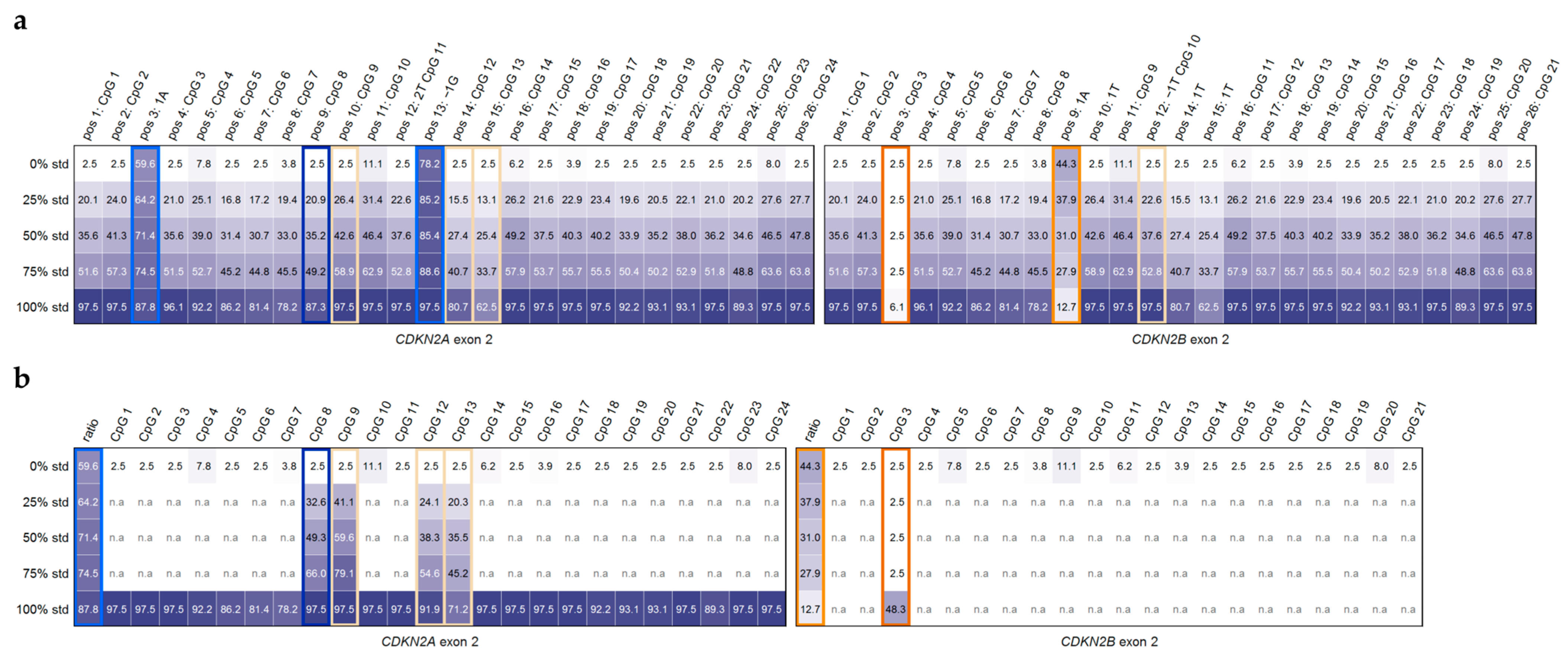
2.1.2. Evaluation of Reliable DNA Methylation Levels Despite High Sequence Identity Between the Second Exons of CDKN2A and CDKN2B
2.2. DNA Methylation Analysis of the CDKN2AINK4a Promoter—Method Development and Validation
2.3. Transcript Variant Analysis of CDKN2A Exon 2—Method Development and Validation
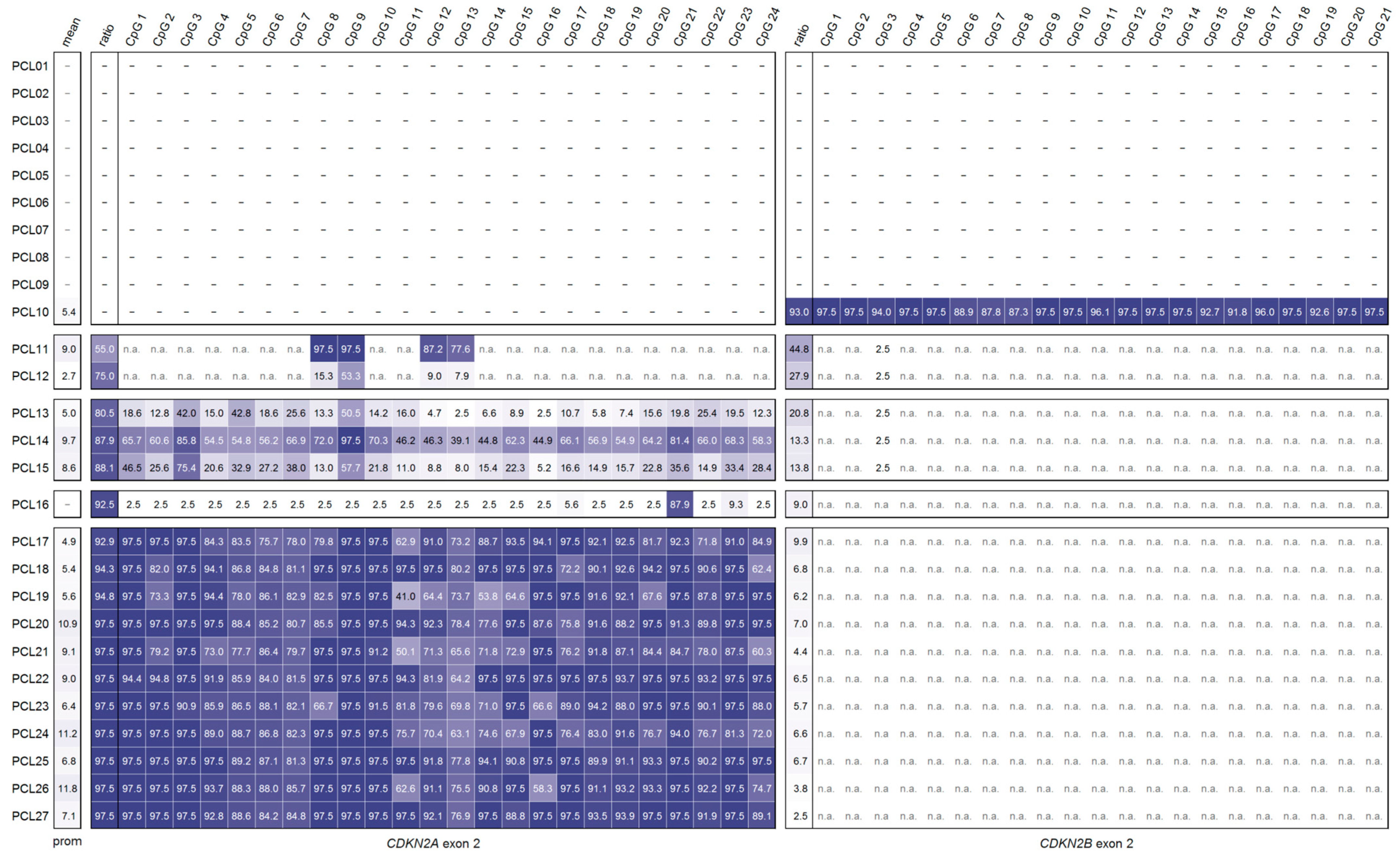
2.4. DNA Methylation Analysis of CDKN2AINK4a Promoter and CDKN2A Exon 2 in Primary Cell Lines from Glioma
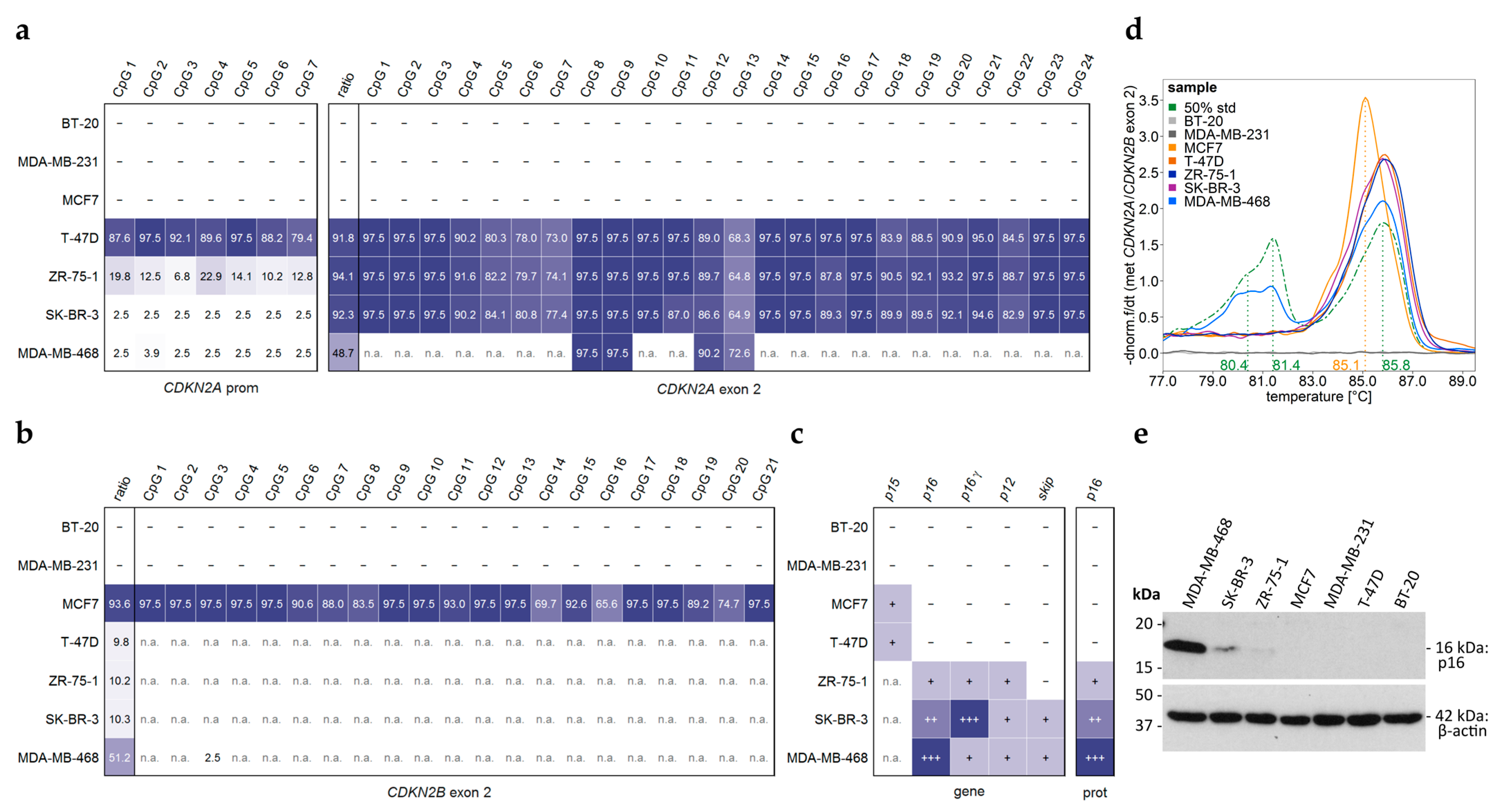
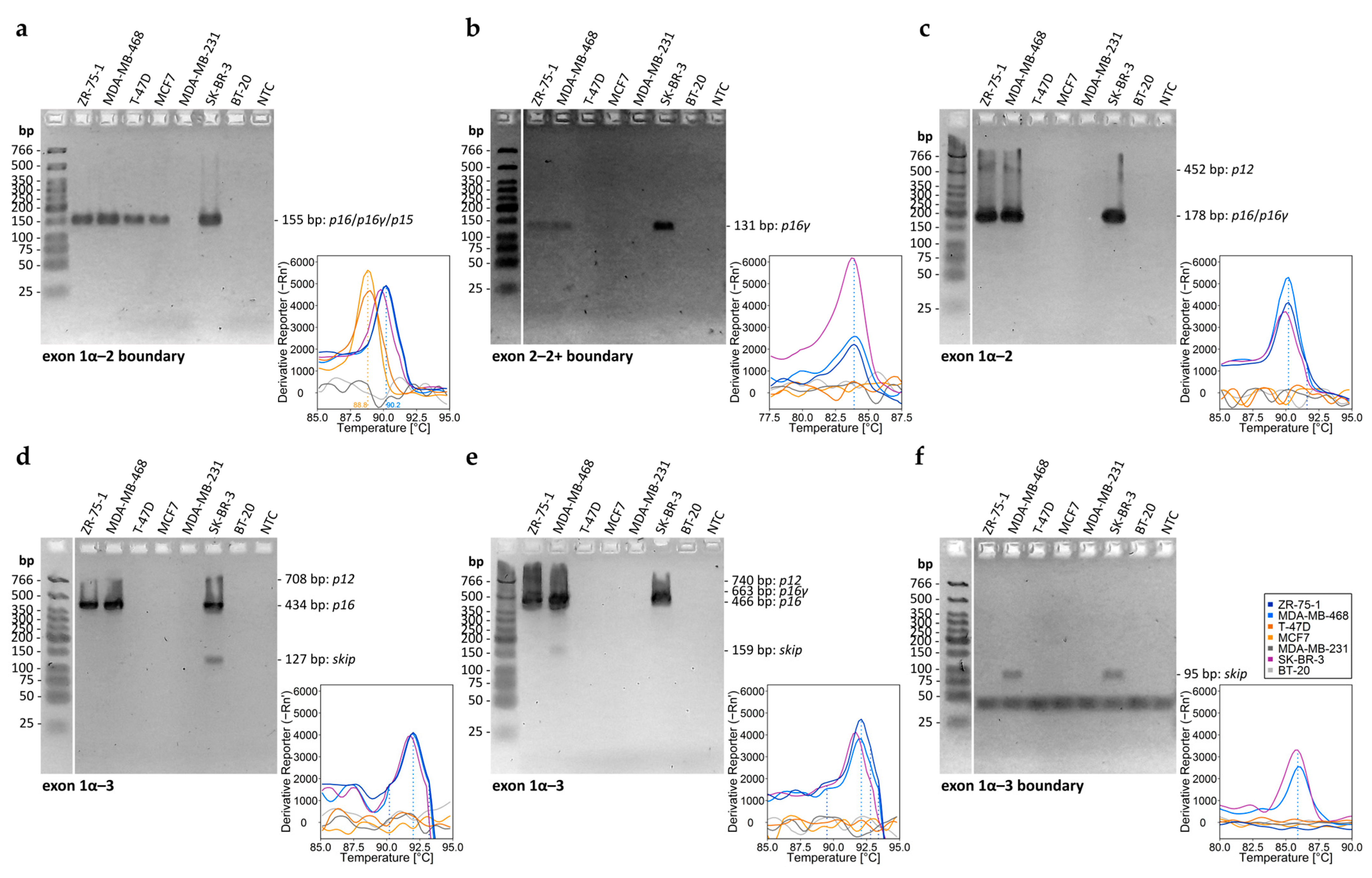
2.5. DNA Methylation Analysis of the CDKN2AINK4a Promoter and CDKN2A Exon 2 and Transcript Variant Analysis of CDKN2A Exons in Commercial Breast Cancer Cell Lines
3. Materials and Methods
3.1. Primary Cell Lines from Glioma Patients
3.2. Commercial Breast Cancer Cell Lines
3.3. DNA/RNA Extraction and Bisulfite Conversion
3.4. Primer Design and PCR Conditions for DNA Methylation Analysis
| Region | Primer Sequence (5′→3′) | Length [bp] | CpGs Analyzed | |
|---|---|---|---|---|
| CDKN2A | CDKN2B | |||
| promoter | F: GAGGGGTTGGTTGGTTATTAG | 75 | ||
| R: [Btn] TACCTACTCTCCCCCTCTC | ||||
| S: GGTTGGTTGGTTATTAGA | 1–7 | - | ||
| exon 2 | F: GTTTTTTTTGGTAGGTTATGATGATGG | 230 | ||
| R: [Btn] ACCCAACTCCTCAACCAAATCC | ||||
| S1: AGGTTATGATGATGGGTA | 1–14 | 1–11 | ||
| S2: GGGAGGGTTTTTTGGATA | 15–24 | 12–21 | ||
3.5. PSQ of PCR Products
3.6. Analysis of Transcript Variants
| Set | Region | Primer Sequence (5′→3′) | Location | Length [bp] |
|---|---|---|---|---|
| 1 | exon 1α–exon 2 | F: GGAGGCCGATCCAGGTCA | boundary | 155 |
| R: CAGCACCACCAGCGTGTC | exon 2 | |||
| 2 | exon 2–exon 2+ | F: GCGGAAGGTCCCTCAGAA | boundary | 131 |
| R: CAGCCAGCTTGCGATAACCA | exon 2+ | |||
| 3 | exon 1α–exon 3 | F: GATCCAGACATCCCCGATTG | boundary | 95 |
| R: CCTGTAGGACCTTCGGTGA | exon 3 |
| Set | Region | Primer Sequence (5′→3′) | Location | Length [bp] |
|---|---|---|---|---|
| 4 | exon 1α–exon 2 | F: CAACGCACCGAATAGTTACG | exon 1α | 178/452 |
| R: CAGCACCACCAGCGTGTC | exon 2 | |||
| 5 | exon 1α–exon 3 | F: CAACGCACCGAATAGTTACG | exon 1α | 434/631/ |
| R: CAGTTGTGGCCCTGTAGGA | exon 3 | 708/127 | ||
| 6 | exon 1α–exon 3 | F: GGTCGGGTAGAGGAGGTG | exon 1α | 466/663/ |
| R: AGGACCTTCGGTGACTGATGA | exon 3 | 740/159 |
3.7. Western Blot Analysis
3.8. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Labuhn, M.; Jones, G.; Speel, E.J.M.; Maier, D.; Zweifel, C.; Gratzl, O.; Van Meir, E.G.; Hegi, M.E.; Merlo, A. Quantitative real-time PCR does not show selective targeting of p14ARF but concomitant inactivation of both p16INK4A and p14ARF in 105 human primary gliomas. Oncogene 2001, 20, 1103–1109. [Google Scholar] [CrossRef][Green Version]
- Aftab, A.; Shahzad, S.; Hussain, H.M.J.; Khan, R.; Irum, S.; Tabassum, S. CDKN2A/P16INK4A variants association with breast cancer and their in-silico analysis. Breast Cancer 2019, 26, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, G.I.; Park, J.E.; Edwards, C.D.; Mao, L.; Rollins, B.J.; Sidransky, D.; Ewen, M.E. Multiple Mechanisms of p16INK4A Inactivation in Non-Small Cell Lung Cancer Cell Lines. Cancer Res. 1995, 55, 6200–6209. [Google Scholar]
- Sharpless, N.E. INK4a/ARF: A multifunctional tumor suppressor locus. Mutat. Res. 2005, 576, 22–38. [Google Scholar] [CrossRef]
- Okamoto, A.; Demetrick, D.J.; Spillare, E.A.; Hagiwara, K.; Hussain, S.P.; Bennett, W.P.; Forrester, K.; Gerwin, B.; Serrano, M.; Beach, D.H.; et al. Mutations and altered expression of p16INK4 in human cancer. Proc. Natl. Acad. Sci. USA 1994, 91, 11045–11049. [Google Scholar] [CrossRef] [PubMed]
- Mitomi, H.; Fukui, N.; Tanaka, N.; Kanazawa, H.; Saito, T.; Matsuoka, T.; Yao, T. Aberrant p16INK4a methylation is a frequent event in colorectal cancers: Prognostic value and relation to mRNA expression and immunoreactivity. J. Cancer Res. Clin. Oncol. 2010, 136, 323–331. [Google Scholar] [CrossRef]
- Bhagat, R.; Kumar, S.S.; Vaderhobli, S.; Premalata, C.S.; Pallavi, V.R.; Ramesh, G.; Krishnamoorthy, L. Epigenetic alteration of p16 and retinoic acid receptor beta genes in the development of epithelial ovarian carcinoma. Tumor Biol. 2014, 35, 9069–9078. [Google Scholar] [CrossRef]
- Woodcock, D.M.; Linsenmeyer, M.E.; Doherty, J.P.; Warren, W.D. DNA methylation in the promoter region of the p16 (CDKN2/MTS-1/INK4A) gene in human breast tumours. Br. J. Cancer 1999, 79, 251–256. [Google Scholar] [CrossRef]
- Krassenstein, R.; Sauter, E.; Dulaimi, E.; Battagli, C.; Ehya, H.; Klein-Szanto, A.; Cairns, P. Detection of Breast Cancer in Nipple Aspirate Fluid by CpG Island Hypermethylation. Clin. Cancer Res. 2004, 10, 28–32. [Google Scholar] [CrossRef]
- Sinha, S.; Chunder, N.; Mukherjee, N.; Alam, N.; Roy, A.; Roychoudhury, S.; Kumar Panda, C. Frequent deletion and methylation in SH3GL2 and CDKN2A loci are associated with early- and late-onset breast carcinoma. Ann. Surg. Oncol. 2008, 15, 1070–1080. [Google Scholar] [CrossRef]
- Tao, M.H.; Shields, P.G.; Nie, J.; Millen, A.; Ambrosone, C.B.; Edge, S.B.; Krishnan, S.S.; Marian, C.; Xie, B.; Winston, J.; et al. DNA hypermethylation and clinicopathological features in breast cancer: The Western New York Exposures and Breast Cancer (WEB) Study. Breast Cancer Res. Treat. 2009, 114, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Askari, M.; Sobti, R.C.; Nikbakht, M.; Sharma, S.C. Promoter hypermethylation of tumour suppressor genes (p14/ARF and p16/INK4a): Case-control study in North Indian population. Mol. Biol. Rep. 2013, 40, 4921–4928. [Google Scholar] [CrossRef]
- Di Vinci, A.; Perdelli, L.; Banelli, B.; Salvi, S.; Casciano, I.; Gelvi, I.; Allemanni, G.; Margallo, E.; Gatteschi, B.; Romani, M. p16INK4a promoter methylation and protein expression in breast fibroadenoma and carcinoma. Int. J. Cancer 2005, 114, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Kempster, S.; Phillips, W.A.; Baindur-Hudson, S.; Thomas, R.J.S.; Dow, C.; Rockman, S.P. Methylation of exon 2 of p16 is associated with late stage oesophageal cancer. Cancer Lett. 2000, 150, 57–62. [Google Scholar] [CrossRef]
- Spitzwieser, M.; Entfellner, E.; Werner, B.; Pulverer, W.; Pfeiler, G.; Hacker, S.; Cichna-Markl, M. Hypermethylation of CDKN2A exon 2 in tumor, tumor-adjacent and tumor-distant tissues from breast cancer patients. BMC Cancer 2017, 17, 260. [Google Scholar] [CrossRef] [PubMed]
- Genbank. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/ (accessed on 20 February 2025).
- Zappe, K.; Pirker, C.; Miedl, H.; Schreiber, M.; Heffeter, P.; Pfeiler, G.; Hacker, S.; Haslik, W.; Spiegl-Kreinecker, S.; Cichna-Markl, M. Discrimination between 34 of 36 possible combinations of three C>T SNP genotypes in the MGMT promoter by high resolution melting analysis coupled with pyrosequencing using a single primer set. Int. J. Mol. Sci. 2021, 22, 12527. [Google Scholar] [CrossRef]
- Appay, R.; Dehais, C.; Maurage, C.-A.; Alentorn, A.; Carpentier, C.; Colin, C.; Ducray, F.; Escande, F.; Idbaih, A.; Kamoun, A.; et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro-Oncol. 2019, 21, 1519–1528. [Google Scholar] [CrossRef]
- Lu, V.M.; O’Connor, K.P.; Shah, A.H.; Eichberg, D.G.; Luther, E.M.; Komotar, R.J.; Ivan, M.E. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: A systematic review of the contemporary literature. J. Neuro-Oncol. 2020, 148, 221–229. [Google Scholar] [CrossRef]
- Noack, D.; Wach, J.; Barrantes-Freer, A.; Nicolay, N.H.; Güresir, E.; Seidel, C. Homozygous CDKN2A/B deletions in low- and high-grade glioma: A meta-analysis of individual patient data and predictive values of p16 immunohistochemistry testing. Acta Neuropathol. Commun. 2024, 12, 180. [Google Scholar] [CrossRef]
- Wang, J.; Xi, Y.F.; Zhao, Q.; Guo, J.-h.; Zhang, Z.; Zhang, M.-b.; Chang, J.; Wu, Y.-q.; Su, W. CDKN2A promoter methylation enhances self-renewal of glioblastoma stem cells and confers resistance to carmustine. Mol. Biol. Rep. 2024, 51, 385. [Google Scholar] [CrossRef] [PubMed]
- Lebok, P.; Roming, M.; Kluth, M.; Koop, C.; Özden, C.; Taskin, B.; Hussein, K.; Lebeau, A.; Witzel, I.; Wölber, L.; et al. p16 overexpression and 9p21 deletion are linked to unfavorable tumor phenotype in breast cancer. Oncotarget 2016, 7, 81322–81331. [Google Scholar] [CrossRef]
- Hui, R.; Macmillan, R.D.; Kenny, F.S.; Musgrove, E.A.; Blamey, R.W.; Nicholson, R.I.; Robertson, J.F.R.; Sutherland, R.L. INK4a gene expression and methylation in primary breast cancer: Overexpression of p16INK4a messenger RNA is a marker of poor prognosis. Clin. Cancer Res. 2000, 6, 2777–2787. [Google Scholar]
- ATCC. American Type Culture Collection. Available online: https://www.atcc.org/ (accessed on 20 February 2025).
- ShahidSales, S.; Mehramiz, M.; Ghasemi, F.; Aledavood, A.; Shamsi, M.; Hassanian, S.M.; Ghayour-Mobarhan, M.; Avan, A. A genetic variant in CDKN2A/B gene is associated with the increased risk of breast cancer. J. Clin. Lab. Anal. 2018, 32, e22190. [Google Scholar] [CrossRef]
- Seifi, S.; Pouya, F.; Rahmani, M.; Mehramiz, M.; Rastgar-Moghadam, A.; Gharib, M.; Rahmani, F.; Shahidsales, S.; Hassanian, S.M.; Khazaei, M.; et al. Association of cyclin-dependent kinase inhibitor 2A/B with increased risk of developing breast cancer. J. Cell Physiol. 2020, 235, 5141–5145. [Google Scholar] [CrossRef]
- Krimpenfort, P.; Ijpenberg, A.; Song, J.-Y.; Van Der Valk, M.; Nawijn, M.; Zevenhoven, J.; Berns, A. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature 2007, 448, 943–946. [Google Scholar] [CrossRef] [PubMed]
- Holm, K.; Hegardt, C.; Staaf, J.; Vallon-Christersson, J.; Jönsson, G.; Olsson, H.; Borg, Å.; Ringnér, M. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010, 12, R36. [Google Scholar] [CrossRef] [PubMed]
- Bediaga, N.G.; Acha-Sagredo, A.; Guerra, I.; Viguri, A.; Albaina, C.; Ruiz Diaz, I.; Rezola, R.; Alberdi, M.J.; Dopazo, J.; Montaner, D.; et al. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010, 12, R77. [Google Scholar] [CrossRef]
- Mohanad, M.; Hamza, H.M.; Bahnassy, A.A.; Shaarawy, S.; Ahmed, O.; El-Mezayen, H.A.; Ayad, E.G.; Tahoun, N.; Abdellateif, M.S. Molecular profiling of breast cancer methylation pattern in triple negative versus non- triple negative breast cancer. Sci. Rep. 2025, 15, 6894. [Google Scholar] [CrossRef]
- Spiegl-Kreinecker, S.; Pirker, C.; Marosi, C.; Buchroithner, J.; Pichler, J.; Silye, R.; Fischer, J.; Micksche, M.; Berger, W. Dynamics of chemosensitivity and chromosomal instability in recurrent glioblastoma. Br. J. Cancer 2007, 96, 960–969. [Google Scholar] [CrossRef][Green Version]
- James Kent, W.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Zappe, K.; Cichna-Markl, M. Temperature-Wise Calibration Increases the Accuracy of DNA Methylation Levels Determined by High-Resolution Melting (HRM). Int. J. Mol. Sci. 2024, 25, 5082. [Google Scholar] [CrossRef] [PubMed]
- Zappe, K.; Pühringer, K.; Pflug, S.; Berger, D.; Böhm, A.; Spiegl-Kreinecker, S.; Cichna-Markl, M. Association between MGMT Enhancer Methylation and MGMT Promoter Methylation, MGMT Protein Expression, and Overall Survival in Glioblastoma. Cells 2023, 12, 1639. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Berger, W.; Elbling, L.; Micksche, M. Expression of the major vault protein LRP in human non-small-cell lung cancer cells: Activation by short-term exposure to antineoplastic drugs. Int. J. Cancer 2000, 88, 293–300. [Google Scholar] [CrossRef]
- R Core Team, R. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 20 February 2025).
- Assigning Values to Non-Detected/Non-Quantified Pesticide Residues in Human Health Food Exposure Assessments; Office of Pesticide Programs, US Environmental Protection Agency: Washington, DC, USA, 2000.


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zappe, K.; Jenik, A.; Berger, D.; Uhlik, L.; Heffeter, P.; Cichna-Markl, M. DNA Methylation and Transcript Variant Analysis of CDKN2A Exon 2 Despite High Sequence Identity with CDKN2B Exon 2. Int. J. Mol. Sci. 2025, 26, 6128. https://doi.org/10.3390/ijms26136128
Zappe K, Jenik A, Berger D, Uhlik L, Heffeter P, Cichna-Markl M. DNA Methylation and Transcript Variant Analysis of CDKN2A Exon 2 Despite High Sequence Identity with CDKN2B Exon 2. International Journal of Molecular Sciences. 2025; 26(13):6128. https://doi.org/10.3390/ijms26136128
Chicago/Turabian StyleZappe, Katja, Andreas Jenik, Daniel Berger, Lukas Uhlik, Petra Heffeter, and Margit Cichna-Markl. 2025. "DNA Methylation and Transcript Variant Analysis of CDKN2A Exon 2 Despite High Sequence Identity with CDKN2B Exon 2" International Journal of Molecular Sciences 26, no. 13: 6128. https://doi.org/10.3390/ijms26136128
APA StyleZappe, K., Jenik, A., Berger, D., Uhlik, L., Heffeter, P., & Cichna-Markl, M. (2025). DNA Methylation and Transcript Variant Analysis of CDKN2A Exon 2 Despite High Sequence Identity with CDKN2B Exon 2. International Journal of Molecular Sciences, 26(13), 6128. https://doi.org/10.3390/ijms26136128






