Uncovering Anti-Melanoma Mechanisms of Bambusa stenostachya Leaf Compounds via Network Pharmacology and Molecular Docking
Abstract
1. Introduction
2. Results
2.1. LCMS Analysis
2.2. Bioactive Compounds from B. stenostachya
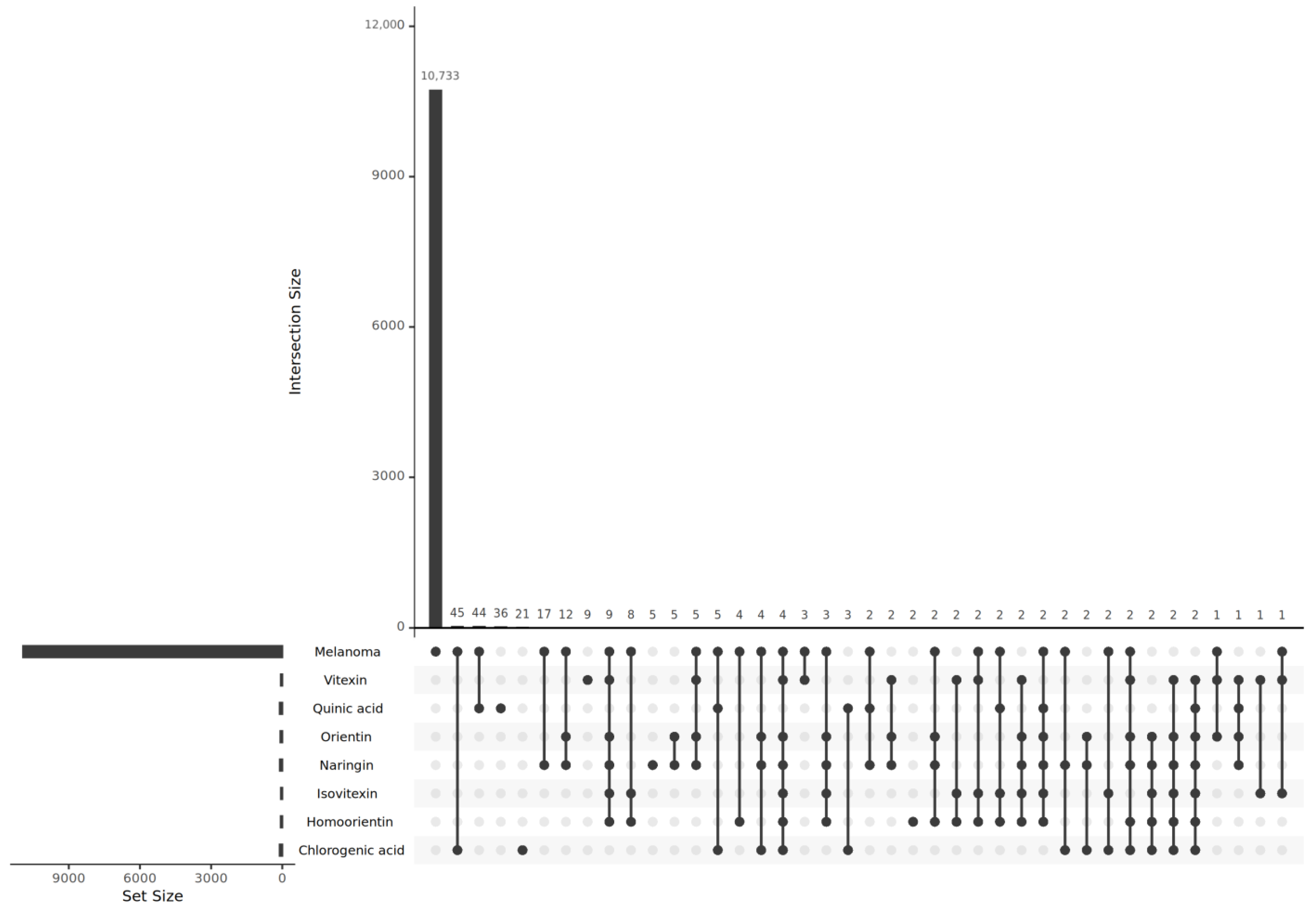
2.3. Predicted Target Genes of B. stenostachya and Related Genes of Melanoma
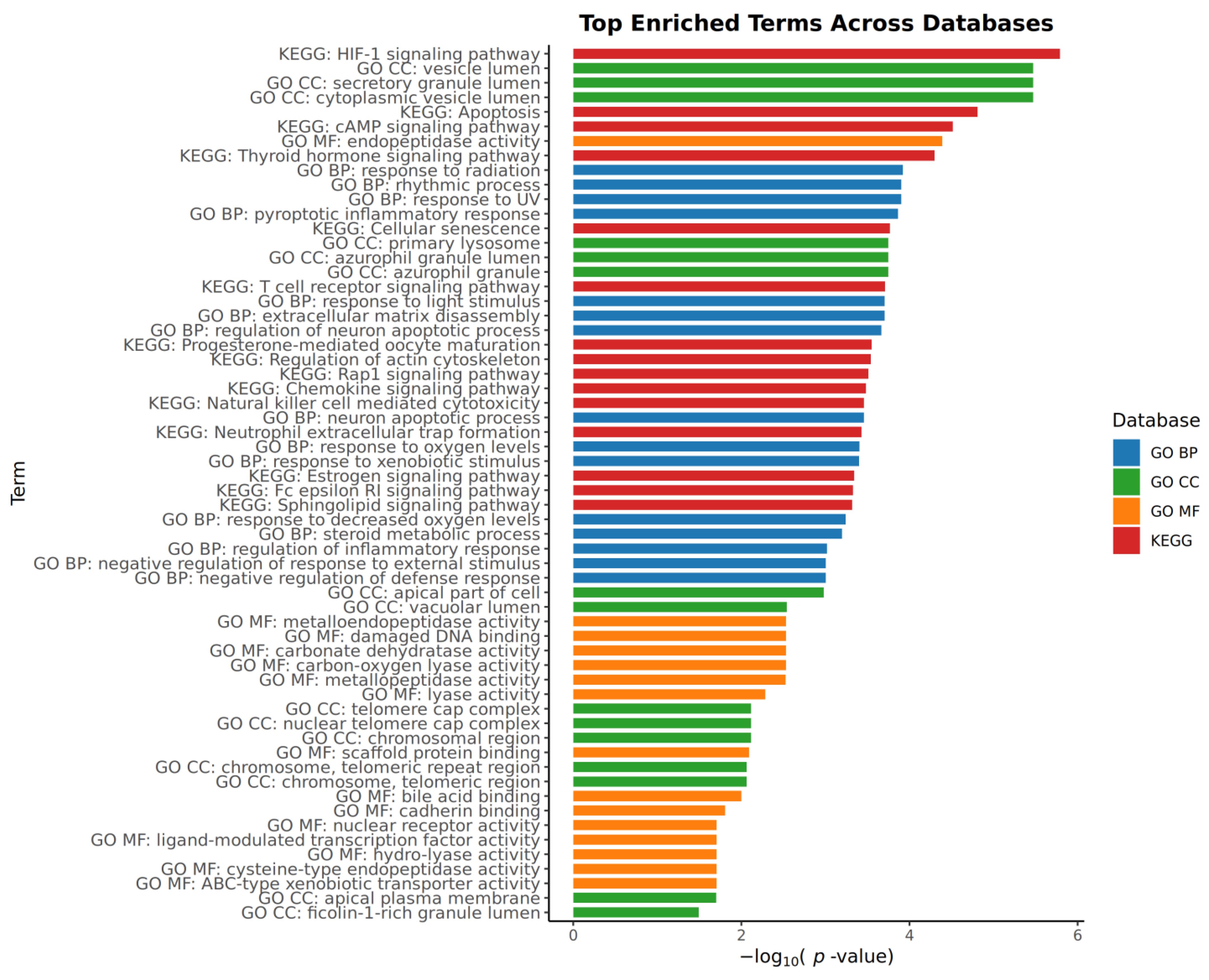
2.4. Pathway and Process Enrichment Analysis
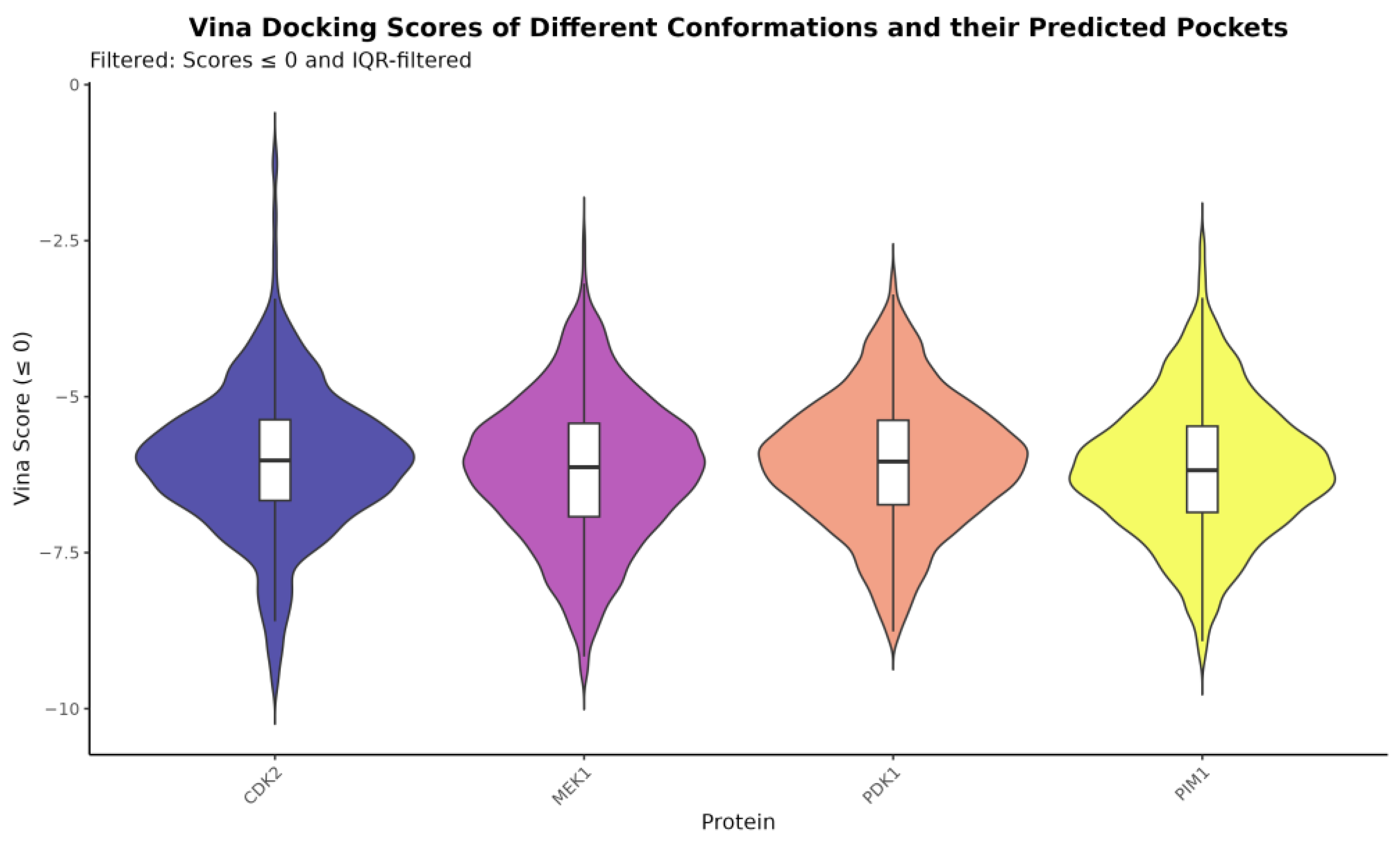
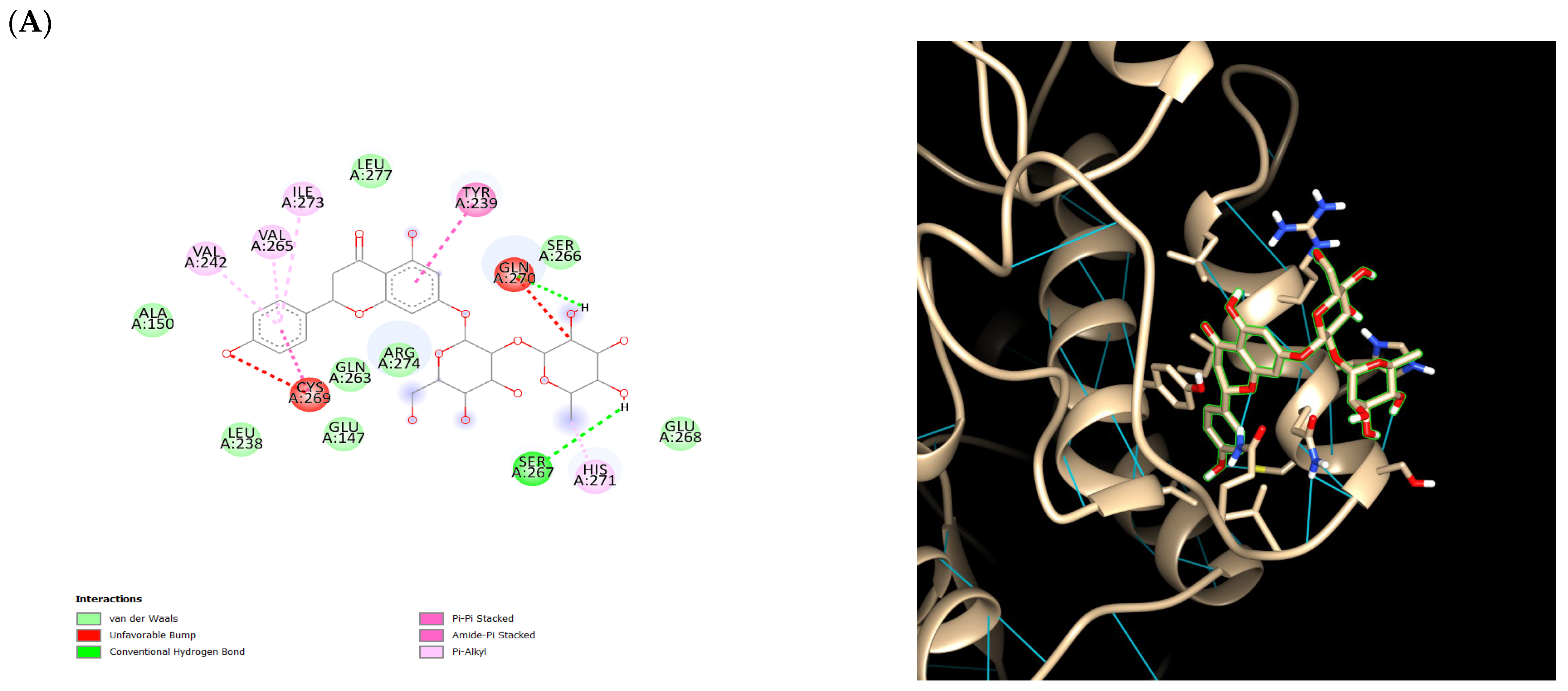
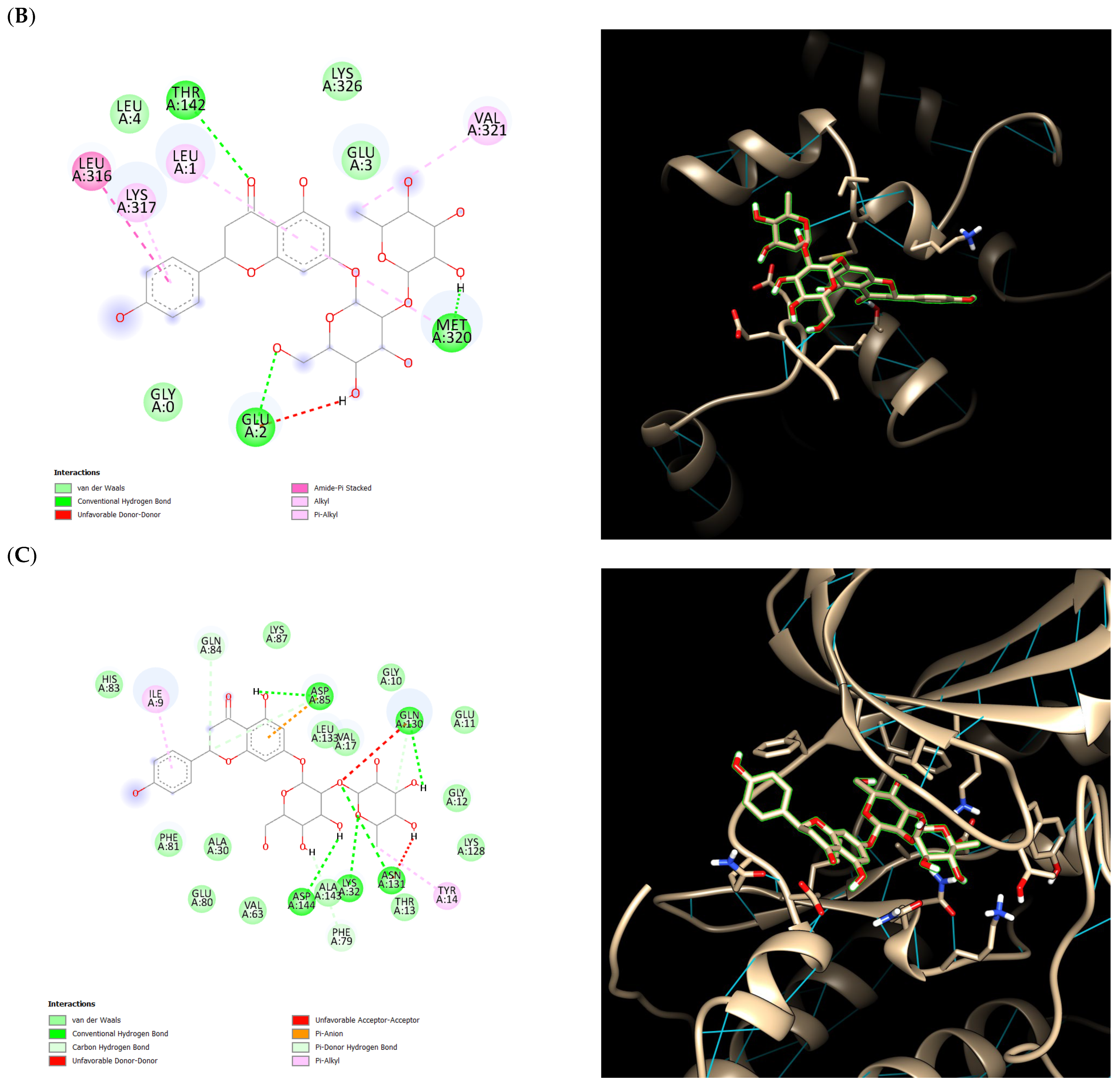
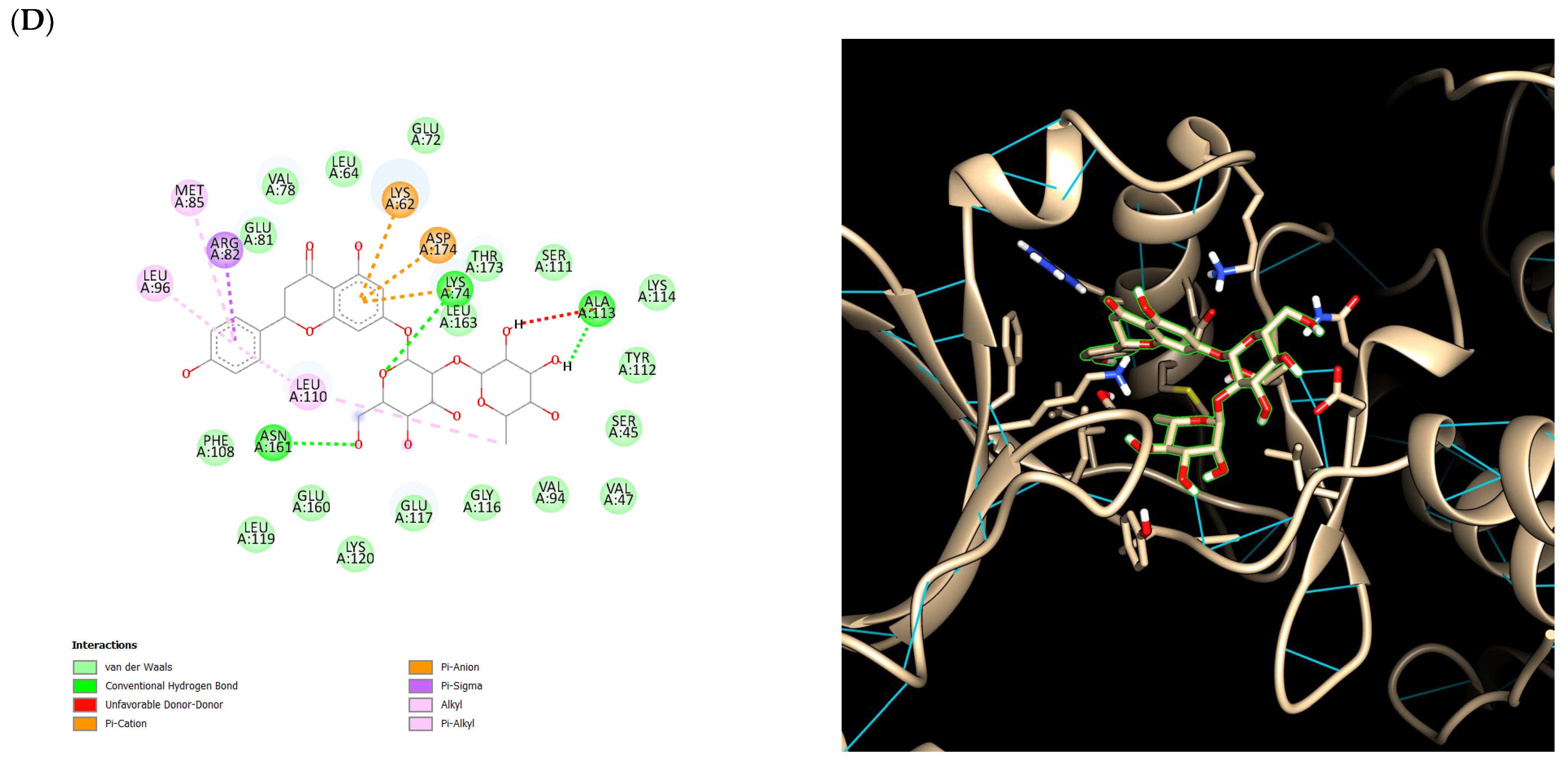
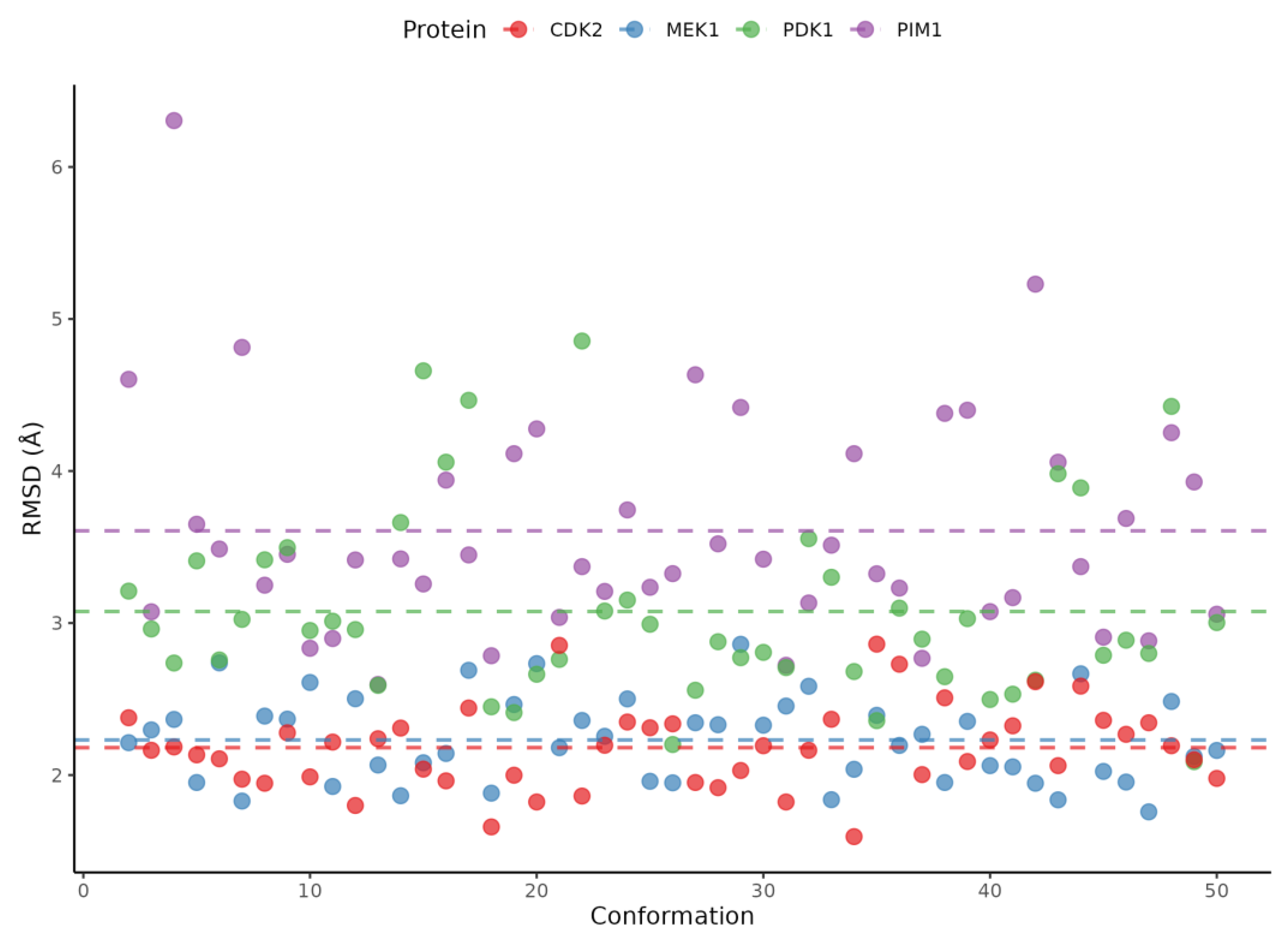
2.5. Ensemble Docking Results
3. Discussion
4. Materials and Methods
4.1. Materials and Sample Preparation
4.2. LCMS Analysis of B. stenostachya Leaf Crude Extract
4.3. Screening of Bioactive Compounds from B. stenostachya
4.4. Target Prediction of Bioactive Compounds from B. stenostachya
4.5. Pathway Enrichment Analysis
4.6. Molecular Docking Preparation
4.7. Ensemble Docking
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Council. Melanoma; Better Health Channel: Melbourne, Australia, 2024. [Google Scholar]
- Brozyna, A.; Zbytek, B.; Granese, J.; Carlson, J.A.; Ross, J.; Slominski, A. Mechanism of UV-related carcinogenesis and its contribution to nevi/melanoma. Expert Rev. Dermatol. 2008, 2, 451–469. [Google Scholar] [CrossRef]
- World Health Organization. Skin Cancer; International Agency for Research on Cancer: Lyon, France, 2022. [Google Scholar]
- American Cancer Society. Treating Melanoma Skin Cancer; American Cancer Society: Atlanta, GA, USA, 2024. [Google Scholar]
- Serrone, L.; Zeuli, M.; Sega, F.M.; Cognetti, F. Dacarbazine-based chemotherapy for metastatic melanoma: Thirty-year experience overview. J. Exp. Clin. Cancer Res. 2000, 19, 21–34. [Google Scholar] [PubMed]
- Lima, I.B.; Alvarenga, B.M.; de Tótaro, P.I.S.; Boratto, F.; Guimaraes, P.P.G. Improved antiproliferative activity of doxorubicin-loaded calcium phosphate nanoparticles against melanoma cells. Braz. Arch. Biol. Technol. 2022, 66, e23220572. [Google Scholar] [CrossRef]
- Nair, R. Herbal Medicine: Connecting Traditional Knowledge with Modern Pharmacology. J. Basic Clin. Pharm. 2024, 15, 360. [Google Scholar]
- Chaachouay, N.; Zidane, Z.L. Plant-Derived Natural Products: A Source for Drug Discovery and Development. Drug Candidates Nat. Sources 2023, 3, 184–207. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene; International Agency for Research on Cancer: Lyon, France, 2002. [Google Scholar]
- Lin, Y.-T.; Whitman, W.B.; Coleman, D.C.; Shiau, Y.-J.; Jien, S.-H.; Chiu, C.-Y. The influences of thorny bamboo growth on the bacterial community in badland soils of southwestern Taiwan. Land Degrad. Dev. 2018, 29, 2728–2738. [Google Scholar] [CrossRef]
- Choi, M.-H.; Jo, H.-G.; Yang, J.H.; Ki, S.H.; Shin, H.-J. Antioxidative and Anti-Melanogenic Activities of Bamboo Stems (Phyllostachys nigra variety henosis) via PKA/CREB-Mediated MITF Downregulation in B16F10 Melanoma Cells. Int. J. Mol. Sci. 2018, 19, 409. [Google Scholar] [CrossRef]
- Ashour, A.; Elbermawi, A.; Amen, Y.; Allam, A.E.; Ikeda, H.; Nagata, M.; Kumagae, K.; Azuma, T.; Taguchi, A.; Takemoto, T.; et al. Melanin Synthesis Inhibition Activity of Compounds Isolated from Bamboo Shoot Skin (Phyllostachys pubescens). Molecules 2023, 28, 23. [Google Scholar] [CrossRef]
- Chang, T.; Ding, H.; Wang, T.; Wu, J.; Tsai, P.; Suratos, K.S.; Tayo, L.L.; Liu, G.; Ting, H. In silico–guided synthesis of a new, highly soluble, and anti-melanoma flavone glucoside: Skullcapflavone II-6′-O-β-glucoside. Biotechnol. Appl. Biochem. 2024, 72, 621–637. [Google Scholar] [CrossRef]
- Liu, H.-C.; Hsieh, C.-Y.; Tsai, P.-W.; Chou, T.-Y.; Yang, S.-C.; Chang, C.-H.; Huang, Y.-P.; Chien, C.-C.; Lee, S.-C.; Shih, H.-D.; et al. Development and Applications of Bambusa stenostachya Leaf Extract in Personal Care Products. Processes 2025, 13, 233. [Google Scholar] [CrossRef]
- Mayo Clinic. Melanoma; Mayo Clinic: Rochester, MN, USA, 2023. [Google Scholar]
- Patel, N.; Neupane, R.; Balaji, S.; Tiwari, A.K.; Ray, S.D. Dacarbazine. In Encyclopedia of Toxicology; Academic Press: Cambridge, MA, USA, 2024; Volume 3. [Google Scholar]
- Kciuk, M.; Gielecińska, A.; Mujwar, S.; Kołat, D.; Kałuzińska-Kołat, Ż.; Celik, I.; Kontek, R. Doxorubicin—An Agent with Multiple Mechanisms of Anticancer Activity. Cells 2023, 12, 659. [Google Scholar] [CrossRef]
- World Health Organization. Traditional Medicine Has a Long History of Contributing to Conventional Medicine and Continues to Hold Promise 10 August 2023; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- Ouhtit, A. Understanding the functional discrepancy of Pim-1 in cancer. Front. Biosci. 2015, 7, 728. [Google Scholar] [CrossRef] [PubMed]
- Tursynbay, Y.; Zhang, J.; Li, Z.; Tokay, T.; Zhumadilov, Z.; Wu, D.; Xie, Y. Pim-1 kinase as cancer drug target: An update. Biomed. Rep. 2016, 4, 140–146. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). MAP2K1 Mitogen-Activated Protein Kinase Kinase 1 [Homo sapiens (Human)]; National Center for Biotechnology Information (NCBI): Bethesda, MD, USA, 2025.
- Mizuno, S.; Ikegami, M.; Koyama, T.; Sunami, K.; Ogata, D.; Kage, H.; Yanagaki, M.; Ikeuchi, H.; Ueno, T.; Tanikawa, M.; et al. High-Throughput Functional Evaluation of MAP2K1 Variants in Cancer. Mol. Cancer Ther. 2023, 22, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gan, Y.; Li, H.; Yin, J.; He, X.; Lin, L.; Xu, S.; Fang, Z.; Kim, B.; Gao, L.; et al. Inhibition of the CDK2 and Cyclin A complex leads to autophagic degradation of CDK2 in cancer cells. Nat. Commun. 2022, 13, 2835. [Google Scholar] [CrossRef]
- Levina, A.; Fleming, K.D.; Burke, J.E.; Leonard, T.A. Activation of the essential kinase PDK1 by phosphoinositide-driven trans-autophosphorylation. Nat. Commun. 2022, 13, 1874. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Wang, W.; Jeon, J.H.; Shen, T.; Huang, X.; Yi, P.; Dong, B.; Yang, F. Cooperative activation of PDK1 and AKT by MAPK4 enhances cancer growth and resistance to therapy. PLoS Biol. 2023, 21, e3002227. [Google Scholar] [CrossRef]
- Motwani, J.; Eccles, M.R. Genetic and Genomic Pathways of Melanoma Development, Invasion and Metastasis. Genes 2021, 12, 1543. [Google Scholar] [CrossRef]
- Molina, J.R.; Adjei, A.A. The Ras/Raf/MAPK Pathway. J. Thorac. Oncol. 2006, 1, 7–9. [Google Scholar] [CrossRef]
- Davies, M.A. The Role of the PI3K-AKT Pathway in Melanoma. Cancer J. 2012, 18, 142–147. [Google Scholar] [CrossRef]
- Blalock, W.L.; Navolanic, P.M.; Steelman, L.S.; Shelton, J.G.; Moye, P.W.; Lee, J.T.; Franklin, R.A.; Mirza, A.; McMahon, M.; White, M.K.; et al. Requirement for the PI3K/Akt pathway in MEK1-mediated growth and prevention of apoptosis: Identification of an Achilles heel in leukemia. Leukemia 2003, 17, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Beale, G.; Haagensen, E.J.; Thomas, H.D.; Wang, L.-Z.; Revill, C.H.; Payne, S.L.; Golding, B.T.; Hardcastle, I.R.; Newell, D.R.; Griffin, R.J.; et al. Combined PI3K and CDK2 inhibition induces cell death and enhances in vivo antitumour activity in colorectal cancer. Br. J. Cancer 2016, 115, 682–690. [Google Scholar] [CrossRef]
- Xia, H.; Huang, Z.; Xu, Y.; Yam, J.W.P.; Cui, Y. Reprogramming of central carbon metabolism in hepatocellular carcinoma. Biomed. Pharmacother. 2022, 153, 113485. [Google Scholar] [CrossRef]
- Choudhury, R.; Bahadi, C.K.; Ray, I.P.; Dash, P.; Pattanaik, I.; Mishra, S.; Mohapatra, S.R.; Patnaik, S.; Nikhil, K. PIM1 kinase and its diverse substrate in solid tumors. Cell Commun. Signal. 2024, 22, 529. [Google Scholar] [CrossRef]
- Liang, C.; Li, Y.-Y. Use of regulators and inhibitors of Pim-1, a serine/threonine kinase, for tumour therapy (Review). Mol. Med. Rep. 2014, 9, 2051–2060. [Google Scholar] [CrossRef]
- Arafah, A.; Rehman, M.U.; Mir, T.M.; Wali, A.F.; Ali, R.; Qamar, W.; Khan, R.; Ahmad, A.; Aga, S.S.; Alqahtani, S.; et al. Multi-Therapeutic Potential of Naringenin (4′,5,7-Trihydroxyflavonone): Experimental Evidence and Mechanisms. Plants 2020, 9, 1784. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, H.-P. Research progress on the anti-tumor effect of Naringin. Front. Pharmacol. 2023, 14, 1217001. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ouyang, S.; Yu, B.; Liu, Y.; Huang, K.; Gong, J.; Zheng, S.; Li, Z.; Li, H.; Jiang, H. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010, 38, W609–W614. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Dunkel, M.; Gunther, S.; Ahmed, J.; Wittig, B.; Preissner, R. SuperPred: Drug classification and target prediction. Nucleic Acids Res. 2008, 36, W55–W59. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Adesina, A.; Ahmad, S.; Bowler-Barnett, E.H.; Bye-A-Jee, H.; Carpentier, D.; Denny, P.; et al. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Weizmann Institute of Science. GeneCards—The Human Gene Database; Weizmann Institute of Science: Rehovot, Israel, 1997. [Google Scholar]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Shreffler, J.; Huecker, M. Hypothesis Testing, P Values, Confidence Intervals, and Significance; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Janson, G.; Jussupow, A.; Feig, M. Deep generative modeling of temperature-dependent structural ensembles of proteins. bioRxiv 2025. [Google Scholar] [CrossRef]
- Schmidtke, P.; Le Guilloux, V.; Maupetit, J.; Tuffery, P. fpocket: Online tools for protein ensemble pocket detection and tracking. Nucleic Acids Res. 2010, 38, W582–W589. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
| PubChem ID | Chemical | Molecular Weight (g/mol) | Molecular Formula |
|---|---|---|---|
| 6508 | Quinic acid | 192.17 | C7H12O6 |
| 1794427 | Chlorogenic acid | 354.31 | C16H18O9 |
| 5280441 | Vitexin | 432.4 | C21H20O10 |
| 162350 | Isovitexin | 432.4 | C21H20O10 |
| 114776 | Homoorientin | 448.38 | C21H20O11 |
| 5281675 | Orientin | 448.4 | C21H20O11 |
| 442428 | Naringin (Naringenin 7-rhamnoglucoside) | 580.5 | C27H32O14 |
| Molecule | GI Absorption | Bioavailability Score | OB% | Structure |
|---|---|---|---|---|
| Chlorogenic acid | Low | 0.11 | 13.61 |  |
| Quinic acid | High | 0.56 | 63.53 | 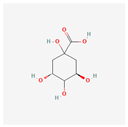 |
| Homoorientin (isoorientin) | Low | 0.17 | 23.3 | 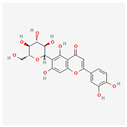 |
| Orientin | Low | 0.17 | 1.79 | 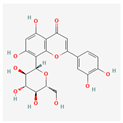 |
| Vitexin | Low | 0.55 | 3.05 | 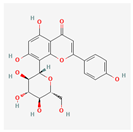 |
| Isovitexin (rutin and ferulic acid) | Low | 0.55 | 31.29 | 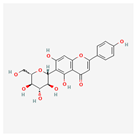 |
| Naringin-7-rhamnoglucoside | Low | 0.17 | 6.92 | 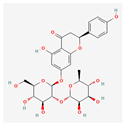 |
| Protein | Ligand | csMIN | csAVG | csMED | csTRIMMEAN |
|---|---|---|---|---|---|
| CDK2 | CHLOROGENIC ACID | −9.43000 | −4.92246 | −5.91150 | −5.18689 |
| CDK2 | DACARBAZINE | −6.11900 | −3.67363 | −4.36200 | −3.87281 |
| CDK2 | DOXORUBICIN | −86.06400 | −5.21502 | −5.88450 | −5.13361 |
| CDK2 | HOMOORIENTIN | −9.63400 | −4.89919 | −5.75800 | −5.11512 |
| CDK2 | ISOVITEXIN | −9.60300 | −4.81899 | −5.75450 | −5.03151 |
| CDK2 | NARINGIN | −10.17400 | −5.26133 | −6.20200 | −5.48337 |
| CDK2 | ORIENTIN | −12.12300 | −5.05633 | −5.98100 | −5.26331 |
| CDK2 | QUINIC ACID | −8.12300 | −4.79348 | −5.66450 | −5.04931 |
| CDK2 | VITEXIN | −10.40400 | −4.96989 | −5.93700 | −5.17481 |
| MEK1 | CHLOROGENIC ACID | −8.99900 | −5.54884 | −6.07900 | −5.94549 |
| MEK1 | DACARBAZINE | −6.33300 | −4.06540 | −4.46100 | −4.37759 |
| MEK1 | DOXORUBICIN | −20.71700 | −5.71248 | −6.14900 | −6.01304 |
| MEK1 | HOMOORIENTIN | −10.53400 | −5.63281 | −6.12200 | −5.99234 |
| MEK1 | ISOVITEXIN | −11.00200 | −5.59132 | −6.09000 | −5.95523 |
| MEK1 | NARINGIN | −15.06100 | −6.07004 | −6.59300 | −6.42142 |
| MEK1 | ORIENTIN | −12.31800 | −5.77236 | −6.25400 | −6.15324 |
| MEK1 | QUINIC ACID | −7.70700 | −5.18088 | −5.67900 | −5.58443 |
| MEK1 | VITEXIN | −10.73900 | −5.69276 | −6.21200 | −6.06392 |
| PDK1 | CHLOROGENIC ACID | −10.69600 | −5.84483 | −6.08850 | −6.12630 |
| PDK1 | DACARBAZINE | −6.55200 | −4.26388 | −4.47700 | −4.47145 |
| PDK1 | DOXORUBICIN | −17.18300 | −5.94061 | −6.16500 | −6.18560 |
| PDK1 | HOMOORIENTIN | −9.87300 | −5.88804 | −6.12450 | −6.14669 |
| PDK1 | ISOVITEXIN | −9.52400 | −5.83747 | −6.06300 | −6.08772 |
| PDK1 | NARINGIN | −10.60100 | −6.29952 | −6.54000 | −6.57658 |
| PDK1 | ORIENTIN | −10.18200 | −6.00952 | −6.27700 | −6.26988 |
| PDK1 | QUINIC ACID | −8.24700 | −5.43671 | −5.70000 | −5.69214 |
| PDK1 | VITEXIN | −10.14200 | −5.94927 | −6.18250 | −6.19803 |
| PIM1 | CHLOROGENIC ACID | −9.54600 | −5.52084 | −6.11100 | −5.92045 |
| PIM1 | DACARBAZINE | −6.72500 | −4.09535 | −4.45800 | −4.39848 |
| PIM1 | DOXORUBICIN | −50.59300 | −5.57615 | −6.08800 | −5.86939 |
| PIM1 | HOMOORIENTIN | −10.45500 | −5.58319 | −6.20750 | −5.94528 |
| PIM1 | ISOVITEXIN | −10.69600 | −5.52624 | −6.13400 | −5.88888 |
| PIM1 | NARINGIN | −17.22900 | −5.91897 | −6.54600 | −6.30309 |
| PIM1 | ORIENTIN | −11.47800 | −5.60748 | −6.28100 | −5.98845 |
| PIM1 | QUINIC ACID | −8.51000 | −5.33340 | −5.89500 | −5.73615 |
| PIM1 | VITEXIN | −10.43800 | −5.53094 | −6.14600 | −5.91048 |
| PDB | PDB ID | Protein Name | Resolution |
|---|---|---|---|
| 3A99 | pdb_00003a99 | proto-oncogene serine/threonine-protein kinase | 1.60 A |
| 2R3Q | pdb_00002r3q | mitogen-activated protein kinase | 1.35 A |
| 5LVO | pdb_00005lvo | cyclin-dependent kinase 2 | 1.09 A |
| 7B7R | pdb_00007b7r | phosphoinositide-dependent kinase-1 | 1.70 A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darilag, G.M.C.; Liu, H.-C.; Hsieh, C.-Y.; Tayo, L.L.; Talubo, N.D.D.; Yang, S.-C.; Chang, C.-H.; Huang, Y.-P.; Lee, S.-C.; Liu, Y.-C.; et al. Uncovering Anti-Melanoma Mechanisms of Bambusa stenostachya Leaf Compounds via Network Pharmacology and Molecular Docking. Int. J. Mol. Sci. 2025, 26, 6120. https://doi.org/10.3390/ijms26136120
Darilag GMC, Liu H-C, Hsieh C-Y, Tayo LL, Talubo NDD, Yang S-C, Chang C-H, Huang Y-P, Lee S-C, Liu Y-C, et al. Uncovering Anti-Melanoma Mechanisms of Bambusa stenostachya Leaf Compounds via Network Pharmacology and Molecular Docking. International Journal of Molecular Sciences. 2025; 26(13):6120. https://doi.org/10.3390/ijms26136120
Chicago/Turabian StyleDarilag, Gen Maxxine C., Hsuan-Chieh Liu, Cheng-Yang Hsieh, Lemmuel L. Tayo, Nicholas Dale D. Talubo, Shu-Ching Yang, Ching-Hui Chang, Ying-Pin Huang, Shih-Chi Lee, Yung-Chuan Liu, and et al. 2025. "Uncovering Anti-Melanoma Mechanisms of Bambusa stenostachya Leaf Compounds via Network Pharmacology and Molecular Docking" International Journal of Molecular Sciences 26, no. 13: 6120. https://doi.org/10.3390/ijms26136120
APA StyleDarilag, G. M. C., Liu, H.-C., Hsieh, C.-Y., Tayo, L. L., Talubo, N. D. D., Yang, S.-C., Chang, C.-H., Huang, Y.-P., Lee, S.-C., Liu, Y.-C., & Tsai, P.-W. (2025). Uncovering Anti-Melanoma Mechanisms of Bambusa stenostachya Leaf Compounds via Network Pharmacology and Molecular Docking. International Journal of Molecular Sciences, 26(13), 6120. https://doi.org/10.3390/ijms26136120










