Identification of Key Genes for Alcoholic Hepatitis Using Integrated Network Analysis of Differential lncRNA and Gene Expression
Abstract
1. Introduction
2. Results
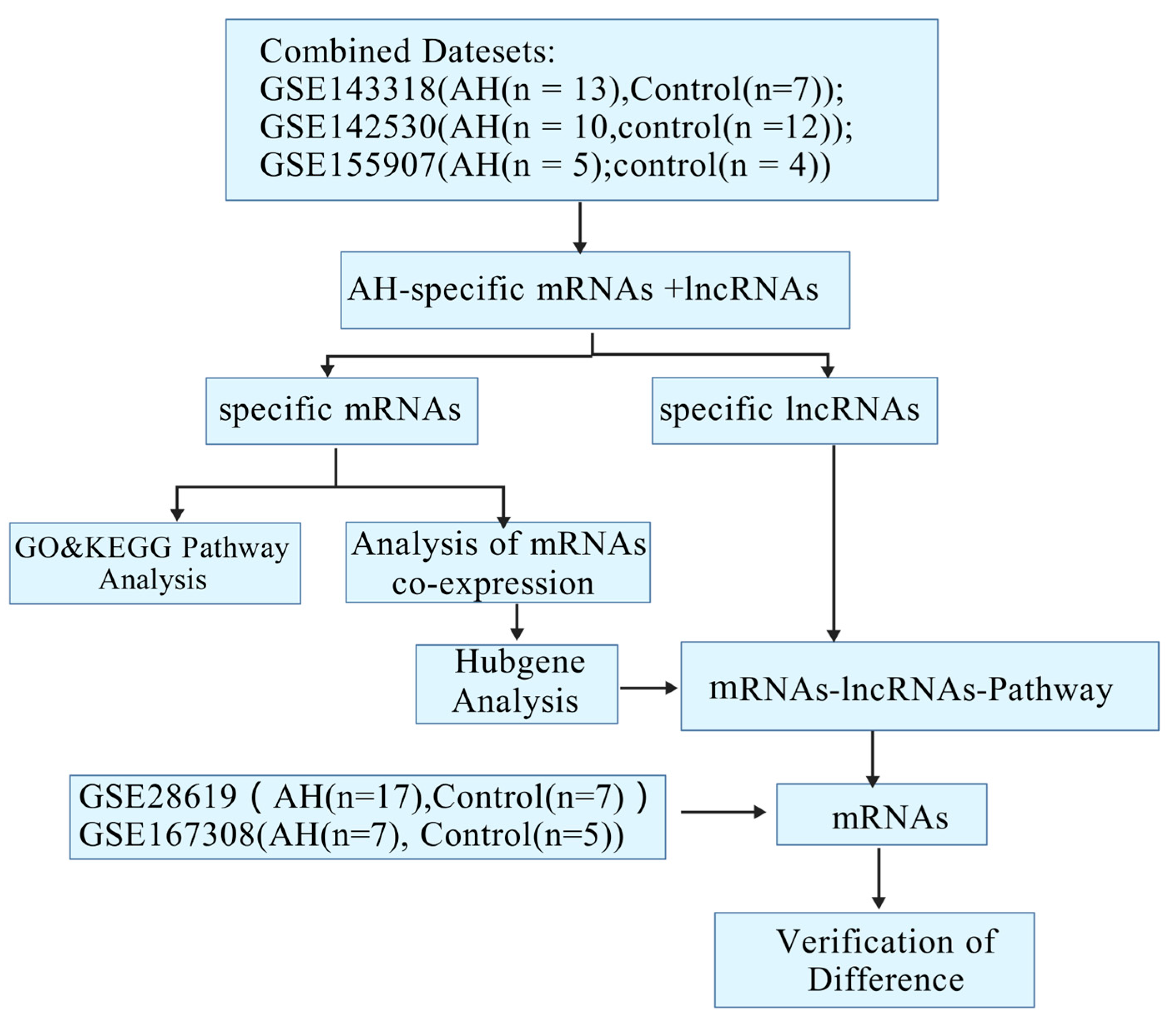
2.1. Identification and Cluster Analysis of Differential Genes in Alcoholic Hepatitis
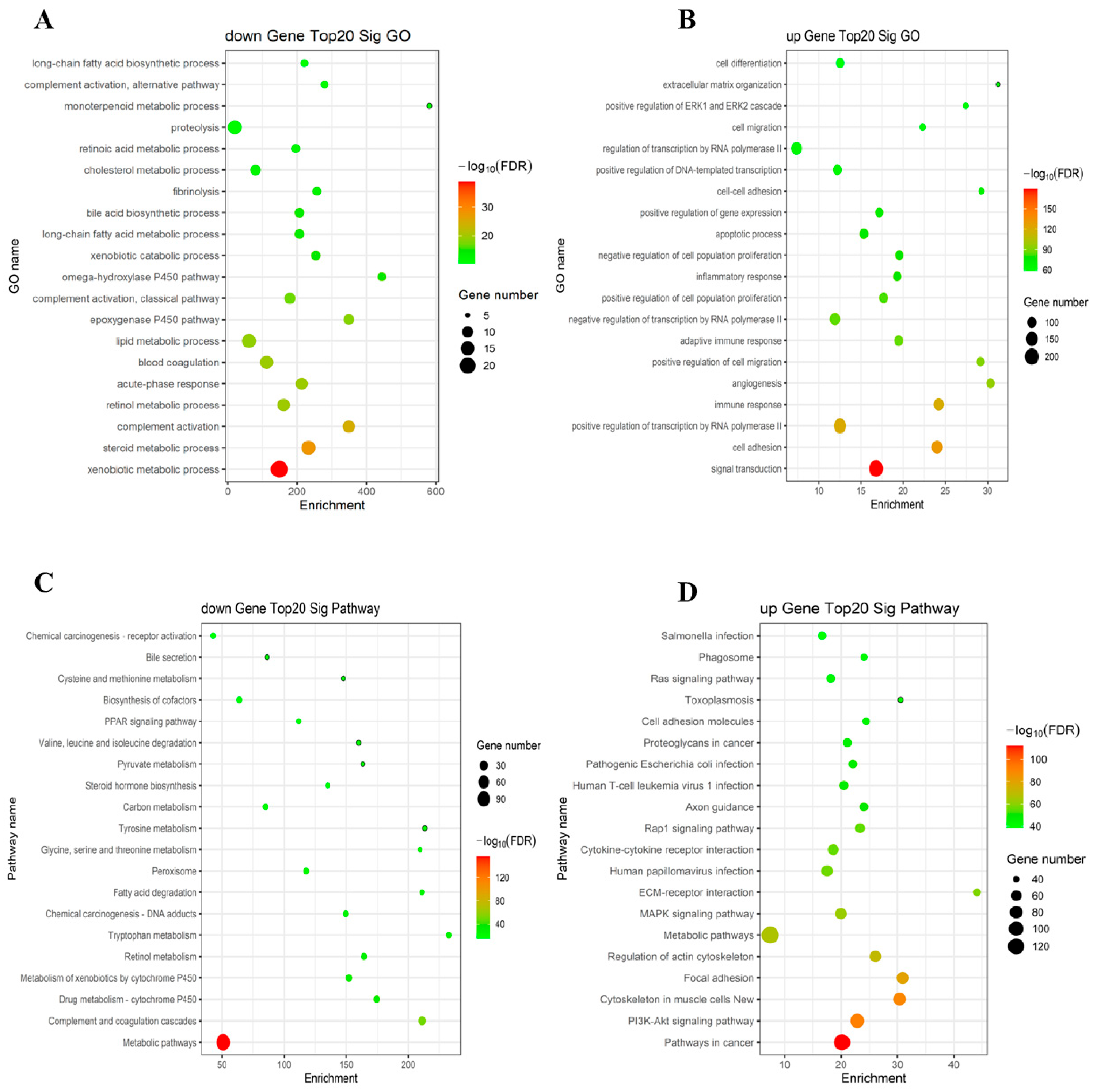
2.2. The GO and KEGG Pathway Analysis
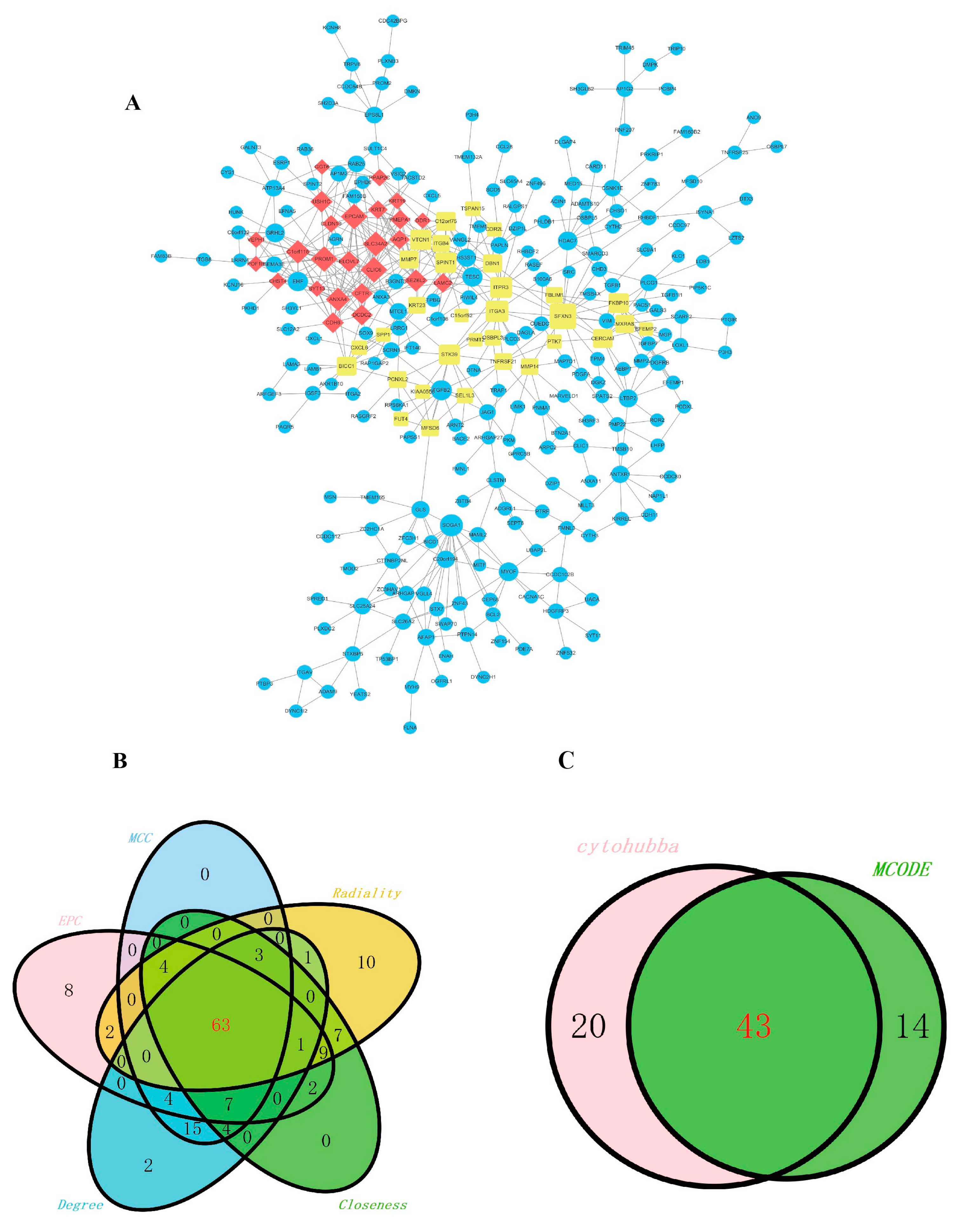
2.3. Construction of the Co-Expression of Differential Genes and the Hub Gene Analysis
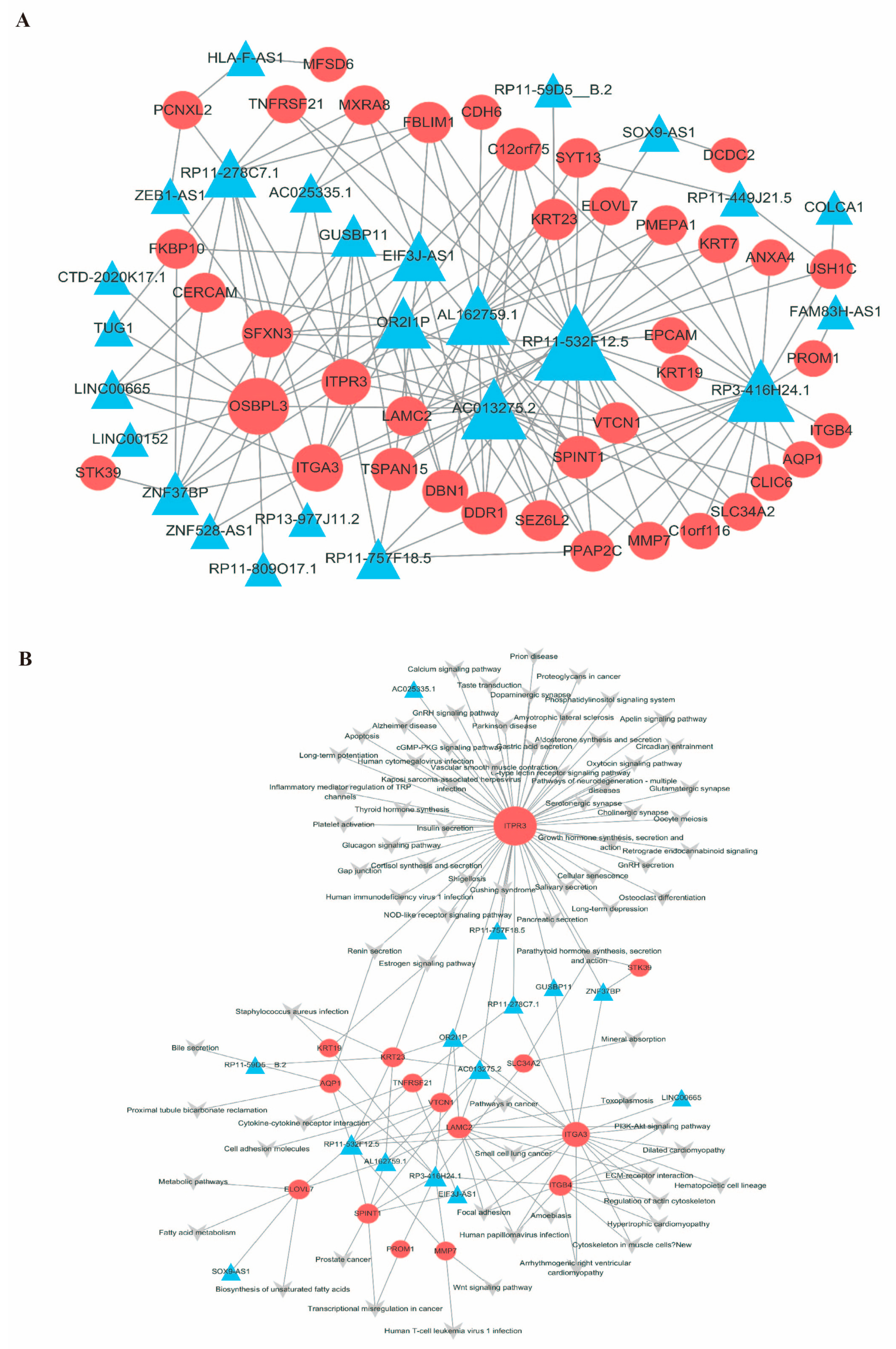
2.4. Construction of the mRNA-LncRNA Co-Expression Network
2.5. Construction of the mRNA-lncRNA–Pathway Co-Expression Network
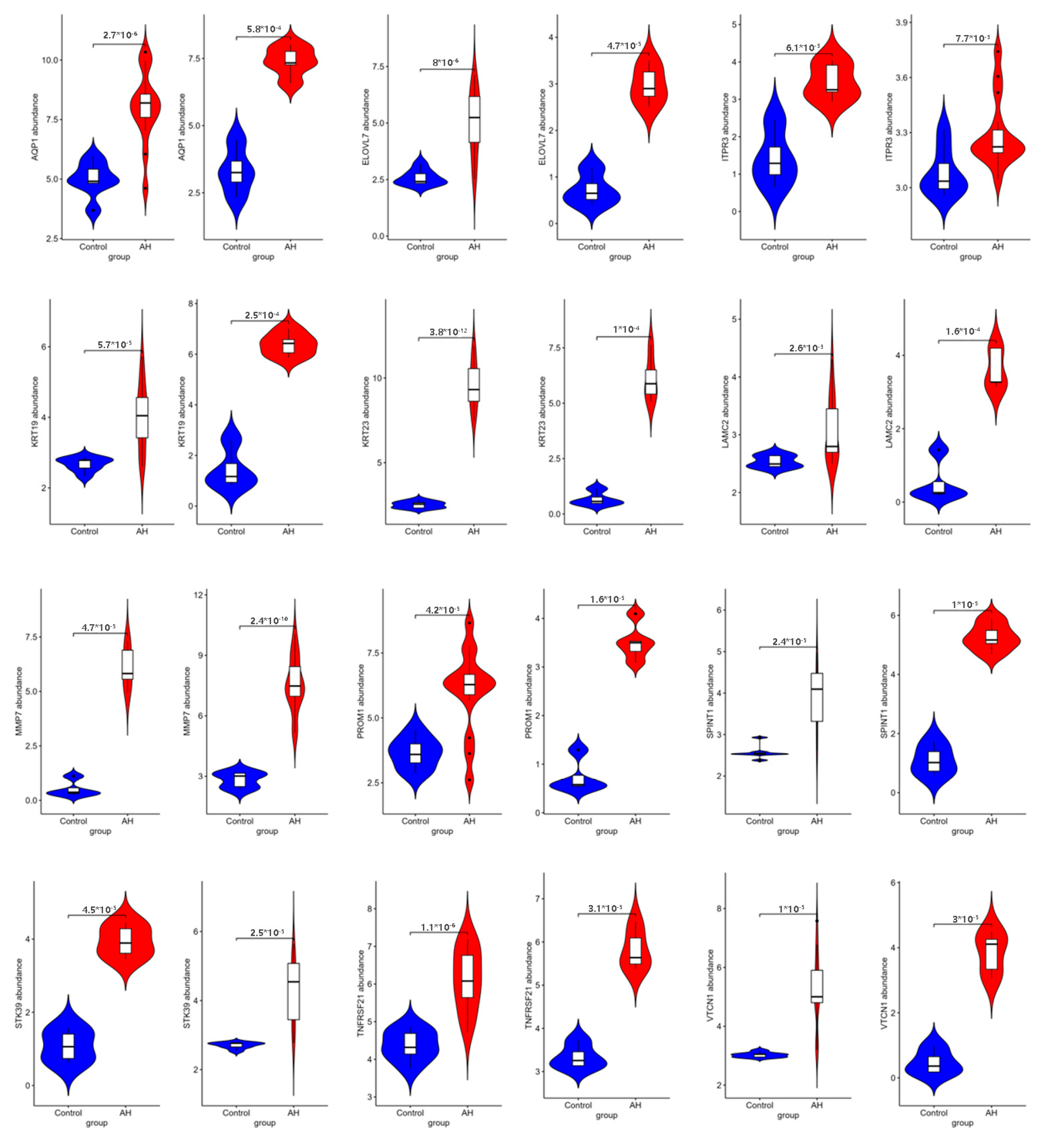
2.6. Verification of Pivotal mRNAs with Diagnostic Value
3. Discussion
4. Materials and Methods
4.1. The Data Collection
4.2. Gene Ontology and Pathway Enrichment Analysis
4.3. A Co-Expression Analysis of Differentially Expressed Genes
4.4. The Hub Gene Analysis
4.5. Construction of the lncRNA-mRNA Pathway Co-Expression Network
4.6. Verification of Hub Genes
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yan, C.; Hu, W.; Tu, J.; Li, J.; Liang, Q.; Han, S. Pathogenic mechanisms and regulatory factors involved in alcoholic liver disease. J. Transl. Med. 2023, 21, 300. [Google Scholar] [CrossRef] [PubMed]
- Kalligeros, M.; Vassilopoulos, A.; Vassilopoulos, S.; Victor, D.W.; Mylonakis, E.; Noureddin, M. Prevalence of Steatotic Liver Disease (MASLD, MetALD, and ALD) in the United States: NHANES 2017–2020. Clin. Gastroenterol. Hepatol. 2024, 22, 1330–1332.e1334. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic liver disease. Nat. Rev. Dis. Primers 2018, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- Safa, A.; Gholipour, M.; Dinger, M.E.; Taheri, M.; Ghafouri-Fard, S. The critical roles of lncRNAs in the pathogenesis of melanoma. Exp. Mol. Pathol. 2020, 117, 104558. [Google Scholar] [CrossRef]
- Yang, J.; Liu, F.; Wang, Y.; Qu, L.; Lin, A. LncRNAs in tumor metabolic reprogramming and immune microenvironment remodeling. Cancer Lett. 2022, 543, 215798. [Google Scholar] [CrossRef]
- Han, J.J. LncRNAs: The missing link to senescence nuclear architecture. Trends Biochem. Sci. 2023, 48, 618–628. [Google Scholar] [CrossRef]
- Pu, S.; Xu, Y.; Li, X.; Yu, Z.; Zhang, Y.; Tong, X.; Shan, Y.; Gao, X. LncRNAS-modulators of neurovascular units in diabetic retinopathy. Eur. J. Pharmacol. 2022, 925, 174937. [Google Scholar] [CrossRef]
- Okuyan, H.M.; Begen, M.A. LncRNAs in Osteoarthritis. Clin. Chim. Acta 2022, 532, 145–163. [Google Scholar] [CrossRef]
- Javed, Z.; Ahmed Shah, F.; Rajabi, S.; Raza, Q.; Iqbal, Z.; Ullah, M.; Ahmad, T.; Salehi, B.; Sharifi-Rad, M.; Pezzani, R.; et al. LncRNAs as Potential Therapeutic Targets in Thyroid Cancer. Asian Pac. J. Cancer Prev. 2020, 21, 281–287. [Google Scholar] [CrossRef]
- Wu, T.; Du, Y. LncRNAs: From Basic Research to Medical Application. Int. J. Biol. Sci. 2017, 13, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Zhang, C.; Yi, H. The dysregulation of lncRNAs by epigenetic factors in human pathologies. Drug Discov. Today 2023, 28, 103664. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Louvet, A.; Mathurin, P. Alcoholic liver disease: Mechanisms of injury and targeted treatment. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, R.; Nan, Y.; Kong, L. Deciphering the role of LncRNA in alcoholic liver disease: Mechanisms and therapeutic potential. Medicine 2024, 103, e40378. [Google Scholar] [CrossRef]
- Wang, Q.; Li, M.; Shen, Z.; Bu, F.; Yu, H.; Pan, X.; Yang, Y.; Meng, X.; Huang, C.; Li, J. The Long Non-coding RNA MEG3/miR-let-7c-5p Axis Regulates Ethanol-Induced Hepatic Steatosis and Apoptosis by Targeting NLRC5. Front. Pharmacol. 2018, 9, 302. [Google Scholar] [CrossRef]
- Shen, S.; Wang, J.; Lin, L.M. Downregulation of long non-coding RNA AIRN promotes mitophagy in alcoholic fatty hepatocytes by promoting ubiquitination of mTOR. Physiol. Res. 2021, 70, 245–253. [Google Scholar] [CrossRef]
- Douds, A.C.; Lim, A.G.; Jazrawi, R.P.; Finlayson, C.; Maxwell, J.D. Serum intercellular adhesion molecule-1 in alcoholic liver disease and its relationship with histological disease severity. J. Hepatol. 1997, 26, 280–286. [Google Scholar] [CrossRef]
- Xiang, H.; Tu, B.; Luo, M.; Hou, P.; Wang, J.; Zhang, R.; Wu, L. Knockdown of UCA1 attenuated the progression of alcoholic fatty disease by sponging miR-214. Mamm. Genome 2022, 33, 534–542. [Google Scholar] [CrossRef]
- Luo, J.; Xie, M.; Hou, Y.; Ma, W.; Jin, Y.; Chen, J.; Li, C.; Zhao, K.; Chen, N.; Xu, L.; et al. A novel epigenetic mechanism unravels hsa-miR-148a-3p-mediated CYP2B6 downregulation in alcoholic hepatitis disease. Biochem. Pharmacol. 2021, 188, 114582. [Google Scholar] [CrossRef]
- Xiong, J.; Ni, J.; Chen, C.; Wang, K. miR-148a-3p regulates alcoholic liver fibrosis through targeting ERBB3. Int. J. Mol. Med. 2020, 46, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Qi, Y.; Bai, Y.; Zhang, H.; Zhu, W.; Zhou, S.; Zhang, Y. LncRNA 1700020I14Rik promotes AKR1B10 expression and activates Erk pathway to induce hepatocyte damage in alcoholic hepatitis. Cell Death Discov. 2022, 8, 374. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Song, Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013, 339, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Shabangu, C.S.; Huang, J.F.; Hsiao, H.H.; Yu, M.L.; Chuang, W.L.; Wang, S.C. Liquid Biopsy for the Diagnosis of Viral Hepatitis, Fatty Liver Steatosis, and Alcoholic Liver Diseases. Int. J. Mol. Sci. 2020, 21, 3732. [Google Scholar] [CrossRef]
- Sehrawat, T.S.; Liu, M.; Shah, V.H. The knowns and unknowns of treatment for alcoholic hepatitis. Lancet Gastroenterol. Hepatol. 2020, 5, 494–506. [Google Scholar] [CrossRef]
- Dukić, M.; Radonjić, T.; Jovanović, I.; Zdravković, M.; Todorović, Z.; Kraišnik, N.; Aranđelović, B.; Mandić, O.; Popadić, V.; Nikolić, N.; et al. Alcohol, Inflammation, and Microbiota in Alcoholic Liver Disease. Int. J. Mol. Sci. 2023, 24, 3735. [Google Scholar] [CrossRef]
- Singal, A.K.; Bataller, R.; Ahn, J.; Kamath, P.S.; Shah, V.H. ACG Clinical Guideline: Alcoholic Liver Disease. Am. J. Gastroenterol. 2018, 113, 175–194. [Google Scholar] [CrossRef]
- Keating, M.; Lardo, O.; Hansell, M. Alcoholic Hepatitis: Diagnosis and Management. Am. Fam. Physician 2022, 105, 412–420. [Google Scholar]
- Pribék, I.K.; Kovács, I.; Kádár, B.K.; Kovács, C.S.; Richman, M.J.; Janka, Z.; Andó, B.; Lázár, B.A. Evaluation of the course and treatment of Alcohol Withdrawal Syndrome with the Clinical Institute Withdrawal Assessment for Alcohol-Revised: A systematic review-based meta-analysis. Drug Alcohol. Depend. 2021, 220, 108536. [Google Scholar] [CrossRef]
- Khalifa, A.; Rockey, D.C. The utility of liver biopsy in 2020. Curr. Opin. Gastroenterol. 2020, 36, 184–191. [Google Scholar] [CrossRef]
- Simonetto, D.A.; Shah, V.H.; Kamath, P.S. Outpatient management of alcohol-related liver disease. Lancet Gastroenterol. Hepatol. 2020, 5, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.Y.; Lee, D.H.; Liu, H.C.; Yang, Y.; Han, Y.H.; Kwon, T. Identifying competing endogenous RNA regulatory networks and hub genes in alcoholic liver disease for early diagnosis and potential therapeutic target insights. Aging 2024, 16, 9147–9167. [Google Scholar] [CrossRef] [PubMed]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Marengo, A.; Rosso, C.; Bugianesi, E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu. Rev. Med. 2016, 67, 103–117. [Google Scholar] [CrossRef]
- Mackowiak, B.; Fu, Y.; Maccioni, L.; Gao, B. Alcohol-associated liver disease. J. Clin. Investig. 2024, 134, e176345. [Google Scholar] [CrossRef]
- Fu, Y.; Maccioni, L.; Wang, X.W.; Greten, T.F.; Gao, B. Alcohol-associated liver cancer. Hepatology 2024, 80, 1462–1479. [Google Scholar] [CrossRef]
- Zhang, D.; Wei, Y.; Huang, Q.; Chen, Y.; Zeng, K.; Yang, W.; Chen, J.; Chen, J. Important Hormones Regulating Lipid Metabolism. Molecules 2022, 27, 7052. [Google Scholar] [CrossRef]
- Tian, L.Y.; Smit, D.J.; Jücker, M. The Role of PI3K/AKT/mTOR Signaling in Hepatocellular Carcinoma Metabolism. Int. J. Mol. Sci. 2023, 24, 2652. [Google Scholar] [CrossRef]
- Zhang, Z.; Shang, J.; Yang, Q.; Dai, Z.; Liang, Y.; Lai, C.; Feng, T.; Zhong, D.; Zou, H.; Sun, L.; et al. Exosomes derived from human adipose mesenchymal stem cells ameliorate hepatic fibrosis by inhibiting PI3K/Akt/mTOR pathway and remodeling choline metabolism. J. Nanobiotechnol. 2023, 21, 29. [Google Scholar] [CrossRef]
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, A.; Ragno, P.; Selleri, C.; Montuori, N. Recent Advances in the Function of the 67 kDa Laminin Receptor and its Targeting for Personalized Therapy in Cancer. Curr. Pharm. Des. 2017, 23, 4745–4757. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Liu, J.X.; Xie, J.; Gao, Z.; Yang, Z. LAMC2 as a prognostic biomarker in human cancer: A systematic review and meta-analysis. BMJ Open 2022, 12, e063682. [Google Scholar] [CrossRef]
- Kim, D.; Brocker, C.N.; Takahashi, S.; Yagai, T.; Kim, T.; Xie, G.; Wang, H.; Qu, A.; Gonzalez, F.J. Keratin 23 Is a Peroxisome Proliferator-Activated Receptor Alpha-Dependent, MYC-Amplified Oncogene That Promotes Hepatocyte Proliferation. Hepatology 2019, 70, 154–167. [Google Scholar] [CrossRef]
- Wang, W.M.; Yang, Z.G.; Liu, C.; Dong, Q. ELOVL fatty acid elongase 7 (ELOVL7), upregulated by Mdr2-knockout, predicts advanced liver fibrosis in patients with chronic hepatitis B. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 1954–1970. [Google Scholar] [CrossRef]
- Tian, F.; Wang, P.; Lin, D.; Dai, J.; Liu, Q.; Guan, Y.; Zhan, Y.; Yang, Y.; Wang, W.; Wang, J.; et al. Exosome-delivered miR-221/222 exacerbates tumor liver metastasis by targeting SPINT1 in colorectal cancer. Cancer Sci. 2021, 112, 3744–3755. [Google Scholar] [CrossRef]
- Liang, L.; Huan, L.; Wang, J.; Wu, Y.; Huang, S.; He, X. LncRNA RP11-295G20.2 regulates hepatocellular carcinoma cell growth and autophagy by targeting PTEN to lysosomal degradation. Cell Discov. 2021, 7, 118. [Google Scholar] [CrossRef]
- Wang, J.; Huang, F.; Shi, Y.; Zhang, Q.; Xu, S.; Yao, Y.; Jiang, R. RP11-323N12.5 promotes the malignancy and immunosuppression of human gastric cancer by increasing YAP1 transcription. Gastric Cancer 2021, 24, 85–102. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, H.; Liu, X.; Xu, L.; Li, T.; Zhou, H.; Zhu, H.; Hao, C.; Lin, C.; Zhang, Y. The RP11-417E7.1/THBS2 signaling pathway promotes colorectal cancer metastasis by activating the Wnt/β-catenin pathway and facilitating exosome-mediated M2 macrophage polarization. J. Exp. Clin. Cancer Res. 2024, 43, 195. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, B.; Xia, H.; Tian, P.; Li, X.; Yang, Y.; Zhu, Z.; Zhou, Y.; Liao, W.; Wang, S.; Yang, L.; et al. Identification of Key Genes for Alcoholic Hepatitis Using Integrated Network Analysis of Differential lncRNA and Gene Expression. Int. J. Mol. Sci. 2025, 26, 6104. https://doi.org/10.3390/ijms26136104
Hu B, Xia H, Tian P, Li X, Yang Y, Zhu Z, Zhou Y, Liao W, Wang S, Yang L, et al. Identification of Key Genes for Alcoholic Hepatitis Using Integrated Network Analysis of Differential lncRNA and Gene Expression. International Journal of Molecular Sciences. 2025; 26(13):6104. https://doi.org/10.3390/ijms26136104
Chicago/Turabian StyleHu, Bihuan, Hui Xia, Peixuan Tian, Xinbao Li, Yu Yang, Zixuan Zhu, Yajie Zhou, Wang Liao, Shoakang Wang, Ligang Yang, and et al. 2025. "Identification of Key Genes for Alcoholic Hepatitis Using Integrated Network Analysis of Differential lncRNA and Gene Expression" International Journal of Molecular Sciences 26, no. 13: 6104. https://doi.org/10.3390/ijms26136104
APA StyleHu, B., Xia, H., Tian, P., Li, X., Yang, Y., Zhu, Z., Zhou, Y., Liao, W., Wang, S., Yang, L., Sun, G., & Sui, J. (2025). Identification of Key Genes for Alcoholic Hepatitis Using Integrated Network Analysis of Differential lncRNA and Gene Expression. International Journal of Molecular Sciences, 26(13), 6104. https://doi.org/10.3390/ijms26136104









