The Link Between Dietary Timing and Exercise Performance Through Adipocyte AMPKα2 Signaling
Abstract
1. Introduction
2. Summary of Key Studies on Temporal Aspects of Fat–Muscle Crosstalk
3. Circadian Rhythms and Adipose Tissue Metabolism
4. Tissue-Specific Roles of AMPKα2 Signaling
5. Molecular Mechanisms of Fat–Muscle Crosstalk
5.1. AMPKα2-Mediated Regulation of Core Clock Genes
5.2. Adipocyte-to-Muscle Signaling Through Metabolic Intermediates
5.3. Circadian–Metabolic Integration Through Acyl-CoA Species
5.4. Tissue-Specific Clock Synchronization
5.5. Integration with Exercise-Induced Signals
6. Interaction Between Dietary Timing and Exercise
7. Multi-Omics Approaches: New Insights
8. Clinical Implications: Temporal Intervention Strategies
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Jakubowicz, D.; Barnea, M.; Wainstein, J.; Froy, O. High caloric intake at breakfast vs. dinner differentially influences weight loss of overweight and obese women. Obesity 2013, 21, 2504–2512. [Google Scholar] [CrossRef]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Laposky, A.; Losee-Olson, S.; Easton, A.; Jensen, D.R.; et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef]
- Zhang, Y.; Proenca, R.; Maffei, M.; Barone, M.; Leopold, L.; Friedman, J.M. Positional cloning of the mouse obese gene and its human homologue. Nature 1994, 372, 425–432. [Google Scholar] [CrossRef]
- Chen, J.; Xiang, J.; Zhou, M.; Huang, R.; Zhang, J.; Cui, Y.; Jiang, X.; Li, Y.; Zhou, R.; Xin, H.; et al. Dietary timing enhances exercise by modulating fat-muscle crosstalk via adipocyte AMPKα2 signaling. Cell Metab. 2025, 34, 1578–1593. [Google Scholar] [CrossRef]
- Yang, J.; Vamvini, M.; Nigro, P.; Ho, L.L.; Galani, K.; Alvarez, M.; Tanigawa, Y.; Renfro, A.; Carbone, N.P.; Laakso, M.; et al. Single-cell dissection of the obesity-exercise axis in adipose-muscle tissues implies a critical role for mesenchymal stem cells. Cell Metab. 2022, 34, 1578–1593. [Google Scholar] [CrossRef] [PubMed]
- Savikj, M.; Stocks, B.; Sato, S.; Caidahl, K.; Krook, A.; Deshmukh, A.S.; Zierath, J.R.; Wallberg-Henriksson, H. Exercise timing influences multi-tissue metabolome and skeletal muscle proteome profiles in type 2 diabetic patients—A randomized crossover trial. Metabolism 2022, 135, 155268. [Google Scholar] [CrossRef]
- Mancilla, R.; Brouwers, B.; Schrauwen-Hinderling, V.B.; Hesselink, M.K.C.; Hoeks, J.; Schrauwen, P. Exercise training elicits superior metabolic effects when performed in the afternoon compared to morning in metabolically compromised humans. Physiol. Rep. 2021, 8, e14669. [Google Scholar] [CrossRef] [PubMed]
- Iwayama, K.; Tanabe, Y.; Tanji, F.; Ohnishi, T.; Takahashi, H. Diurnal variations in muscle and liver glycogen differ depending on the timing of exercise. J. Physiol. Sci. 2021, 71, 35. [Google Scholar] [CrossRef]
- Ezagouri, S.; Zwighaft, Z.; Sobel, J.; Baillieul, S.; Doutreleau, S.; Ladeuix, B.; Golik, M.; Verges, S.; Asher, G. Physiological and molecular dissection of daily variance in exercise capacity. Cell Metab. 2019, 30, 78–91.e4. [Google Scholar] [CrossRef]
- Oishi, K.; Hashimoto, C. Short-term time-restricted feeding during the resting phase is sufficient to induce leptin resistance that contributes to development of obesity and metabolic disorders in mice. Chronobiol. Int. 2018, 35, 1576–1594. [Google Scholar] [CrossRef] [PubMed]
- Basse, A.L.; Dalbram, E.; Larsson, L.; Gerhart-Hines, Z.; Zierath, J.R.; Treebak, J.T. Skeletal muscle insulin sensitivity show circadian rhythmicity which is independent of exercise training status. Front. Physiol. 2018, 9, 1198. [Google Scholar] [CrossRef]
- Noshiro, M.; Kawamoto, T.; Nakashima, A.; Ozaki, N.; Saeki, M.; Honda, K.; Fujimoto, K.; Kato, Y. DEC1 regulates the rhythmic expression of PPARγ target genes involved in lipid metabolism in white adipose tissue. Genes Cells 2020, 25, 232–241. [Google Scholar] [CrossRef]
- Ma, D.; Qu, Y.; Wu, T.; Liu, X.; Cai, L.; Wang, Y. Excessive fat expenditure in MCT-induced heart failure rats is associated with BMAL1/REV-ERBα circadian rhythmic loop disruption. Sci. Rep. 2024, 14, 8128. [Google Scholar] [CrossRef]
- Zvonic, S.; Ptitsyn, A.A.; Conrad, S.A.; Scott, L.K.; Floyd, Z.E.; Kilroy, G.; Wu, X.; Goh, B.C.; Mynatt, R.L.; Gimble, J.M. Characterization of peripheral circadian clocks in adipose tissues. Diabetes 2006, 55, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Damiola, F.; Le Minh, N.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef]
- Kadiri, S.; Monnier, C.; Ganbold, M.; Ledent, T.; Capeau, J.; Antoine, B. The nuclear retinoid-related orphan receptor-α regulates adipose tissue glyceroneogenesis in addition to hepatic gluconeogenesis. Am. J. Physiol. Endocrinol. Metab. 2015, 309, E105–E114. [Google Scholar] [CrossRef]
- Adamovich, Y.; Rousso-Noori, L.; Zwighaft, Z.; Neufeld-Cohen, A.; Golik, M.; Kraut-Cohen, J.; Wang, M.; Han, X.; Asher, G. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014, 19, 319–330. [Google Scholar] [CrossRef]
- Dhamrait, G.K.; Panchal, K.; Fleury, N.J.; Abel, T.N.; Ancliffe, M.K.; Crew, R.C.; Croft, K.; Fernandez, B.O.; Minnion, M.; Hart, P.H.; et al. Characterising nitric oxide-mediated metabolic benefits of low-dose ultraviolet radiation in the mouse: A focus on brown adipose tissue. Diabetologia 2020, 63, 179–193. [Google Scholar] [CrossRef]
- Paschos, G.K.; Ibrahim, S.; Song, W.L.; Kunieda, T.; Grant, G.; Reyes, T.M.; Bradfield, C.A.; Vaughan, C.H.; Eiden, M.; Masoodi, M.; et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 2012, 18, 1768–1777. [Google Scholar] [CrossRef]
- van der Veen, D.R.; Shao, J.; Chapman, S.; Leevy, W.M.; Duffield, G.E. A diurnal rhythm in glucose uptake in brown adipose tissue revealed by in vivo PET-FDG imaging. Obesity 2012, 20, 1527–1529. [Google Scholar] [CrossRef]
- Viollet, B.; Andreelli, F.; Jørgensen, S.B.; Perrin, C.; Geloen, A.; Flamez, D.; Mu, J.; Lenzner, C.; Baud, O.; Bennoun, M.; et al. The AMP-activated protein kinase alpha2 catalytic subunit controls whole-body insulin sensitivity. J. Clin. Investig. 2003, 111, 91–98. [Google Scholar] [CrossRef]
- Davies, S.P.; Hawley, S.A.; Woods, A.; Carling, D.; Haystead, T.A.; Hardie, D.G. Purification of the AMP-activated protein kinase on ATP-gamma-Sepharose and analysis of its subunit structure. Eur. J. Biochem. 1994, 223, 351–357. [Google Scholar] [CrossRef]
- Salt, I.; Celler, J.W.; Hawley, S.A.; Prescott, A.; Woods, A.; Carling, D.; Hardie, D.G. AMP-activated protein kinase: Greater AMP dependence, and preferential nuclear localization, of complexes containing the alpha2 isoform. Biochem. J. 1998, 334, 177–187. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef]
- Liang, J.; Xu, Z.X.; Ding, Z.; Lu, Y.; Yu, Q.; Werle, K.D.; Zhou, G.; Park, Y.Y.; Peng, G.; Gambello, M.J.; et al. Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nat. Commun. 2015, 6, 7926. [Google Scholar] [CrossRef]
- Xiao, B.; Heath, R.; Saiu, P.; Leiper, F.C.; Leone, P.; Jing, C.; Walker, P.A.; Haire, L.; Eccleston, J.F.; Davis, C.T.; et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 2007, 449, 496–500. [Google Scholar] [CrossRef]
- Stapleton, D.; Mitchelhill, K.I.; Gao, G.; Widmer, J.; Michell, B.J.; Teh, T.; House, C.M.; Fernandez, C.S.; Cox, T.; Witters, L.A.; et al. Mammalian AMP-activated protein kinase subfamily. J. Biol. Chem. 1996, 271, 611–614. [Google Scholar] [CrossRef]
- Hawley, S.A.; Davison, M.; Woods, A.; Davies, S.P.; Beri, R.K.; Carling, D.; Hardie, D.G. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 1996, 271, 27879–27887. [Google Scholar] [CrossRef]
- Woods, A.; Johnstone, S.R.; Dickerson, K.; Leiper, F.C.; Fryer, L.G.D.; Neumann, D.; Schlattner, U.; Wallimann, T.; Carlson, M.; Carling, D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 2003, 13, 2004–2008. [Google Scholar] [CrossRef]
- McGee, S.L.; Howlett, K.F.; Starkie, R.L.; Cameron-Smith, D.; Kemp, B.E.; Hargreaves, M. Exercise increases nuclear AMPK alpha2 in human skeletal muscle. Diabetes 2003, 52, 926–928. [Google Scholar] [CrossRef]
- Bergeron, R.; Ren, J.M.; Cadman, K.S.; Moore, I.K.; Perret, P.; Pypaert, M.; Young, L.H.; Semenkovich, C.F.; Shulman, G.I. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E1340–E1346. [Google Scholar] [CrossRef]
- Treebak, J.T.; Glund, S.; Deshmukh, A.; Klein, D.K.; Long, Y.C.; Jensen, T.E.; Jørgensen, S.B.; Viollet, B.; Andersson, L.; Neumann, D.; et al. AMPK-mediated AS160 phosphorylation in skeletal muscle is dependent on AMPK catalytic and regulatory subunits. Diabetes 2006, 55, 2051–2058. [Google Scholar] [CrossRef]
- Li, Y.; Xu, S.; Mihaylova, M.M.; Zheng, B.; Hou, X.; Jiang, B.; Park, O.; Luo, Z.; Castro, E.; Lan, F.; et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011, 13, 376–388. [Google Scholar] [CrossRef]
- Gauthier, M.S.; Miyoshi, H.; Souza, S.C.; Cacicedo, J.M.; Saha, A.K.; Greenberg, A.S.; Ruderman, N.B. AMP-activated protein kinase is activated as a consequence of lipolysis in the adipocyte: Potential mechanism and physiological relevance. J. Biol. Chem. 2008, 283, 16514–16524. [Google Scholar] [CrossRef]
- Villena, J.A.; Viollet, B.; Andreelli, F.; Kahn, A.; Vaulont, S.; Sul, H.S. Induced adiposity and adipocyte hypertrophy in mice lacking the AMP-activated protein kinase-alpha2 subunit. Diabetes 2004, 53, 2242–2249. [Google Scholar] [CrossRef]
- Manieri, E.; Herrera-Melle, L.; Mora, A.; Tomás-Loba, A.; Leiva-Vega, L.; Fernández, D.I.; López-Alcalá, J.; Macías-González, M.; Frühbeck, G.; Muntané, J.; et al. Adiponectin accounts for gender differences in hepatocellular carcinoma incidence. J. Exp. Med. 2019, 216, 1108–1119. [Google Scholar] [CrossRef]
- Norheim, F.; Langleite, T.M.; Hjorth, M.; Holen, T.; Kielland, A.; Stadheim, H.K.; Gulseth, H.L.; Birkeland, K.I.; Jensen, J.; Drevon, C.A. The effects of acute and chronic exercise on PGC-1α, irisin and browning of subcutaneous adipose tissue in humans. FEBS J. 2014, 281, 739–749. [Google Scholar] [CrossRef]
- Minokoshi, Y.; Kim, Y.B.; Peroni, O.D.; Fryer, L.G.; Müller, C.; Carling, D.; Kahn, B.B. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature 2002, 415, 339–343. [Google Scholar] [CrossRef]
- Bostrom, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostrom, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Sachdeva, U.M.; DiTacchio, L.; Williams, E.C.; Alvarez, J.G.; Egan, D.F.; Vasquez, D.S.; Juguilon, H.; Panda, S.; Shaw, R.J.; et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 2009, 326, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Godinho, S.I.; Maywood, E.S.; Shaw, L.; Tucci, V.; Barnard, A.R.; Busino, L.; Hastings, M.H.; Mamalaki, C.; Dillon, N. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 2007, 316, 897–900. [Google Scholar] [CrossRef]
- Hirota, T.; Lewis, W.G.; Liu, A.C.; Lee, J.W.; Schultz, P.G.; Kay, S.A. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. Proc. Natl. Acad. Sci. USA 2008, 105, 20746–20751. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef]
- Yang, J.; Ruchti, E.; Petit, J.M.; Jourdain, P.; Grenningloh, G.; Allaman, I.; Magistretti, P.J. Lactate promotes plasticity gene expression by potentiating NMDA signaling in neurons. Proc. Natl. Acad. Sci. USA 2014, 111, 12228–12233. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Pell, V.R.; Gaude, E.; Aksentijević, D.; Sundier, S.Y.; Robb, E.L.; Logan, A.; Nadtochiy, S.M.; Ord, E.N.J.; Smith, A.C.; et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014, 515, 431–435. [Google Scholar] [CrossRef]
- He, W.; Miao, F.J.; Lin, D.C.; Schwandner, R.T.; Wang, Z.; Gao, J.; Chen, J.L.; Tian, H.; Ling, L. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature 2004, 429, 188–193. [Google Scholar] [CrossRef]
- Wayman, G.A.; Kaech, S.; Grant, W.F.; Davare, M.; Impey, S.; Tokumitsu, H.; Nozaki, N.; Soderling, T.R. Regulation of axonal extension and growth cone motility by calmodulin-dependent protein kinase I. J. Neurosci. 2004, 24, 3786–3794. [Google Scholar] [CrossRef] [PubMed]
- Pinkosky, S.L.; Scott, J.W.; Desjardins, E.M.; Smith, B.K.; Day, E.A.; Ford, R.J.; Langendorf, C.G.; Ling, N.X.Y.; Nero, T.L.; Loh, K.; et al. Long-chain fatty acyl-CoA esters regulate metabolism via allosteric control of AMPK β1 isoforms. Nat. Metab. 2020, 2, 873–881. [Google Scholar] [CrossRef]
- Doi, M.; Hirayama, J.; Sassone-Corsi, P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 2006, 125, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Perelis, M.; Marcheva, B.; Ramsey, K.M.; Schipma, M.J.; Hutchison, A.L.; Taguchi, A.; Rudic, R.D.; Peek, C.B.; McNaughton, K.K.; Allada, R.; et al. Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 2015, 350, aac4250. [Google Scholar] [CrossRef] [PubMed]
- Scheer, F.A.; Hilton, M.F.; Mantzoros, C.S.; Shea, S.A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. USA 2009, 106, 4453–4458. [Google Scholar] [CrossRef]
- Iwabu, M.; Yamauchi, T.; Okada-Iwabu, M.; Sato, K.; Nakagawa, T.; Funata, M.; Yamaguchi, M.; Namiki, K.; Nakayama, R.; Tabata, M.; et al. Adiponectin and AdipoR1 regulate PGC-1α and mitochondria by Ca2+ and AMPK/SIRT1. Nature 2010, 464, 1313–1319. [Google Scholar] [CrossRef]
- Bourlier, V.; Zakaroff-Girard, A.; Miranville, A.; De Barros, S.; Maumus, M.; Sengenes, C.; Galitzky, J.; Lafontan, M.; Karpe, F.; Frayn, K.N.; et al. Remodeling phenotype of human subcutaneous adipose tissue macrophages. Circulation 2008, 117, 806–815. [Google Scholar] [CrossRef]
- Sato, S.; Basse, A.L.; Schönke, M.; Chen, S.; Samad, M.; Altıntaş, A.; Laker, R.C.; Dalbram, E.; Barrès, R.; Baldi, P.; et al. Time of exercise specifies the impact on muscle metabolic pathways and systemic energy homeostasis. Cell Metab. 2019, 30, 92–110.e4. [Google Scholar] [CrossRef]
- Garaulet, M.; Gómez-Abellán, P.; Alburquerque-Béjar, J.J.; Lee, Y.C.; Ordovás, J.M.; Scheer, F.A. Timing of food intake predicts weight loss effectiveness. Int. J. Obes. 2013, 37, 604–611. [Google Scholar] [CrossRef]
- Dyar, K.A.; Ciciliot, S.; Wright, L.E.; Biensø, R.S.; Malagoli Tagliazucchi, G.; Patel, V.R.; Forcato, M.; Paz, M.I.P.; Gudiksen, A.; Solagna, F.; et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 2013, 3, 29–41. [Google Scholar] [CrossRef]
- Yasumoto, Y.; Hashimoto, C.; Nakao, R.; Yamazaki, H.; Hiroyama, H.; Nemoto, T.; Yamamoto, H.; Sakurai, M.; Oike, H.; Wada, N.; et al. Short-term feeding at the wrong time is sufficient to desynchronize peripheral clocks and induce obesity with hyperphagia, physical inactivity and metabolic disorders in mice. Metabolism 2016, 65, 714–727. [Google Scholar] [CrossRef] [PubMed]
- Sutton, E.F.; Beyl, R.; Early, K.S.; Cefalu, W.T.; Ravussin, E.; Peterson, C.M. Early time-restricted feeding improves insulin sensitivity, blood pressure, and oxidative stress even without weight loss in men with prediabetes. Cell Metab. 2018, 27, 1212–1221.e3. [Google Scholar] [CrossRef]
- Eckel-Mahan, K.L.; Patel, V.R.; de Mateo, S.; Orozco-Solis, R.; Ceglia, N.J.; Sahar, S.; Dilag-Penilla, S.A.; Dyar, K.A.; Baldi, P.; Sassone-Corsi, P. Reprogramming of the circadian clock by nutritional challenge. Cell 2013, 155, 1464–1478. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.R.; Pissios, P.; Otu, H.; Roberson, R.; Xue, B.; Asakura, K.; Furukawa, N.; Marino, F.E.; Liu, F.F.; Kahn, B.B.; et al. A high-fat, ketogenic diet induces a unique metabolic state in mice. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E1724–E1739. [Google Scholar] [CrossRef]
- Perrin, L.; Loizides-Mangold, U.; Chanon, S.; Gobet, C.; Hulo, N.; Isenegger, L.; Sagot, B.; Riezman, H.; Lefai, E.; Dibner, C.; et al. Transcriptomic analyses reveal rhythmic and CLOCK-driven pathways in human skeletal muscle. eLife 2018, 7, e34114. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 2016, 534, 213–217. [Google Scholar] [CrossRef]
- Vollmers, C.; Gill, S.; DiTacchio, L.; Pulivarthy, S.R.; Le, H.D.; Panda, S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 21453–21458. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Creek, D.J.; Jankevics, A.; Burgess, K.E.; Breitling, R.; Barrett, M.P. IDEOM: An Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics 2012, 28, 1048–1049. [Google Scholar] [CrossRef]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef]
- Emont, M.P.; Jacobs, C.; Essene, A.L.; Pant, D.; Tenen, D.; Colleluori, G.; Di Vincenzo, A.; Jørgensen, A.M.; Dashti, H.; Stefek, A.; et al. A single-cell atlas of human and mouse white adipose tissue. Nature 2022, 603, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yao, K.; Yu, H.; Zhang, L.; Xu, Y.; Chen, L.; Qin, J.; Xiong, J.; Wang, Y.; Yao, Z.; et al. Metabolic remodelling during early mouse embryo development. Nature 2021, 598, 651–655. [Google Scholar] [CrossRef]
- Li, M.D.; Wang, Y.; Zhu, X.; DiTacchio, L.; Samad, M.; Jørgensen, M.S.; Menon, A.S.; Chen, S.; Bass, J.; Panda, S. Time-restricted feeding mitigates obesity through adipocyte thermogenesis. Science 2022, 376, 399–407. [Google Scholar] [CrossRef]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Kato, T.; Atsumi, A.; Yamamoto, T.; Inoue, N.; Ishikawa, M.; Okada, S.; Ishigaki, N.; et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 2007, 13, 1193–1202. [Google Scholar] [CrossRef]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef]
- Lowden, A.; Moreno, C.; Holmbäck, U.; Lennernäs, M.; Tucker, P. Eating and shift work—Effects on habits, metabolism and performance. Scand. J. Work. Environ. Health 2010, 36, 150–162. [Google Scholar] [CrossRef]
- Crispim, C.A.; Zimberg, I.Z.; dos Reis, B.G.; Diniz, R.M.; Tufik, S.; de Mello, M.T. Relationship between food intake and sleep pattern in healthy individuals. J. Clin. Sleep Med. 2011, 7, 659–664. [Google Scholar] [CrossRef]
- Depner, C.M.; Cheng, P.C.; Devine, J.K.; Khosla, S.; de Zambotti, M.; Robillard, R.; Vakulin, A.; Drummond, S.P.A.; Paillard, T.; Goldsworthy, M.R.; et al. Wearable technologies for developing sleep and circadian biomarkers: A summary, comparison, and next steps. Sleep 2020, 43, zsaa003. [Google Scholar]
- Huseinovic, E.; Winkvist, A.; Freisling, H.; Slimani, N.; Boeing, H.; Buckland, G.; Schwingshackl, L.; Weiderpass, E.; Rostgaard-Hansen, A.L.; Tjønneland, A.; et al. Meal patterns across ten European countries -- results from the European Prospective Investigation into Cancer and Nutrition (EPIC) calibration study. Public Health Nutr. 2016, 19, 2769–2780. [Google Scholar] [CrossRef]
- Cadmus-Bertram, L.A.; Marcus, B.H.; Patterson, R.E.; Parker, B.A.; Morey, B.L. Randomized trial of a Fitbit-based physical activity intervention for women. Am. J. Prev. Med. 2015, 49, 414–418. [Google Scholar] [CrossRef]
- Thamrin, S.A.; Arsyad, D.S.; Kuswanto, H.; Lawi, A.; Nasir, S. Predicting obesity in adults using machine learning techniques: An analysis of Indonesian Basic Health Research 2018. Front. Nutr. 2021, 8, 669155. [Google Scholar] [CrossRef]
- Frie, K.; Hartmann-Boyce, J.; Jebb, S.; Aveyard, P. Effectiveness of a self-regulation intervention for weight loss: A randomized controlled trial. Br. J. Health Psychol. 2020, 25, 652–676. [Google Scholar] [CrossRef]
- Kurauti, M.A.; Costa-Júnior, J.M.; Ferreira, S.M.; Santos, G.J.; Protzek, A.O.; Nardelli, T.R.; de Souza, A.H.; Boschero, A.C.; Carneiro, E.M.; Rezende, L.F. Acute exercise restores insulin clearance in diet-induced obese mice. J. Endocrinol. 2016, 229, 221–232. [Google Scholar] [CrossRef]
- Um, J.H.; Yang, S.; Yamazaki, S.; Kang, H.; Viollet, B.; Foretz, M.; Chung, J.H. Activation of 5’-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J. Biol. Chem. 2007, 282, 20794–20798. [Google Scholar] [CrossRef] [PubMed]
- Vieira, E.; Nilsson, E.C.; Nerstedt, A.; Ormestad, M.; Long, Y.C.; Garcia-Roves, P.M.; Zierath, J.R.; Mahlapuu, M. Relationship between AMPK and the transcriptional balance of clock-related genes in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E1032–E1037. [Google Scholar] [CrossRef] [PubMed]
- Coomans, C.P.; van den Berg, S.A.; Houben, T.; van Klinken, J.B.; van den Berg, R.; Pronk, A.C.; Havekes, L.M.; Romijn, J.A.; van Dijk, K.W.; Biermasz, N.R.; et al. Detrimental effects of constant light exposure and high-fat diet on circadian energy metabolism and insulin sensitivity. FASEB J. 2013, 27, 1721–1732. [Google Scholar] [CrossRef] [PubMed]
- Shea, S.A.; Hilton, M.F.; Orlova, C.; Ayers, R.T.; Mantzoros, C.S. Independent circadian and sleep/wake regulation of adipokines and glucose in humans. J. Clin. Endocrinol. Metab. 2005, 90, 2537–2544. [Google Scholar] [CrossRef]
- Woldt, E.; Sebti, Y.; Solt, L.A.; Duhem, C.; Lancel, S.; Eeckhoute, J.; Hesselink, M.K.; Paquet, C.; van der Horst, G.T.; Leval, L.; et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 2013, 19, 1039–1046. [Google Scholar] [CrossRef]
- Hirten, R.P.; Danieletto, M.; Lewis, J.; Mayer, L.; Mehta, M.; Nakamura, A.; Somers, K.; Grinspan, L.; Glicksberg, B.; Bottles, K.; et al. Use of physiological data from a wearable device to identify SARS-CoV-2 infection and symptoms and predict COVID-19 diagnosis: Observational study. J. Med. Internet Res. 2021, 23, e26107. [Google Scholar] [CrossRef]
- Reid, K.J.; Santostasi, G.; Baron, K.G.; Wilson, J.; Kang, J.; Zee, P.C. Timing and intensity of light correlate with body weight in adults. PLoS ONE 2014, 9, e92251. [Google Scholar] [CrossRef]
- Ebbers, T.; Takes, R.P.; Honings, J.; Smeele, L.E.; Kool, R.B.; van den Broek, G.B. Development and validation of automated electronic health record data reuse for a multidisciplinary quality dashboard. Digit. Health 2023, 9, 20552076231191007. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, J.S.; Branecky, K.L.; Yang, W.; Ellacott, K.L.; Niswender, K.D.; Yamazaki, S. High-fat diet acutely affects circadian organisation and eating behavior. Eur. J. Neurosci. 2013, 37, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Sherman, H.; Genzer, Y.; Cohen, R.; Chapnik, N.; Madar, Z.; Froy, O. Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB J. 2012, 26, 3493–3502. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.; Lindgren, C.M.; Li, S.; Wheeler, E.; Zhao, J.H.; Prokopenko, I.; Inouye, M.; Freathy, R.M.; Attwood, A.P.; Beckmann, J.S.; et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008, 40, 768–775. [Google Scholar] [CrossRef]
- Wang, L.; Jin, Q.; Lee, J.E.; Su, I.H.; Ge, K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 7317–7322. [Google Scholar] [CrossRef]
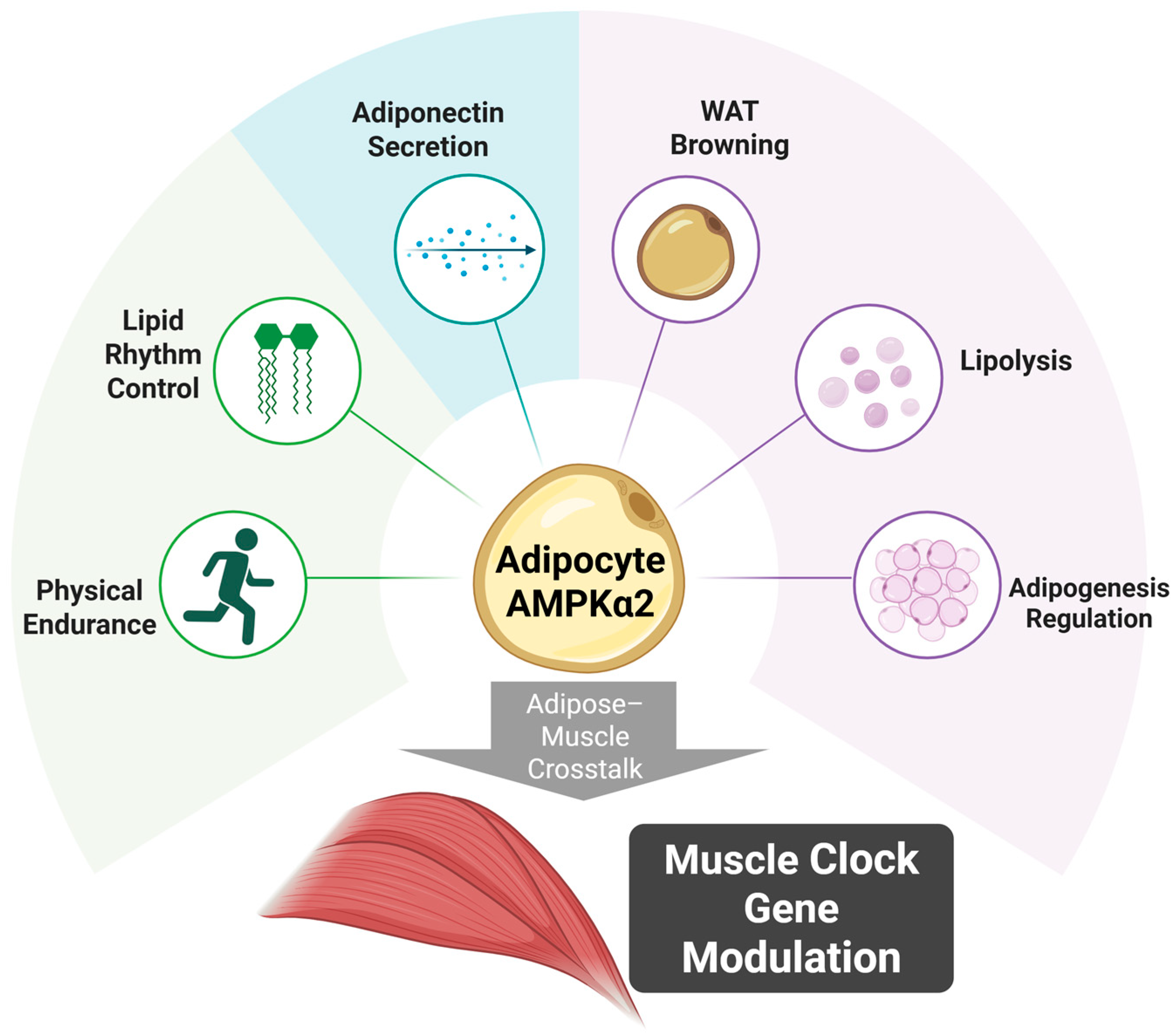

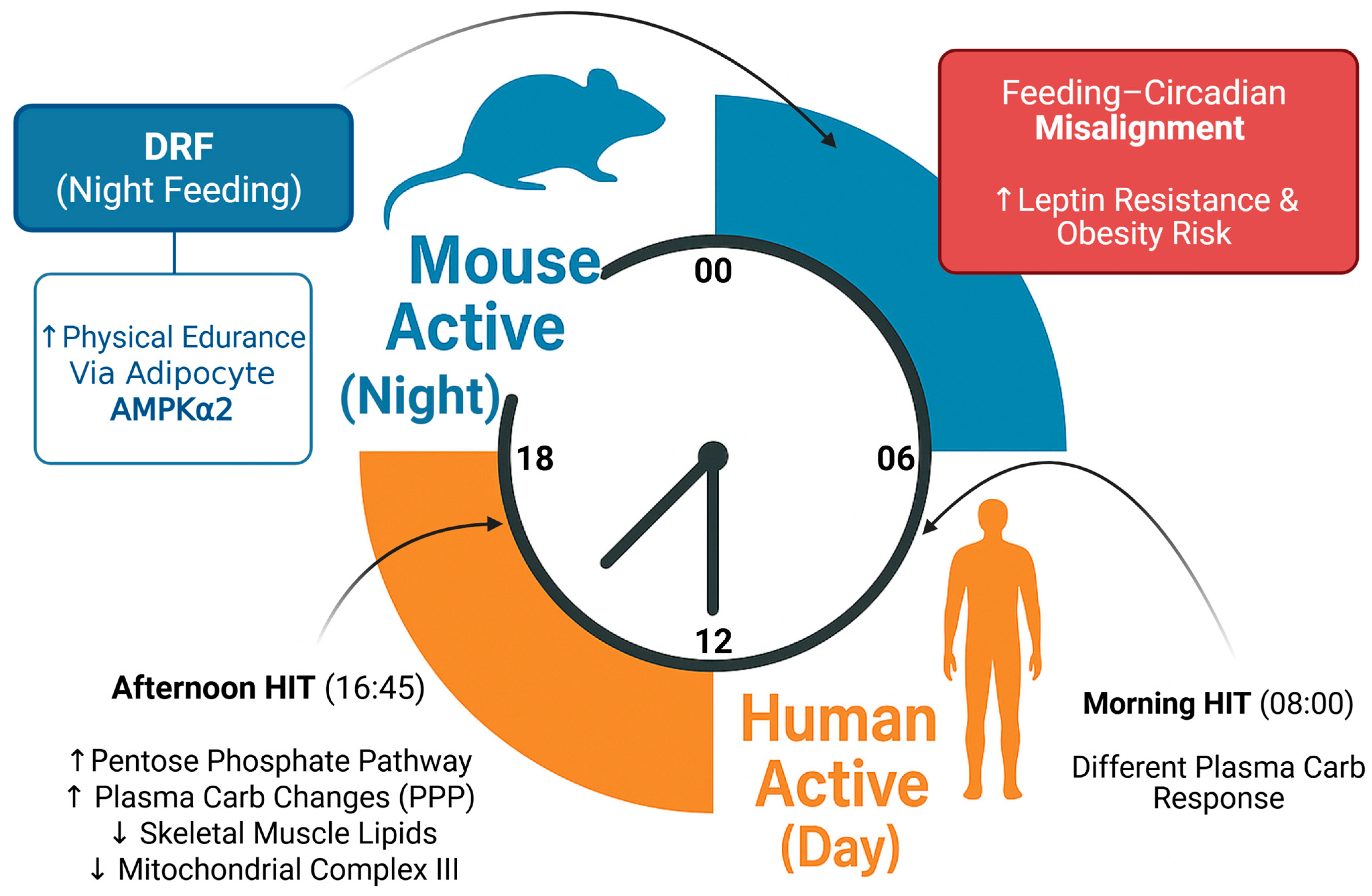
| Study | Model | Intervention | Key Findings | Significance |
|---|---|---|---|---|
| Chen et al. (2025) [5] | Mouse (GWAT-specific Prkaa2 knockout) | Day-restricted feeding (DRF), AMPK activator (C29) | Adipocyte AMPKα2 regulates adipose metabolism, modulates muscle clock | Adipose acts as circadian regulator of muscle |
| Yang et al. (2022) [6] | Mouse (single-cell analysis) | Obesity + exercise | Circadian genes altered in stromal cells | Fat–muscle circadian crosstalk |
| Savikj et al. (2022) [7] | Human (T2DM patients) | Morning vs. afternoon exercise | PM exercise ↑ mitochondria and lipids | Exercise timing impacts metabolism |
| Mancilla et al. (2021) [8] | Human (metabolically compromised) | Morning vs. afternoon exercise training | PM training ↑ insulin sensitivity and performance | Superior metabolic benefits from PM training |
| Ezagouri et al. (2019) [10] | Mouse and Human | Exercise at different times | Time-dependent exercise capacity via PER1/2 | Circadian control of exercise performance |
| Oishi & Hashimoto (2018) [11] | Mouse | Time-restricted feeding during resting period | Leptin resistance, obesity | Feeding timing affects metabolism |
| Basse et al. (2018) [12] | Human | Circadian rhythm vs. exercise training | Circadian insulin sensitivity | Clock-regulated sensitivity independent of exercise |
| Molecule | Origin | Target Tissue | Function | Circadian Regulation | Molecular Mechanism |
|---|---|---|---|---|---|
| AMPKα2 | Adipose tissue | Adipose, Skeletal muscle | Energy sensor, regulates metabolic homeostasis, controls muscle clock genes | Yes | Activates downstream targets like ACC and PGC-1α; regulates Bmal1 and Per2 expression [5] |
| Lactate | Muscle, Adipose | Liver, Muscle, Adipose | Energy substrate, signaling molecule, gene expression regulation | Yes | Binds to GPR81; regulates gene expression via HIF-1α and CREB pathways [29] |
| Succinate | Multiple tissues | Liver, Muscle, Adipose | TCA cycle intermediate, signaling molecule | Yes | Binds to SUCNR1 receptor; activates MAPK signaling and gene transcription [30] |
| Irisin | Muscle | White Adipose tissue | Promotes browning of white adipose tissue, enhances thermogenesis | Unclear | Derived from FNDC5 cleavage; activates UCP1 expression via PGC-1α pathway [33] |
| IL-6 | Muscle, Adipose | Liver, Muscle, Adipose, Immune cells | Pro/anti-inflammatory cytokine, regulates glucose metabolism | Partial | Activates JAK/STAT3 and AMPK pathways; enhances glucose uptake in muscle [32] |
| Adiponectin | Adipose tissue | Muscle, Liver | Enhances insulin sensitivity, fatty acid oxidation | Yes | Activates AMPK and PPARα pathways; increases GLUT4 translocation [24] |
| Leptin | Adipose tissue | CNS, Muscle | Regulates energy balance, influences muscle metabolism | Yes | Binds to leptin receptor; activates JAK2/STAT3 and PI3K signaling [24] |
| Acyl-CoA | Multiple tissues | Liver, Muscle, Adipose | Metabolic intermediate, involved in fatty acid metabolism | Yes | Serves as substrate for β-oxidation; regulates CPT1 and energy flux [25] |
| Intervention | Mechanism | Potential Benefits | Target Conditions | Implementation Considerations |
|---|---|---|---|---|
| Time-restricted eating (TRE) | Aligns feeding with active phase; optimizes adipocyte AMPKα2 signaling | Enhanced exercise performance; improved adipose–muscle communication; metabolic benefits | Obesity, Type 2 diabetes, sarcopenia | 8–12 h eating window during active phase; consistent daily timing essential |
| Time-optimized exercise | Utilizes circadian variations in metabolic responses to exercise stimuli | Improved lipid metabolism, mitochondrial function, insulin sensitivity | Type 2 diabetes, metabolic syndrome | Afternoon (16:00–18:00) sessions may be superior for certain outcomes |
| Chronopharma-cological AMPK targeting | Time-specific activation of AMPK pathways via pharmacological agents | Enhanced muscle endurance, metabolic regulation, reduced side effects | Metabolic disorders, physical fatigue syndromes | Requires synchronized drug timing; dependent on functional AMPKα2 in adipose |
| Time-optimized antidiabetic medication | Aligns drug action with peak insulin resistance periods | Greater glycemic stability; higher medication efficacy | Type 2 diabetes | Consider circadian rhythm of insulin sensitivity; personalized schedules recommended |
| Digital-assisted temporal interventions | Wearable monitoring with real-time coaching and adaptive recommendations | Improved compliance; personalized protocols; remote monitoring capabilities | Complex metabolic disorders, shift work-related dysfunction | Integration with healthcare systems; requires digital literacy |
| Integrated temporal approach | Combines timing of feeding, exercise, and medication for synergistic impact | Comprehensive metabolic improvement; potential adherence boost | Complex metabolic disorders, lifestyle disease management | Needs coordination across interventions; potential for digital support tools |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Baek, J.; Kim, M.S. The Link Between Dietary Timing and Exercise Performance Through Adipocyte AMPKα2 Signaling. Int. J. Mol. Sci. 2025, 26, 6061. https://doi.org/10.3390/ijms26136061
Kim S, Baek J, Kim MS. The Link Between Dietary Timing and Exercise Performance Through Adipocyte AMPKα2 Signaling. International Journal of Molecular Sciences. 2025; 26(13):6061. https://doi.org/10.3390/ijms26136061
Chicago/Turabian StyleKim, Sohyun, Jihyun Baek, and Man S. Kim. 2025. "The Link Between Dietary Timing and Exercise Performance Through Adipocyte AMPKα2 Signaling" International Journal of Molecular Sciences 26, no. 13: 6061. https://doi.org/10.3390/ijms26136061
APA StyleKim, S., Baek, J., & Kim, M. S. (2025). The Link Between Dietary Timing and Exercise Performance Through Adipocyte AMPKα2 Signaling. International Journal of Molecular Sciences, 26(13), 6061. https://doi.org/10.3390/ijms26136061






