Genome-Wide Identification and Expression Profiling of SlGeBP Gene Family in Response to Hormone and Abiotic Stresses in Solanum lycopersicum L.
Abstract
1. Introduction
2. Results
2.1. Identification and Physical and Chemical Parameters of SlGeBP Gene Family in Tomato
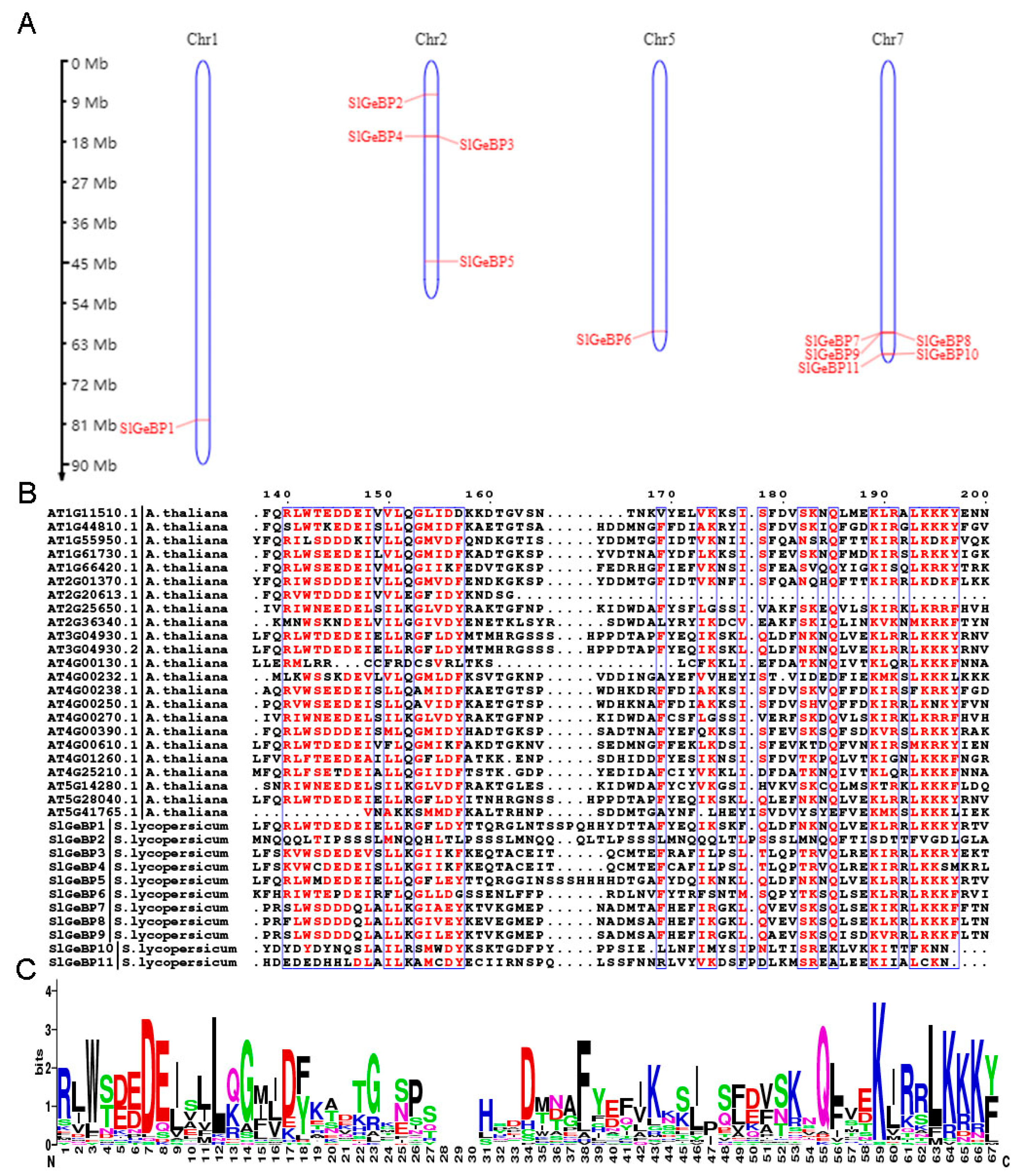
2.2. Chromosome Distribution and Multiple Sequence Alignment Analysis of SlGeBP Genes
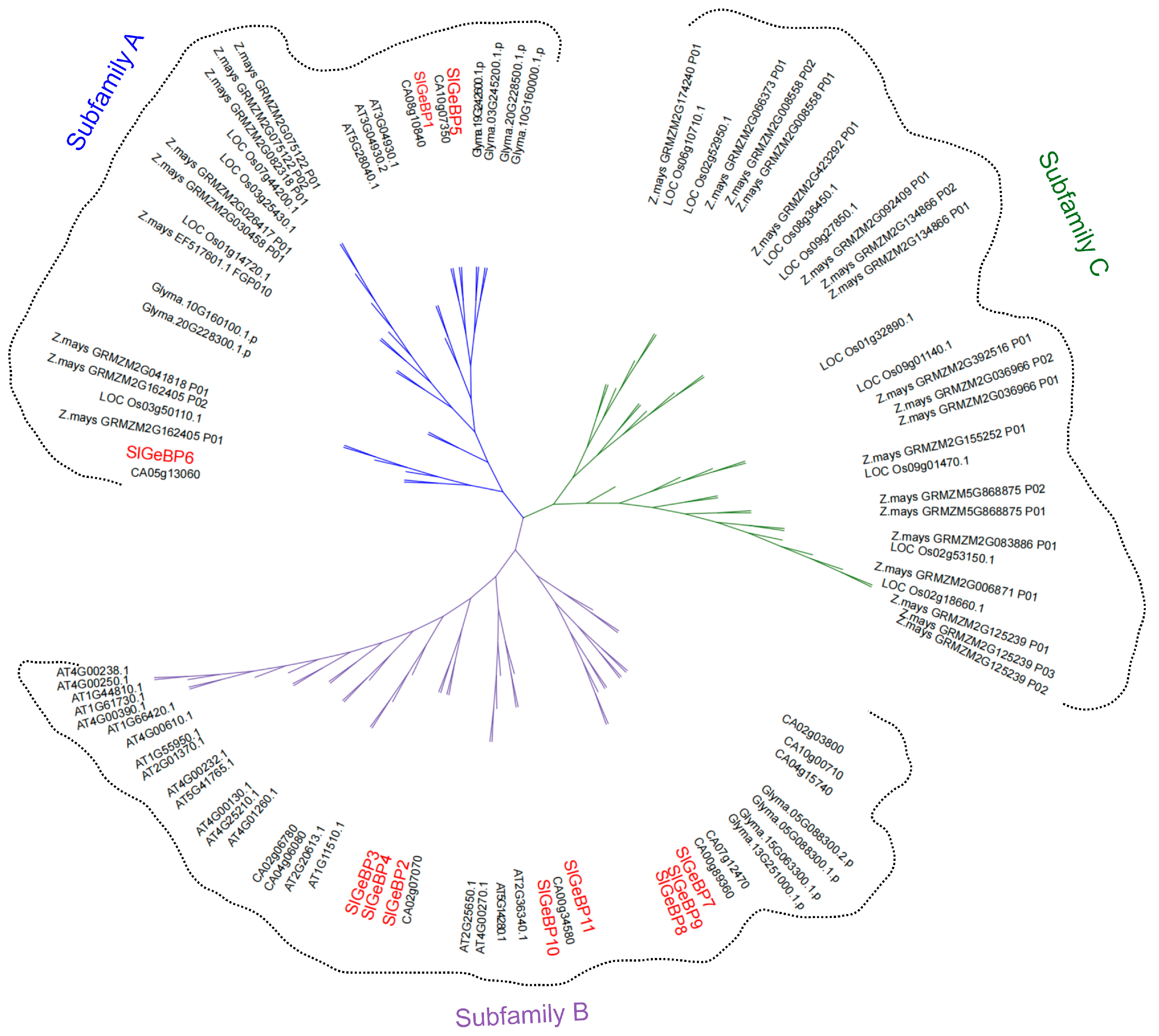
2.3. Spatial Structure and Phylogenetic Analysis of SlGeBP Genes
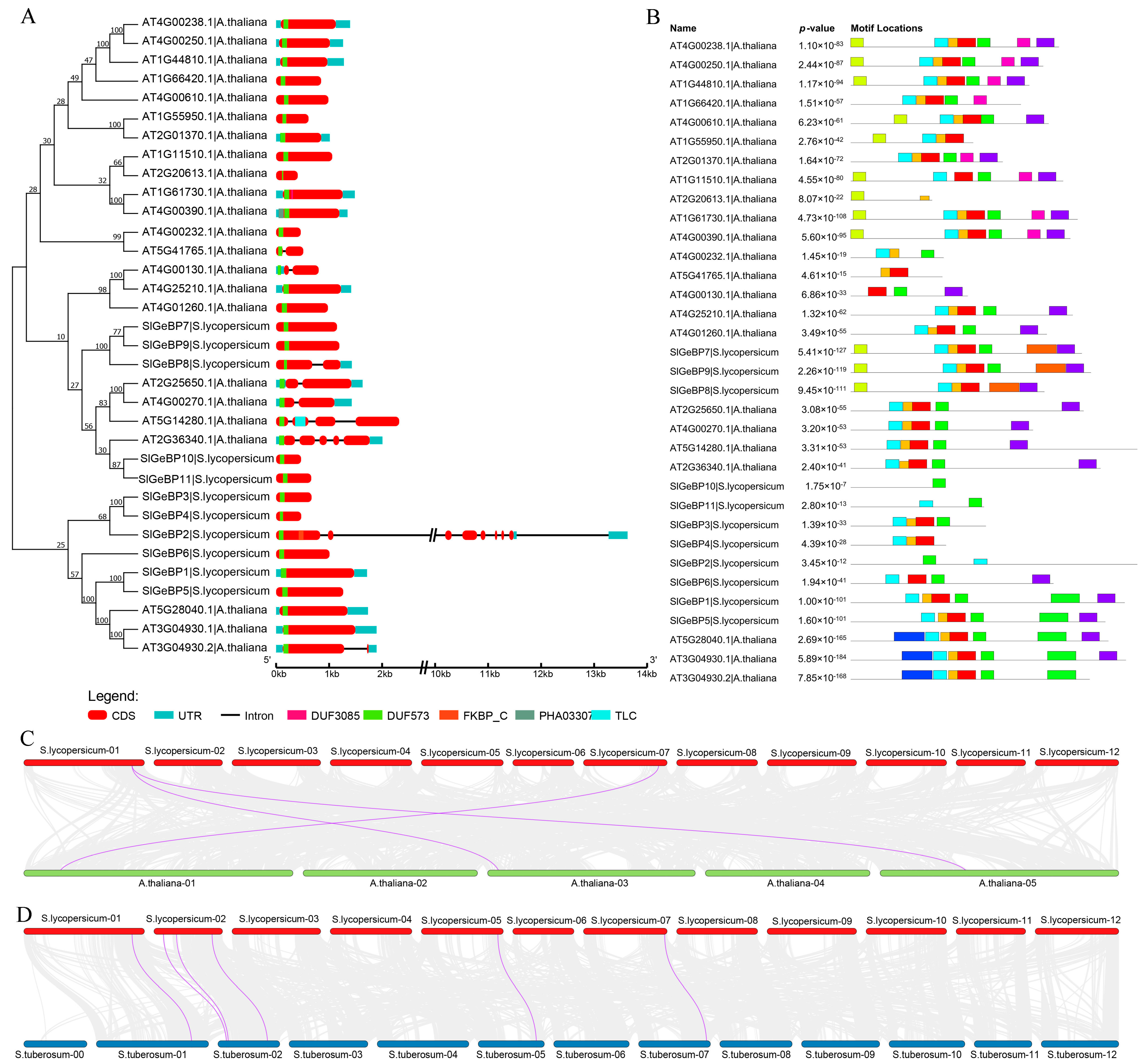
2.4. Gene Structure, Conserved Motif, and Collinearity Analysis of SlGeBP Genes
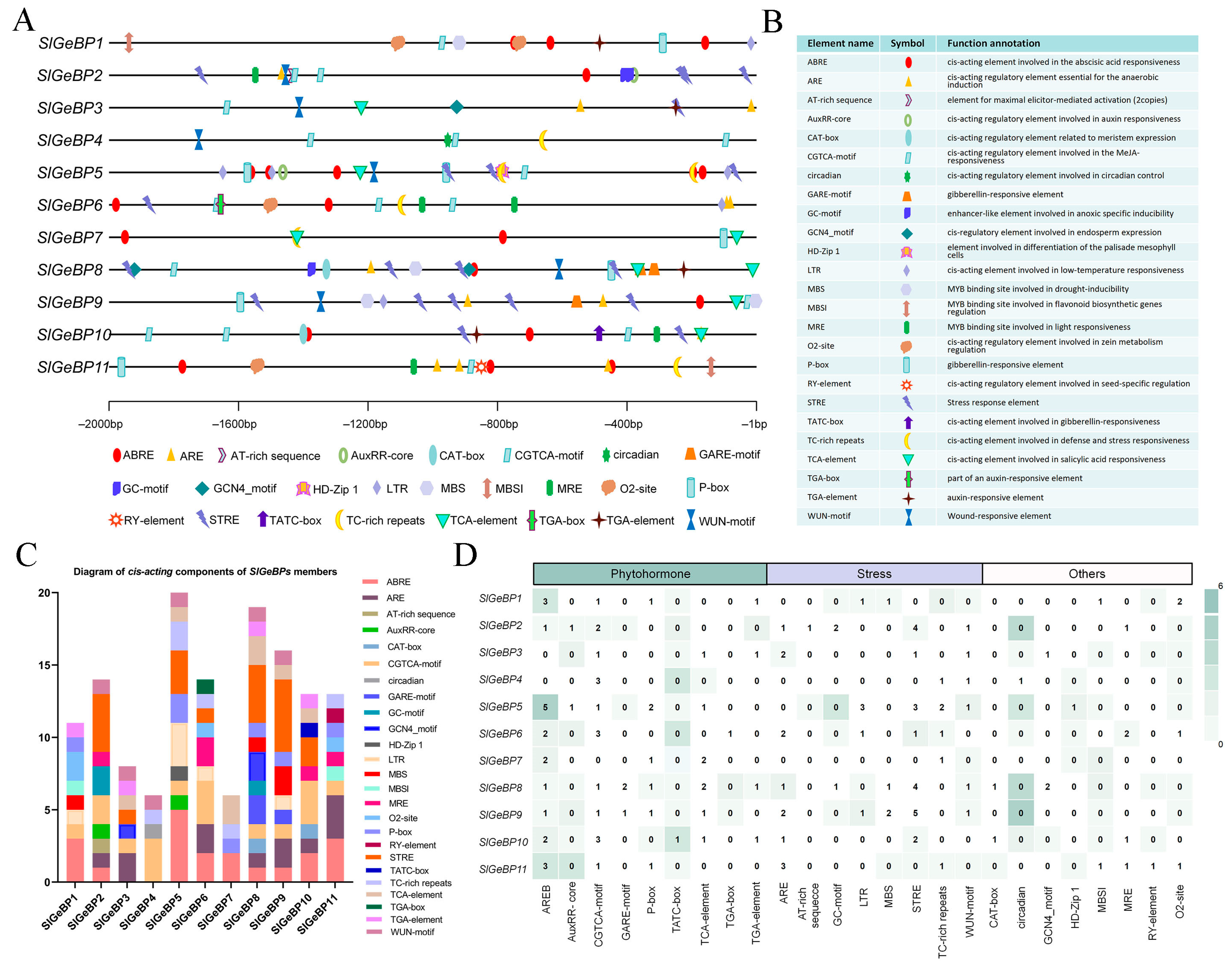
2.5. Analysis of the cis-Acting Regulatory Elements in the SlGeBP Gene Promoters
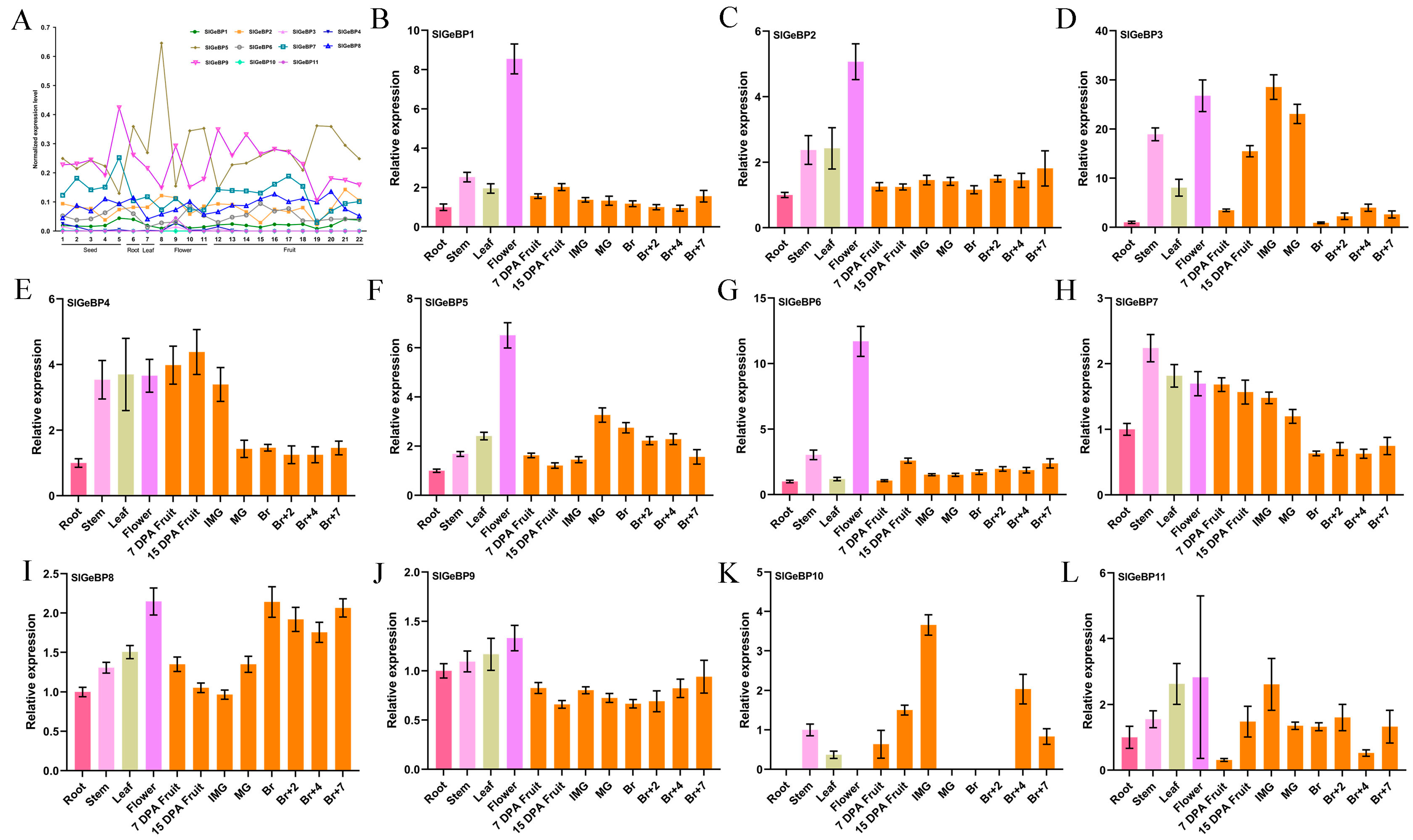
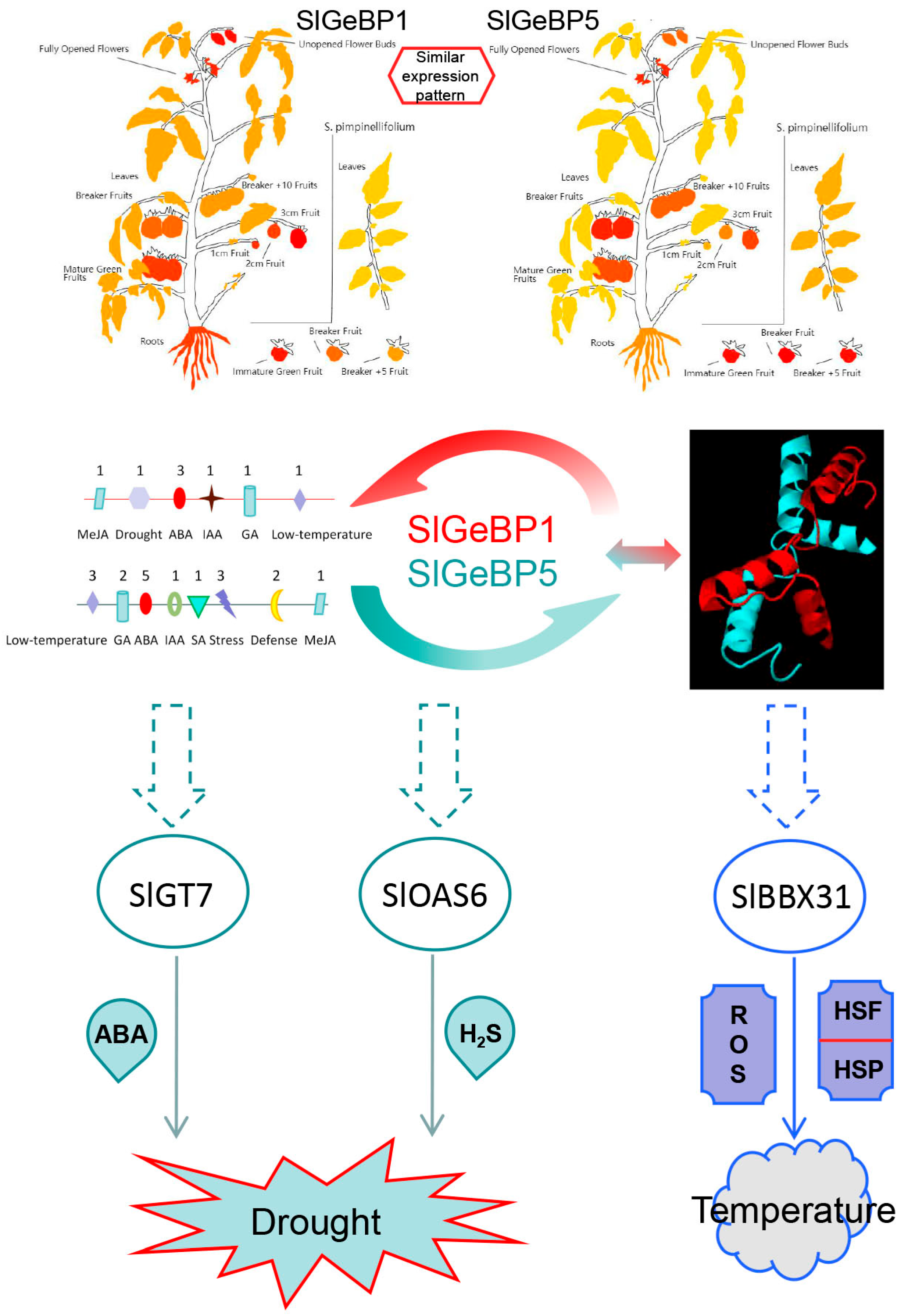
2.6. Tissue-Specific Expression Profiles Analysis of SlGeBPs
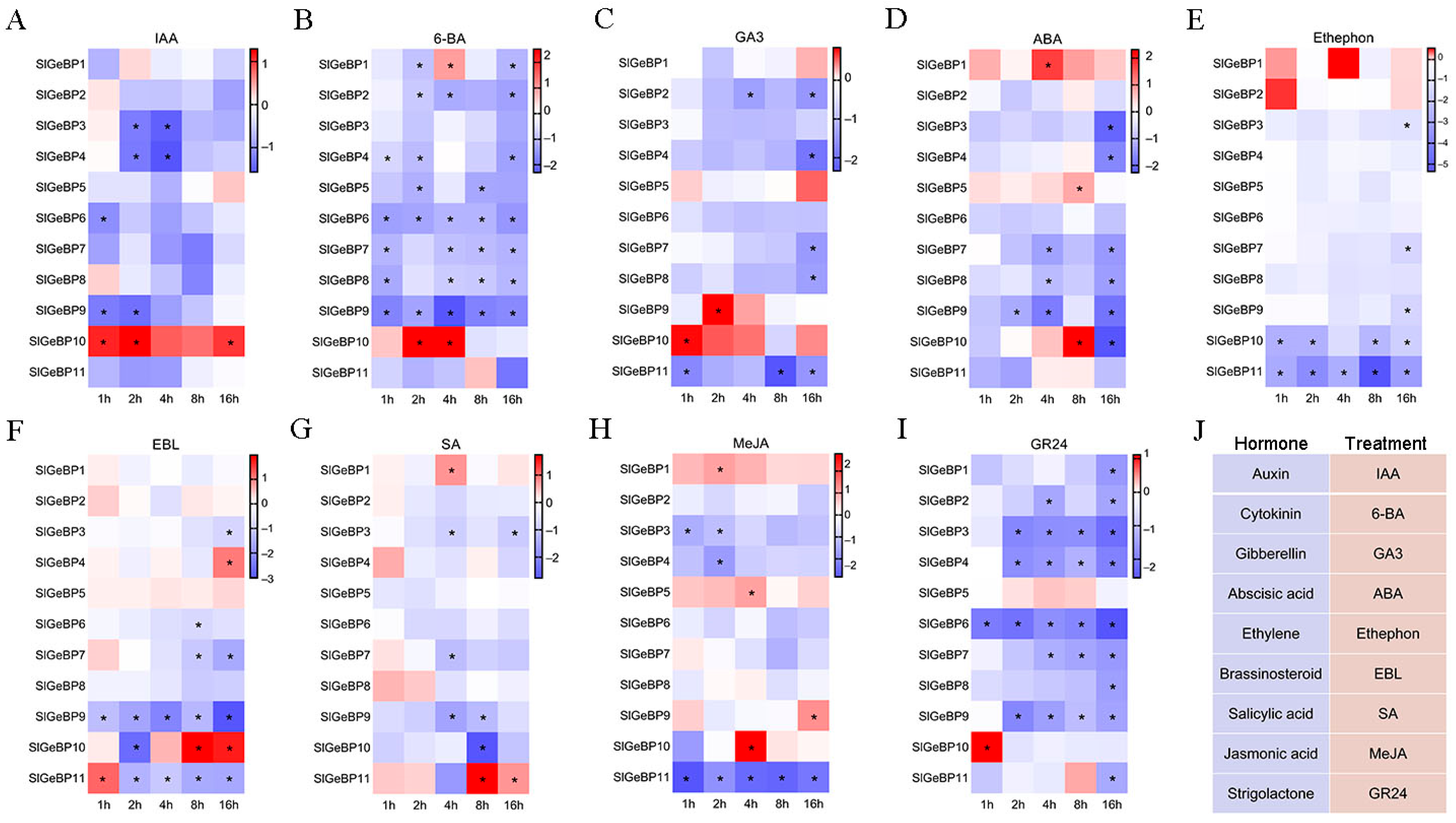
2.7. Expression Profiles of SlGeBP Genes in Response to Plant Hormones
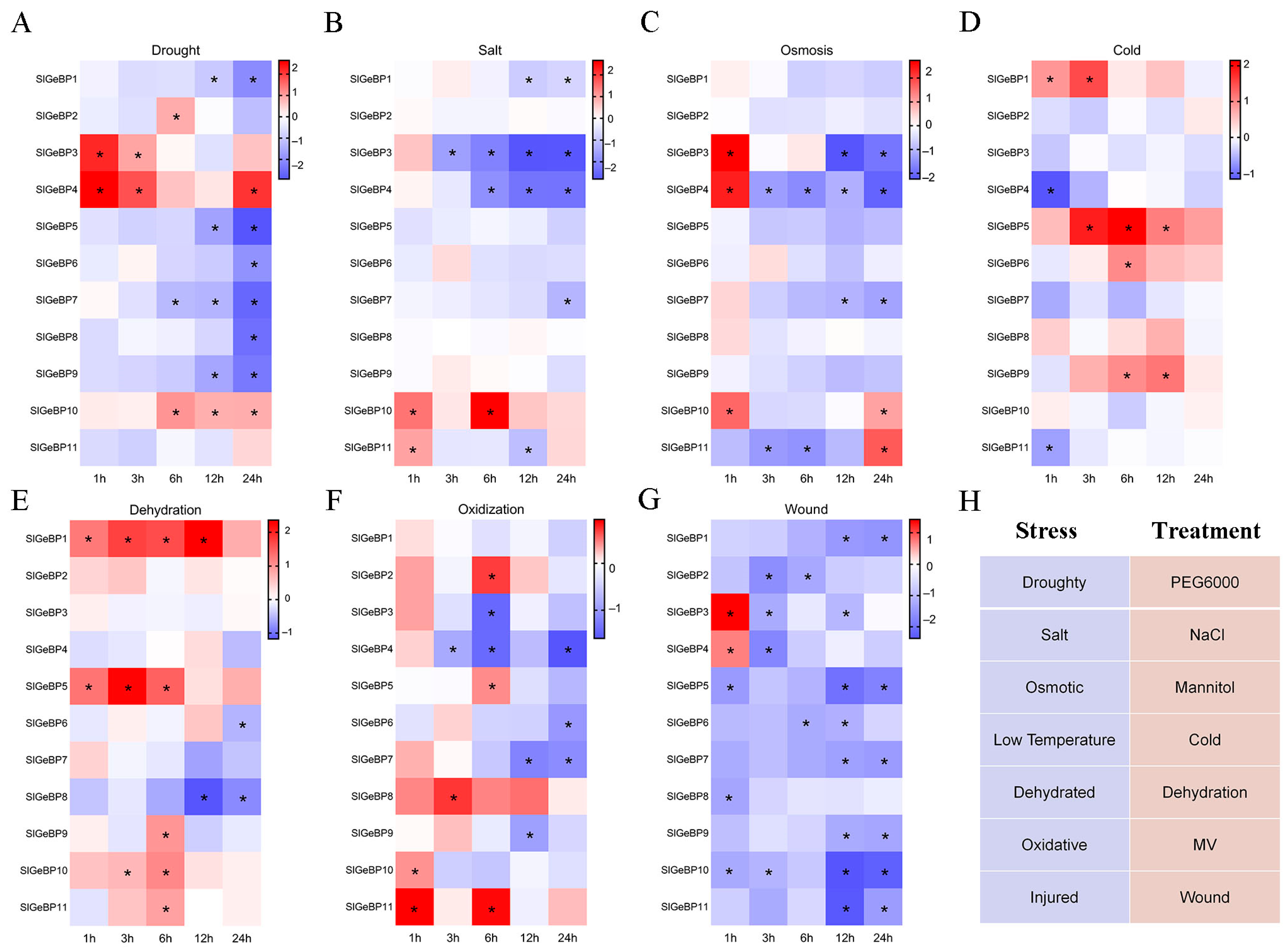
2.8. Expression Profiles of SlGeBP Genes in Response to Stresses
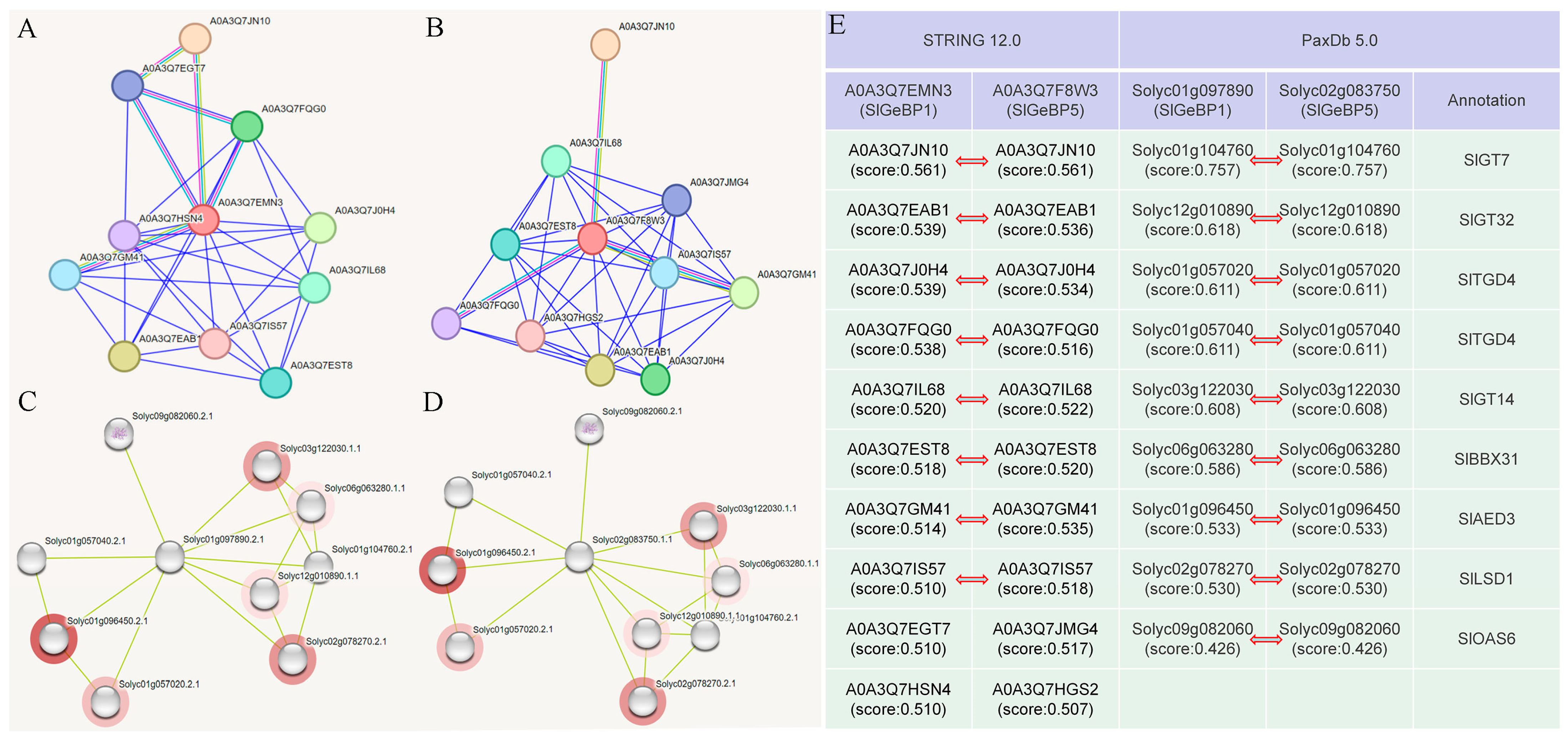
2.9. Prediction of the Protein–Protein Interaction with SlGeBP Proteins
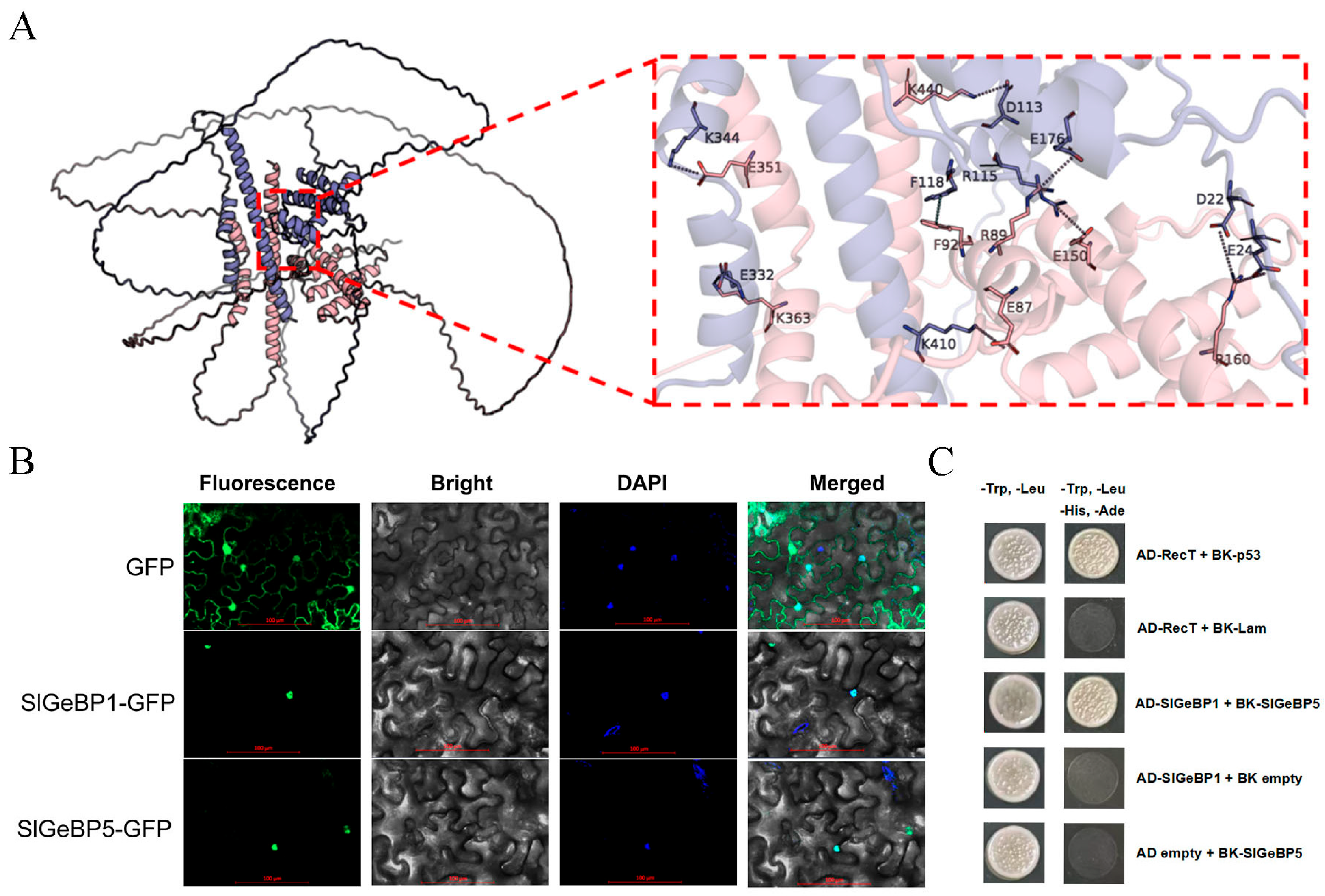
2.10. The Interaction Between and Subcellular Localization of SlGeBP1 and SlGeBP5
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of SlGeBP in Tomato
4.3. Physicochemical Characterization and Multiple Sequence Alignment of SlGeBP Proteins
4.4. Three-Dimensional Structure, Secondary Structure, and Subcellular Localization Prediction
4.5. Phylogenetic Analysis
4.6. Gene Structure, Conserved Motifs, and Collinearity Analysis of SlGeBP Proteins
4.7. cis-Acting Element Analysis
4.8. Hormone and Stress Treatments
4.9. RNA Extraction and qRT-PCR
4.10. Predicting the Interactors with SlGeBP Proteins
4.11. Subcellular Localization of SlGeBP1 and SlGeBP5
4.12. Yeast Two-Hybrid Assay (Y2H)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| bZIP | Basic region leucine zipper |
| CD-Search | Conserved domain search |
| DPA | Day post anthesis |
| DUF573 | Domain of Unknown Function 573 |
| GeBP | The GLABROUS1 enhancer-binding protein |
| GPL | GeBP-like |
| HHM | Hidden Markov model |
| MV | Methyl viologen |
| NCBI | National Center for Biotechnology Information |
| NJ | Neighbor joining method |
| NLS | Nuclear localization signal |
| PaxDb | Protein Abundance Database |
| PEPPI | Pipeline for the Extraction of Predicted Protein-protein Interactions |
| pLDDT | Per-residue model confidence score |
| PPI | protein-protein interaction |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| RRs | response regulators |
| TAIR | The Arabidopsis Information Resource |
| TF | Transcription factor |
References
- Wang, X.; Shen, C.; Meng, P.; Tan, G.; Lv, L. Analysis and review of trichomes in plants. BMC Plant Biol. 2021, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Schuurink, R.; Tissier, A. Glandular trichomes: Micro-organs with model status? New Phytol. 2020, 225, 2251–2266. [Google Scholar] [CrossRef] [PubMed]
- Huchelmann, A.; Boutry, M.; Hachez, C. Plant glandular trichomes: Natural cell factories of high biotechnological interest. Plant Physiol. 2017, 175, 6–22. [Google Scholar] [CrossRef] [PubMed]
- Kortbeek, R.; Galland, M.; Muras, A.; Therezan, R.; Maia, S.; Haring, M.; Schuurink, R.; Bleeker, P. Genetic and physiological requirements for high-level sesquiterpene-production in tomato glandular trichomes. Front. Plant Sci. 2023, 14, 1139274. [Google Scholar] [CrossRef]
- Berhin, A.; Nawrath, C.; Hachez, C. Subtle interplay between trichome development and cuticle formation in plants. New Phytol. 2022, 233, 2036–2046. [Google Scholar] [CrossRef]
- Chalvin, C.; Drevensek, S.; Dron, M.; Bendahmane, A.; Boualen, A. Genetic control of glandular trichome development. Trends Plant Sci. 2020, 25, 477–487. [Google Scholar] [CrossRef]
- Chevalier, F.; Perazza, D.; Laporte, F.; Le Hénanff, G.; Hornitschek, P.; Bonneville, J.; Herzog, M.; Vachon, G. GeBP and GeBP-Like proteins are noncanonical leucine-zipper transcription factors that regulate Cytokinin response in Arabidopsis. Plant Physiol. 2008, 146, 1142–1154. [Google Scholar] [CrossRef][Green Version]
- Khare, D.; Mitsuda, N.; Lee, S.; Song, W.; Hwang, D.; Ohme-Takagi, M.; Martinoia, E.; Lee, Y.; Hwang, J. Root avoidance of toxic metals requires the GeBP-LIKE 4 transcription factor in Arabidopsis thaliana. New Phytol. 2017, 213, 1257–1273. [Google Scholar] [CrossRef]
- Liu, R.; Li, H.; Qiao, Z.; Liu, H.; Zhao, L.; Wang, X.; Zhang, Z.; Zhang, S.; Song, L.; You, C. Genome-wide analysis of MdGeBP family and functional identification of MdGeBP3 in Malus domestica. Environ. Exp. Bot. 2023, 208, 105262. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Liu, C.; Zhang, F.; Wei, J.; Li, B. Genome-wide characterization and expression analysis of GeBP family genes in soybean. Plants 2022, 11, 1848. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, Q.; He, Y.; Liu, W.; Xu, Y.; Liu, K.; Xian, F.; Li, J.; Hu, J. Genome-wide identification, expansion mechanism and expression profiling analysis of GLABROUS1 enhancer-binding protein (GeBP) gene family in Gramineae crops. Int. J. Mol. Sci. 2021, 22, 8758. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wu, X.; Wang, Z.; Zhang, X.; Chen, L.; Duan, Q.; Huang, J. Genome-wide identification and expression analysis of BrGeBP genes reveal their potential roles in cold and drought stress tolerance in Brassica rapa. Int. J. Mol. Sci. 2023, 24, 13597. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhou, W.; Yao, X.; Zhao, Q.; Lu, L. Genome-wide investigation and functional analysis reveal that CsGeBP4 is required for Tea Plant trichome formation. Int. J. Mol. Sci. 2023, 24, 5207. [Google Scholar] [CrossRef] [PubMed]
- Curaba, J.; Herzog, M.; Vachon, G. GeBP, the first member of a new gene family in Arabidopsis, encodes a nuclear protein with DNA-binding activity and is regulated by KNAT1. Plant J. 2003, 33, 305–317. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Z.; Bao, Z.; Li, H.; Lyu, Y.; Zan, Y.; Wu, Y.; Cheng, L.; Fang, Y.; Wu, K.; et al. Graph pangenome captures missing heritability and empowers tomato breeding. Nature 2022, 606, 527–534. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, C.; Ye, Z.; Li, C.; Huang, S.; Lin, T. The genomic route to tomato breeding: Past, present, and future. Plant Physiol. 2024, 195, 2500–2514. [Google Scholar] [CrossRef]
- Fonseca, R.; Capel, C.; Nieto-Canseco, R.; Ortiz-Atienza, A.; Bretones, S.; López-Fábregas, J.D.; Quevedo-Colmena, A.S.; Lebrón, R.; Barragán-Lozano, T.; Villalobos-Ramírez, V.; et al. A tomato EMS-mutagenized population provides new valuable resources for gene discovery and breeding of developmental traits. Plants 2022, 11, 2453. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Egea, I.; Estrada, Y.; Flores, F.B.; Bolarín, M.C. Improving production and fruit quality of tomato under abiotic stress: Genes for the future of tomato breeding for a sustainable agriculture. Environ. Exp. Bot. 2022, 204, 105086. [Google Scholar] [CrossRef]
- Tripodi, P.; Soler, S.; Campanelli, G.; Díez, M.; Esposito, S.; Sestili, S.; Figàs, M.R.; Leteo, F.; Casanova, C.; Platani, C.; et al. Genome wide association mapping for agronomic, fruit quality, and root architectural traits in tomato under organic farming conditions. BMC Plant Biol. 2021, 21, 481. [Google Scholar] [CrossRef]
- Ahmed, N.; Zhang, B.; Bozdar, B.; Chachar, S.; Rai, M.; Li, J.; Li, Y.; Hayat, F.; Chachar, Z.; Tu, P. The power of magnesium: Unlocking the potential for increased yield, quality, and stress tolerance of horticultural crops. Front. Plant Sci. 2023, 14, 1285512. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Kissoudis, C.; Yan, Z.; Visser, R.; van der Linden, G. Plant behaviour under combined stress: Tomato responses to combined salinity and pathogen stress. Plant J. 2018, 93, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Mauxion, J.; Chevalier, C.; Gonzalez, N. Complex cellular and molecular events determining fruit size. Trends Plant Sci. 2021, 26, 1023–1038. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, X.; Fei, Z.; Giovannoni, J. Fruit ripening and postharvest changes in very early-harvested tomatoes. Hortic. Res. 2024, 11, uhae199. [Google Scholar] [CrossRef]
- Dong, H.; Wang, J.; Song, X.; Hu, C.; Zhu, C.; Sun, T.; Zhou, Z.; Hu, Z.; Xia, X.; Zhou, J.; et al. HY5 functions as a systemic signal by integrating BRC1-dependent hormone signaling in tomato bud outgrowth. Proc. Natl. Acad. Sci. USA 2023, 120, e2301879120. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.; Meng, Y.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Santnerr, A.; Esteller, M. Recent advances and emerging trends in plant hormone signalling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef]
- Santnerr, A.; Calderon-Villalobosr, L.; Esteller, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef]
- Wengr, J.; Yer, M.; Lir, B.; Noelr, J. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 2016, 166, 881–893. [Google Scholar] [CrossRef]
- Ali, M.; Luo, D.; Khan, A.; Haq, S.; Gai, W.; Zhang, H.; Cheng, G.; Muhammad, I.; Gong, Z. Classification and genome-wide analysis of chitin-binding proteins gene family in pepper (Capsicum annuum L.) and transcriptional regulation to Phytophthora capsici, abiotic stresses and hormonal applications. Int. J. Mol. Sci. 2018, 19, 2216. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Cai, X.; Ye, Z.; Li, H. Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem. Biophys. Res. Commun. 2015, 468, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.; Yu, M.; Bai, J.; Zhu, Z. SlbHLH22-induced hypertrophy development is related to the salt stress response of the GTgamma gene in tomatoes. Metabolites 2023, 13, 1195. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, C.; Benning, C. TGD4 involved in endoplasmic reticulum-to-chloroplast lipid trafficking is a phosphatidic acid binding protein. Plant J. 2012, 70, 614–623. [Google Scholar] [CrossRef]
- Fan, J.; Zhai, Z.; Yan, C.; Xu, C. Arabidopsis TRIGALACTOSYLDIACYLGLYCEROL5 interacts with TGD1, TGD2, and TGD4 to facilitate lipid transfer from the endoplasmic reticulum to plastids. The Plant Cell 2015, 27, 2941–2955. [Google Scholar] [CrossRef]
- Wang, Q.; Zhan, X. Elucidating the role of SlBBX31 in plant growth and heat-stress resistance in tomato. Int. J. Mol. Sci. 2024, 25, 9289. [Google Scholar] [CrossRef]
- Fang, H.; Zhang, X.; Chen, W.; Xu, L.; Yao, J.; Pei, Y.; Zhang, Y. Dynamic changes of endogenous H2S generation during responding to developmental and environmental signals in Solanum lycopersicum L. Sci. Hortic. 2024, 335, 113346. [Google Scholar] [CrossRef]
- Rowarth, N.; Curtis, B.; Einfeldt, A.; Archibald, J.; Lacroix, C.; Gunawardena, A. RNA-Seq analysis reveals potential regulators of programmed cell death and leaf remodelling in lace plant (Aponogeton madagascariensis). BMC Plant Biol. 2021, 21, 375. [Google Scholar] [CrossRef]
- Alves, M.; Dadalto, S.; Gonçalves, A.; de Souza, G.; Barros, V.; Fietto, L. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2014, 2, 85–106. [Google Scholar] [CrossRef]
- Muhlenbock, P.; Plaszczyca, M.; Plaszczyca, M.; Mellerowicz, E.; Karpinsk, S. Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell 2007, 19, 3819–3830. [Google Scholar] [CrossRef]
- Kaminaka, H.; Näke, C.; Epple, P.; Dittgen, J.; Schütze, K.; Chaban, C.; Holt, B., III; Merkle, T.; Schäfer, E.; Harter, K.; et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006, 25, 4400–4411. [Google Scholar] [CrossRef] [PubMed]
- Perazza, D.; Laporte, F.; Balagué, C.; Chevalier, F.; Remo, S.; Bourge, M.; Larkin, J.; Herzog, M.; Vachon, G. GeBP/GPL transcription factors regulate a subset of CPR5-dependent processes. Plant Physiol. 2011, 157, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Chen, Q.; Wang, S.; Lers, A. Downregulation of GeBP-like α factor by MiR827 suggests their involvement in senescence and phosphate homeostasis. BMC Biol. 2021, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, M.; Zhang, L.; Wu, N.; Tang, X.; Yang, X.; Ghanem, H.; Wu, M.; Wu, G.; Qing, L. Insights into geminiviral pathogenesis: Interaction between βC1 protein and GLABROUS1 enhancer binding protein (GeBP) in Solanaceae. Phytopathol. Res. 2025, 7, 30. [Google Scholar] [CrossRef]
- García-Cano, E.; Hak, H.; Magori, S.; Lazarowitz, S.; Citovsky, V. The Agrobacterium F-box protein effector VirF destabilizes the Arabidopsis GLABROUS1 enhancer/binding protein-like transcription factor VFP4, a transcriptional activator of defense response genes. Mol. Plant-Microbe Interact. 2018, 31, 576–586. [Google Scholar] [CrossRef]
- Wu, J.; Liu, R.; Xie, Y.; Zhao, S.; Yan, M.; Sun, N.; Zhan, Y.; Li, F.; Yu, S.; Feng, Z.; et al. Association of GhGeBP genes with fiber quality and early maturity related traits in upland cotton. BMC Genom. 2024, 25, 1058. [Google Scholar] [CrossRef]
- Simm, S.; Fragkostefanakis, S.; Paul, P.; Keller, M.; Einloft, J.; Scharf, K.; Schleiff, E. Identification and expression analysis of ribosome biogenesis factor co-orthologs in Solanum lycopersicum. Bioinform. Biol. Insights 2015, 9, S20751. [Google Scholar] [CrossRef]
- Lu, Y.; Ma, L.; Cheng, K.; Li, J.; Tang, H.; Zhu, G.; Wen, H.; Zhu, B.; Fu, D.; Qu, G.; et al. Comprehensive identification of ripening-related RNA-binding proteins in tomatoes using improved plant phase extraction. Plant J. 2025, 122, e70215. [Google Scholar] [CrossRef]
- Xiong, F.; Dong, P.; Liu, M.; Xie, G.; Wang, K.; Zhuo, F.; Feng, L.; Yang, L.; Li, Z.; Ren, M. Tomato FK506 binding protein 12KD (FKBP12) mediates the interaction between rapamycin and target of rapamycin (TOR). Front. Plant Sci. 2016, 7, 1746. [Google Scholar] [CrossRef]
- Khan, I.; Lubna; Asaf, S.; Jan, R.; Bilal, S.; Khan, A.; Kim, K.; Al-Harrasi, A. Dynamic interplay of WRKY, GRAS, and ERF transcription factor families in tomato-endophytic fungal symbiosis: Insights from transcriptome and genome-wide analysis. Front. Plant Sci. 2023, 14, 1181227. [Google Scholar] [CrossRef]
- Ahouvi, Y.; Haber, Z.; Zach, Y.; Rosental, L.; Toubiana, D.; Sharma, D.; Alseekh, S.; Tajima, H.; Fernie, A.; Brotman, Y.; et al. The alteration of tomato chloroplast vesiculation positively affects whole-plant source–sink relations and fruit metabolism under stress conditions. Plant Cell Physiol. 2022, 63, 2008–2026. [Google Scholar] [CrossRef] [PubMed]
- Chew, B.; Fisk, I.; Fray, R.; Tucker, G.; Bodi, Z.; Ferguson, A.; Xia, W.; Seymour, G. The effect of adenosine monophosphate deaminase overexpression on the accumulation of umami-related metabolites in tomatoes. Plant Cell Rep. 2017, 36, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A. Role of ABA in integrating plant responses to drought and salt stresses. Field Crop Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Huber, A.; Melcher, P.; Piñeros, M.; Setter, T.; Bauerle, T. Signal coordination before, during and after stomatal closure in response to drought stress. New Phytol. 2019, 224, 675–688. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef]
- Cao, H.; Chen, J.; Yue, M.; Xu, C.; Jian, W.; Liu, Y.; Song, B.; Gao, Y.; Cheng, Y.; Li, Z. Tomato transcriptional repressor MYB70 directly regulates ethylene-dependent fruit ripening. Plant J. 2020, 104, 1568–1581. [Google Scholar] [CrossRef]
| Gene Name | Gene Accession | Genomic Locus | Exon | AA | MW(Da) | pI | Strand | A.I | GRAVY | Best Hit in Arabidopsis |
|---|---|---|---|---|---|---|---|---|---|---|
| SlGeBP1 | Solyc01g097890 | ch01:88581415..88583129 | 1 | 454 | 48,609.88 | 5.83 | - | 73.60 | −0.371 | AT3G04930.2 |
| SlGeBP2 | Solyc02g005290 | ch02:7633701..7645348 | 8 | 519 | 57,645.42 | 9.45 | - | 85.08 | −0.491 | AT1G61730.1 |
| SlGeBP3 | Solyc02g014740 | ch02:18023947..18024618 | 1 | 224 | 26,187.87 | 9.24 | + | 69.46 | −1.073 | AT4G00390.1 |
| SlGeBP4 | Solyc02g014750 | ch02:18044460..18044933 | 1 | 158 | 18,184.97 | 8.66 | + | 71.97 | −0.617 | AT4G00390.1 |
| SlGeBP5 | Solyc02g083750 | ch02:47039175..47040441 | 1 | 422 | 46,853.96 | 4.71 | - | 71.28 | −0.687 | AT5G28040.1 |
| SlGeBP6 | Solyc05g051330 | ch05:61607815..61608822 | 1 | 336 | 37,761.07 | 5.00 | - | 72.96 | −0.692 | AT5G28040.1 |
| SlGeBP7 | Solyc07g052760 | ch07:61209111..61210259 | 1 | 383 | 42,923.35 | 6.42 | + | 59.48 | −1.000 | AT4G00390.1 |
| SlGeBP8 | Solyc07g052830 | ch07:61287331..61288758 | 2 | 321 | 35,723.94 | 5.12 | + | 65.88 | −0.909 | AT4G00390.1 |
| SlGeBP9 | Solyc07g052900 | ch07:61314239..61315432 | 1 | 398 | 44,075.41 | 5.80 | + | 64.13 | −0.972 | AT4G00390.1 |
| SlGeBP10 | Solyc07g063840 | ch07:66179566..66180037 | 1 | 157 | 18,606.58 | 5.20 | + | 58.14 | −1.295 | AT2G25650.1 |
| SlGeBP11 | Solyc07g064000 | ch07:66308723..66309386 | 1 | 221 | 25,221.15 | 4.26 | - | 63.82 | −1.167 | AT2G25650.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, H.; Wang, D.; Li, X.; Zhang, Y.; Su, D.; Lu, W.; Xu, K.; Li, Z. Genome-Wide Identification and Expression Profiling of SlGeBP Gene Family in Response to Hormone and Abiotic Stresses in Solanum lycopersicum L. Int. J. Mol. Sci. 2025, 26, 6008. https://doi.org/10.3390/ijms26136008
Cao H, Wang D, Li X, Zhang Y, Su D, Lu W, Xu K, Li Z. Genome-Wide Identification and Expression Profiling of SlGeBP Gene Family in Response to Hormone and Abiotic Stresses in Solanum lycopersicum L. International Journal of Molecular Sciences. 2025; 26(13):6008. https://doi.org/10.3390/ijms26136008
Chicago/Turabian StyleCao, Haohao, Danfeng Wang, Xiaoli Li, Yi Zhang, Deding Su, Wang Lu, Kedong Xu, and Zhengguo Li. 2025. "Genome-Wide Identification and Expression Profiling of SlGeBP Gene Family in Response to Hormone and Abiotic Stresses in Solanum lycopersicum L." International Journal of Molecular Sciences 26, no. 13: 6008. https://doi.org/10.3390/ijms26136008
APA StyleCao, H., Wang, D., Li, X., Zhang, Y., Su, D., Lu, W., Xu, K., & Li, Z. (2025). Genome-Wide Identification and Expression Profiling of SlGeBP Gene Family in Response to Hormone and Abiotic Stresses in Solanum lycopersicum L. International Journal of Molecular Sciences, 26(13), 6008. https://doi.org/10.3390/ijms26136008







