Analysis of GEN1 as a Breast Cancer Susceptibility Gene in Polish Women
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Participants
4.1.1. Hereditary Breast Cancer Cases
4.1.2. Unselected Breast Cancer Cases
4.1.3. Controls
4.2. Methods
4.2.1. Sequencing of GEN1
4.2.2. Genotyping
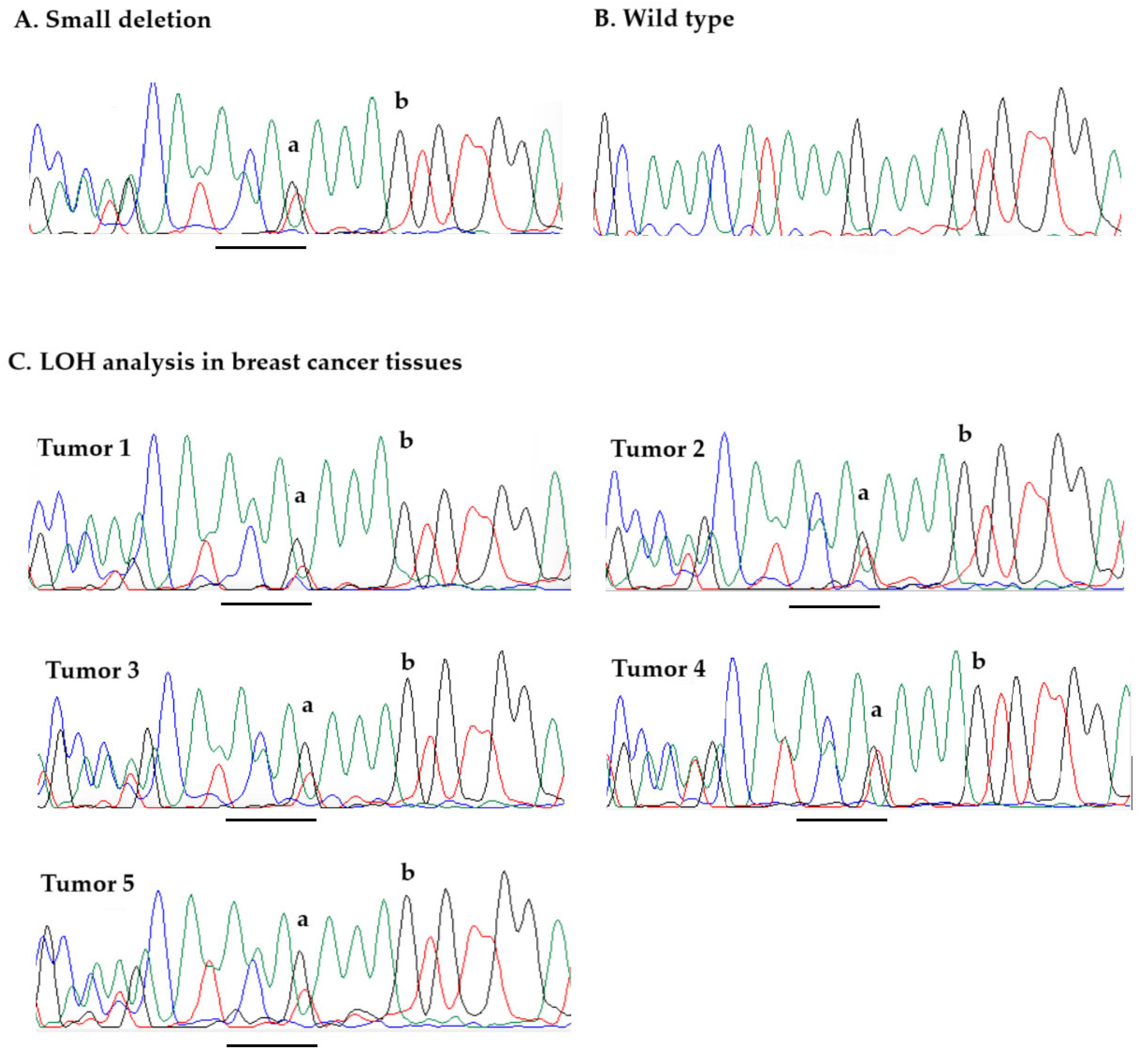
4.2.3. Loss of Heterozygosity Analysis
4.2.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HBC | Hereditary Brest Cancer |
| FFPE | Formalin-fixed Paraffin-embedded |
| BCAC | Breast Cancer Association Consortium |
| LOH | Loss of Heterozygosity |
| ER | Estrogen Receptor |
| PR | Progesterone Receptor |
| HER2 | Human epidermal growth factor receptor 2 |
| UK | United Kingdom |
| DNA | Deoxyribonucleic acid |
| LCIS | Lobular carcinoma in situ |
| NGS | Next-generation sequencing |
References
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and Future Burden of Breast Cancer: Global Statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Narod, S.A.; Foulkes, W.D. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer 2004, 4, 665–676. [Google Scholar] [CrossRef]
- Nelson, H.D.; Pappas, M.; Zakher, B.; Mitchell, J.P.; Okinaka-Hu, L.; Fu, R. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: A systematic review to update the U.S. Preventive Services Task Force recommendation. Ann. Intern. Med. 2014, 160, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Goldgar, D.E.; Healey, S.; Dowty, J.G.; Da Silva, L.; Chen, X.; Hutter, P. Rare variants in the ATM gene and risk of breast cancer. Breast Cancer Res. 2011, 13, R73. [Google Scholar] [CrossRef] [PubMed]
- Ratajska, M.; Antoszewska, E.; Piskorz, A.; Brozek, I.; Borg, Å.; Kusmierek, H. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res. Treat. 2012, 131, 89–97. [Google Scholar] [CrossRef]
- Seal, S.; Thompson, D.; Renwick, A.; Elliott, A.M.; Kelly, P.; Barfoot, R.; Chagtai, T.; Jayatilake, H.; Ahmed, M.; Spanova, K.; et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 2006, 38, 1239–1241. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, F.; Wang, Y.; Liu, S.C. CHEK2 1100delC variant and breast cancer risk in Caucasians: A meta-analysis based on 25 studies with 29,154 cases and 37,064 controls. Asian Pac. J. Cancer Prev. 2012, 13, 3501–3505. [Google Scholar] [CrossRef]
- Rahman, N.; Seal, S.; Thompson, D.; Kelly, P.; Renwick, A.; Elliott, A.M.; Reid, S.; Spanova, K.; Barfoot, R.; Chagtai, T.; et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 2007, 39, 165–167. [Google Scholar] [CrossRef]
- Foulkes, W.; Ghadirian, P.; Akbari, M.R.; Hamel, N.; Giroux, S.; Sabbaghian, N.; Darnel, A.; Royer, R.; Poll, A.; Fafard, E.; et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007, 9, R83. [Google Scholar] [CrossRef]
- Blanco, A.; Graña, B.; Fachal, L.; Santamariña, M.; Cameselle-Teijeiro, J.; Ruíz-Ponte, C.; Carracedo, A.; Vega, A. Beyond BRCA1 and BRCA2 wild-type breast and/or ovarian cancer families: Germline mutations in TP53 and PTEN. Clin. Genet. 2010, 77, 193. [Google Scholar] [CrossRef]
- Osher, D.J.; De Leeneer, K.; Michils, G.; Hamel, N.; Tomiak, E.; Poppe, B.; Leunen, K.; Legius, E.; Shuen, A.; Smith, E.; et al. Mutation analysis of RAD51D in non-BRCA1/2 ovarian and breast cancer families. Br. J. Cancer 2012, 106, 1460–1463. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guénard, F.; Pedneault, C.S.; Ouellette, G.; Labrie, Y.; Simard, J.; INHERIT; Durocher, F. Evaluation of the contribution of the three breast cancer susceptibility genes CHEK2, STK11, and PALB2 in non-BRCA1/2 French Canadian families with high risk of breast cancer. Genet. Test. Mol. Biomarkers 2010, 14, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, C.; Achatz, M.I.; Balmaña, J.; Castro, E.; Curigliano, G.; Cybulski, C.; Domchek, S.M.; Evans, D.G.; Hanson, H.; Hoogerbrugge, N.; et al. Breast cancer multigene germline panel testing in mainstream oncology based on clinical-public health utility: ESMO Precision Oncology Working Group recommendations. Ann. Oncol. 2025; in press. [Google Scholar] [CrossRef] [PubMed]
- Kotsopoulos, J.; Gronwald, J.; Huzarski, T.; Møller, P.; Pal, T.; McCuaig, J.M.; Singer, C.F.; Karlan, B.Y.; Aeilts, A.; Eng, C.; et al. Bilateral Oophorectomy and All-Cause Mortality in Women with BRCA1 and BRCA2 Sequence Variations. JAMA Oncol. 2024, 10, 484–492. [Google Scholar] [CrossRef]
- Turnbull, C.; Rahman, N. Genetic predisposition to breast cancer: Past, present, and future. Annu. Rev. Genom. Hum. Genet. 2008, 9, 321–345. [Google Scholar] [CrossRef]
- Venkitaraman, A.R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002, 108, 171–182. [Google Scholar] [CrossRef]
- Zhang, F.; Ma, J.; Wu, J.; Ye, L.; Cai, H.; Xia, B.; Yu, X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 2009, 19, 524–529. [Google Scholar] [CrossRef]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef]
- Lilley, D.M.; White, M.F. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2001, 2, 433–443. [Google Scholar] [CrossRef]
- Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 1964, 5, 282–304. [Google Scholar] [CrossRef]
- Modrich, P.; Lahue, R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996, 65, 101–133. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Galván, S.; Tous, C.; Blanco, M.G.; Aragon, L.; Alonso-Ramos, A.; Aguilera, A. Distinct roles of Mus81, Yen1, Slx1-Slx4 and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol. Cell Biol. 2012, 32, 1592–1603. [Google Scholar] [CrossRef]
- Agmon, N.; Yovel, M.; Harari, Y.; Shahar, O.; Friedler, A.; Kupiec, M. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011, 39, 7009–7019. [Google Scholar] [CrossRef] [PubMed]
- Ip, S.C.; Rass, U.; Blanco, M.G.; Bai, X.; Flynn, H.R.; Skehel, J.M.; West, S.C. Identification of Holliday junction resolvases from humans and yeast. Nature 2008, 456, 357–361. [Google Scholar] [CrossRef]
- West, S.C. Formation, translocation and resolution of Holliday junctions during homologous genetic recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995, 347, 21. [Google Scholar] [CrossRef]
- Schwacha, A.; Kleckner, N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 1995, 83, 783. [Google Scholar] [CrossRef]
- Bzymek, M.; Thayer, N.H.; Oh, S.D.; Kleckner, N.; Hunter, N. Double Holliday junctions are intermediates of DNA break repair. Nature 2010, 464, 937. [Google Scholar] [CrossRef]
- Wu, Y.; Qian, Y.; Zhou, G.; Lv, J.; Yan, Q.; Dong, X. Effect of GEN1 interference on the chemosensitivity of the breast cancer MCF-7 and SKBR3 cell lines. Oncol. Lett. 2016, 11, 3597–3604. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Guo, B.; Zhang, Y.; Gong, Y.; Zhang, C.; Xu, H.; Wu, X. Gen1 and Eme1 play redundant roles in DNA repair and meiotic recombination in mice. DNA Cell Biol. 2016, 35, 585–590. [Google Scholar] [CrossRef]
- Thaker, P.H.; Borys, N.; Fewell, J.; Anwer, K. GEN-1 immunotherapy for the treatment of ovarian cancer. Future Oncol. 2019, 15, 421–438. [Google Scholar] [CrossRef]
- Arter, M.; Hurtado-Nieves, V.; Oke, A.; Zhuge, T.; Wettstein, R.; Fung, J.C.; Blanco, M.G.; Matos, J. Regulated crossing-over requires inactivation of Yen1/GEN1 resolvase during meiotic prophase I. Dev. Cell 2018, 45, 785.e6. [Google Scholar] [CrossRef] [PubMed]
- Falquet, B.; Rass, U. Structure-specific endonucleases and the resolution of chromosome underreplication. Genes 2019, 10, 232. [Google Scholar] [CrossRef]
- West, S.C. The search for a human Holliday junction resolvase. Biochem. Soc. Trans. 2009, 37, 519–526. [Google Scholar] [CrossRef]
- Ashton, T.M.; Mankouri, H.W.; Heidenblut, A.; McHugh, P.J.; Hickson, I.D. Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol. Cell Biol. 2011, 31, 1921–1933. [Google Scholar] [CrossRef] [PubMed]
- Sobhy, M.A.; Bralić, A.; Raducanu, V.S.; Takahashi, M.; Tehseen, M.; Rashid, F.; Zaher, M.S.; Hamdan, S.M. Resolution of the Holliday junction recombination intermediate by human GEN1 at the single-molecule level. Nucleic Acids Res. 2019, 47, 1935–1949. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.W.; West, S.C. GEN1 Endonuclease: Purification and Nuclease Assays. Methods Enzymol. 2018, 600, 527–542. [Google Scholar] [CrossRef]
- Forbes, S.A.; Bhamra, G.; Bamford, S.; Dawson, E.; Kok, C.; Clements, J.; Menzies, A.; Teague, J.; Futreal, P.; Stratton, M. The catalogue of somatic mutations in cancer (COSMIC). Curr. Protoc. Hum. Genet. 2008, 57, 10.11.1–10.11.26. [Google Scholar] [CrossRef]
- Wood, L.D.; Parsons, D.W.; Jones, S.; Lin, J.; Sjoblom, T.; Leary, R.J.; Shen, D.; Boca, S.M.; Barber, T.; Ptak, J.; et al. The genomic landscapes of human breast and colorectal cancers. Science 2007, 318, 1108–1113. [Google Scholar] [CrossRef]
- Turnbull, C.; Hines, S.; Renwick, A.; Seal, S.; Parker, A.; Elliott, A.; Seal, S.; Warren-Perry, M.; Evans, D.G.; Eccles, D.; et al. Mutation and association analysis of GEN1 in breast cancer susceptibility. Breast Cancer Res. Treat. 2010, 124, 283–288. [Google Scholar] [CrossRef]
- Rass, U.; Compton, S.A.; Matos, J.; Singleton, M.R.; Ip, S.C.; Blanco, M.G.; Griffith, J.D.; West, S.C. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 2010, 24, 1559–1569. [Google Scholar] [CrossRef][Green Version]
- Breast Cancer Association Consortium; Dorling, L.; Carvalho, S.; Allen, J.; González-Neira, A.; Luccarini, C.; Wahlström, C.; Pooley, K.A.; Parsons, M.T.; Fortuno, C.; et al. Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Cybulski, C.; Kluźniak, W.; Huzarski, T.; Wokołorczyk, D.; Kashyap, A.; Rusak, B.; Stempa, K.; Gronwald, J.; Szymiczek, A.; Bagherzadeh, M.; et al. The Spectrum of Mutations Predisposing to Familial Breast Cancer in Poland. Int. J. Cancer 2019, 145, 3311–3320. [Google Scholar] [CrossRef]
- Gliniewicz, K.; Kluźniak, W.; Wokołorczyk, D.; Huzarski, T.; Stempa, K.; Rudnicka, H.; Jakubowska, A.; Szwiec, M.; Jarkiewicz-Tretyn, J.; Naczk, M.; et al. The APOBEC3B c.783delG Truncating Mutation Is Not Associated with an Increased Risk of Breast Cancer in the Polish Population. Genes 2023, 14, 1329. [Google Scholar] [CrossRef] [PubMed]
- Wokołorczyk, D.; Kluźniak, W.; Huzarski, T.; Gronwald, J.; Szymiczek, A.; Rusak, B.; Stempa, K.; Gliniewicz, K.; Kashyap, A.; Morawska, S.; et al. Mutations in ATM, NBN and BRCA2 predispose to aggressive prostate cancer in Poland. Int. J. Cancer 2020, 147, 2793–2800. [Google Scholar] [CrossRef]
- Gronwald, J.; Raczyński, A.; Tarhoni, M.; Blachowski, M.; Huzarski, T.; Byrski, T.; Tołoczko-Grabarek, A.; Dębniak, T.; Cybulski, C.; Huzarska, J.; et al. Population screening for cancer family syndromes in the West Pomeranian region of Poland. Hered. Cancer Clin. Pract. 2006, 4, 56. [Google Scholar] [CrossRef] [PubMed]
- Feszak, S.; Feszak, I.J.; Kluźniak, W.; Wokołorczyk, D.; Stempa, K.; Gliniewicz, K.; Uciński, J.; Huzarski, T.; Dębniak, T.; Gronwald, J.; et al. BRCA1 and BRCA2 Mutations in Polish Women with Ductal Carcinoma In Situ. Cancers 2025, 17, 613. [Google Scholar] [CrossRef]
- Lahiri, D.K.; Schnabel, B. DNA isolation by a rapid method from human blood samples: Effects of MgCl2, EDTA, storage time, and temperature on DNA yield and quality. Biochem. Genet. 1993, 31, 321–328. [Google Scholar] [CrossRef]
- Cybulski, C.; Górski, B.; Dębniak, T.; Gliniewicz, B.; Mierzejewski, M.; Masojć, B.; Jakubowska, A.; Matyjasik, J.; Złowocka, E.; Sikorski, A.; et al. NBS1 is a prostate cancer susceptibility gene. Cancer Res. 2004, 64, 1215–1219. [Google Scholar] [CrossRef]

| Variant a | Exon | Genotype b | Frequency Among Cases | Frequency Among Controls | p Value |
|---|---|---|---|---|---|
| c.1929_1932delAAAG (p.Lys645Cysfs*29) | 13 | del/wt | 4/617 (0.6%) | 1/299 (0.3%) | 1.00 |
| c.2515_2519delAAGTT (p.Lys839Glufs*2) | 13 | del/del | 5/616 (0.8%) | 4/299 (1.3%) | 0.48 |
| del/wt | 125/616 (20.3%) | 51/299 (17.1%) | 0.28 | ||
| del/del or del/wt | 130/616 (21.1%) | 55/299 (18.4%) | 0.38 |
| Total (n) | GEN1 Variant-Positive | Prevalence (%) | OR (CI 95%) | p-Value | |
|---|---|---|---|---|---|
| Patients with Breast Cancer | |||||
| All cases | 15,930 | 38 | 0.24% | 1.40 (0.66–3.01) | p = 0.49 |
| Age (years) | |||||
| ≤50 | 5723 | 9 | 0.16% | 0.92 (0.36–2.40) | p = 0.87 |
| 51–60 | 4909 | 15 | 0.30% | 1.80 (0.76–4.25) | p = 0.25 |
| >60 | 5298 | 14 | 0.26% | 1.55 (0.65–3.71) | p = 0.43 |
| Number of Relatives with Breast Cancer * | |||||
| 0 | 14,592 | 28 | 0.19% | 1.13 (0.51–2.48) | p = 0.92 |
| 1 | 1797 | 5 | 0.28% | 1.64 (0.53–5.01) | p = 0.57 |
| ≥2 | 510 | 2 | 0.39% | 2.31 (0.49–10.91) | p = 0.58 |
| Reference | |||||
| Cancer-free controls | 4702 | 8 | 0.17% | ref | ref |
| GEN1 Variant Positive Cases n = 38 | GEN1 Variant Negative Cases n = 15,892 | p-Value | |
|---|---|---|---|
| Age at diagnosis (years) | 56.8 (31–81) | 56.0 (18–94) | p = 0.64 |
| Histological Features: | |||
| Ductal, grade 3 | 5/32 (15.6%) | 2422/12,452 (19.4%) | p = 0.59 |
| Ductal, grade 1–2 | 15/32 (46.9%) | 5324/12,452 (42.7%) | p = 0.64 |
| Ductal, grade unknown | 2/32 (6.2%) | 1014/12,452 (8.1%) | p = 0.70 |
| Medullary | 2/32 (6.2%) | 259/12,452 (2.1%) | p = 0.12 |
| Lobular | 2/32 (6.2%) | 1528/12,452 (12.3%) | p = 0.31 |
| Tubulolobular | 1/32 (3.1%) | 263/12,452 (2.1%) | p = 0.69 |
| DCIS | 2/32 (6.2%) | 475/12,452 (3.8%) | p = 0.48 |
| Other or undefined | 3/32 (9.4%) | 1165/12,452 (9.4%) | p = 1.00 |
| Receptor status: | |||
| Estrogen receptor-positive | 17/23 (73.9%) | 7485/10,366 (72.2%) | p = 0.86 |
| Progesterone receptor-positive | 18/23 (78.3%) | 7353/10,221 (71.9%) | p = 0.50 |
| HER2-positive | 3/19 (15.8%) | 1601/8619 (18.6%) | p = 0.76 |
| Size (cm): | |||
| <1 | 5/22 (22.7%) | 1081/9966 (10.8%) | p = 0.08 |
| 1–1.9 | 8/22 (36.4%) | 3999/9966 (40.1%) | p = 0.72 |
| 2–4.9 | 7/22 (31.8%) | 4447/9966 (44.6%) | p = 0.23 |
| ≥5 | 2/22 (9.1%) | 439/9966 (4.4%) | p = 0.30 |
| Lymph node-positive | 8/22 (36.4%) | 4547/10,332 (44.0%) | p = 0.47 |
| Bilateral | 2/28 (7.1%) | 496/11,755 (4.2%) | p = 0.45 |
| Chemotherapy (yes) | 10/25 (40.0%) | 4631/8678 (53.4%) | p = 0.19 |
| Vital status (deceased) | 5/30 (16.7%) | 2036/12,037 (16.9%) | p = 0.97 |
| Cancer Site | Number (%) of Cancers in Relatives of GEN1 Variant-Positive Patients (n = 35 Families) | Number (%) of Cancers in Relatives of GEN1 Variant-Negative Patients (n = 14,592 Families) | p-Value | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Breast | 7 | 20.0% | 2431 | 16.7% | 0.76 |
| Colon | 2 | 5.7% | 1112 | 7.6% | 0.92 |
| Kidney | 0 | 0.0% | 411 | 2.8% | 0.62 |
| Larynx | 2 | 5.7% | 554 | 3.8% | 0.88 |
| Lung | 5 | 14.3% | 2156 | 14.8% | 0.93 |
| Leukemia or Lymphoma | 1 | 2.9% | 561 | 3.8% | 0.76 |
| Pancreas | 0 | 0.0% | 402 | 2.8% | 0.63 |
| Prostate | 1 | 2.9% | 991 | 6.8% | 0.57 |
| Stomach | 2 | 5.7% | 1243 | 8.5% | 0.77 |
| Cervix/endometrium | 3 | 8.6% | 1513 | 10.4% | 0.94 |
| Ovary | 2 | 5.7% | 538 | 3.7% | 0.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gliniewicz, K.; Wokołorczyk, D.; Kluźniak, W.; Stempa, K.; Huzarski, T.; Rudnicka, H.; Jakubowska, A.; Szwiec, M.; Jarkiewicz-Tretyn, J.; Cechowska, M.; et al. Analysis of GEN1 as a Breast Cancer Susceptibility Gene in Polish Women. Int. J. Mol. Sci. 2025, 26, 5991. https://doi.org/10.3390/ijms26135991
Gliniewicz K, Wokołorczyk D, Kluźniak W, Stempa K, Huzarski T, Rudnicka H, Jakubowska A, Szwiec M, Jarkiewicz-Tretyn J, Cechowska M, et al. Analysis of GEN1 as a Breast Cancer Susceptibility Gene in Polish Women. International Journal of Molecular Sciences. 2025; 26(13):5991. https://doi.org/10.3390/ijms26135991
Chicago/Turabian StyleGliniewicz, Katarzyna, Dominika Wokołorczyk, Wojciech Kluźniak, Klaudia Stempa, Tomasz Huzarski, Helena Rudnicka, Anna Jakubowska, Marek Szwiec, Joanna Jarkiewicz-Tretyn, Magdalena Cechowska, and et al. 2025. "Analysis of GEN1 as a Breast Cancer Susceptibility Gene in Polish Women" International Journal of Molecular Sciences 26, no. 13: 5991. https://doi.org/10.3390/ijms26135991
APA StyleGliniewicz, K., Wokołorczyk, D., Kluźniak, W., Stempa, K., Huzarski, T., Rudnicka, H., Jakubowska, A., Szwiec, M., Jarkiewicz-Tretyn, J., Cechowska, M., Domagała, P., Dębniak, T., Lener, M., Gronwald, J., Lubiński, J., Narod, S. A., Akbari, M. R., & Cybulski, C. (2025). Analysis of GEN1 as a Breast Cancer Susceptibility Gene in Polish Women. International Journal of Molecular Sciences, 26(13), 5991. https://doi.org/10.3390/ijms26135991







