Abstract
The objectives of this study were to design, synthesize, and evaluate the antibacterial activity of a series of novel aminoguanidine-tetralone derivatives. Thirty-four new compounds were effectively synthesized through nucleophilic substitution reaction and guanidinylation reaction. Chemical structures of all the desired compounds were identified by NMR and HR-MS spectroscopy. Most of the synthesized compounds showed significant antibacterial activity against ESKAPE pathogens and clinically resistant Staphylococcus aureus (S. aureus) isolates. S. aureus is an important pathogen that has the capacity to cause a variety of diseases, including skin infections, pneumonia, and sepsis. The most active compound, 2D, showed rapid bactericidal activity against S. aureus ATCC 29213 and MRSA-2 with MIC/MBC values of 0.5/4 µg/mL and 1/4 µg/mL, respectively. The hemolytic activity and cytotoxicity of 2D was low, with HC50 and IC50(HEK 293-T) values of 50.65 µg/mL and 13.09 µg/mL, respectively. Compound 2D induced the depolarization of the bacterial membrane and disrupted bacterial membrane integrity, ultimately leading to death. Molecular docking revealed that dihydrofolate reductase (DHFR) may be a potential target for 2D. In the mouse skin abscess model caused by MRSA-2, 2D reduced the abscess volume, decreased bacterial load, and alleviated tissue pathological damage at doses of 5 and 10 mg/kg. Therefore, compound 2D may be a promising drug candidate for antibacterial purposes against S. aureus.
1. Introduction
The emergence and dissemination of antimicrobial resistance represents a significant global public health concern [1]. The resistance of the six pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Escherichia coli) known as ESKAPE pathogens has been particularly serious [2,3,4]. Staphylococcus aureus (S. aureus) is a widely distributed opportunistic pathogen that has the capacity to cause a variety of diseases, including skin infections, pneumonia, and sepsis [5]. As a significant clinical pathogen, drug-resistant Staphylococcus aureus (S. aureus) has attracted widespread attention [1,2,3,4,5]. In particular, methicillin-resistant S. aureus (MRSA) has attracted much attention since it was first identification in 1961 [6]. MRSA can be resistant to multiple antibiotics [7], and its recurrent bloodstream infections are associated with high mortality rates [8]. In addition to the rational use of existing antimicrobial agents, novel antimicrobial agents should be actively developed to combat S. aureus resistance.
Tetralone and tetralone derivatives, as crucial structural scaffolds of potential novel drugs, are normally found in several natural compounds and can be used as parental scaffolds and/or intermediates for the synthesis of a series of pharmacologically active compounds with a broad spectrum of bioactivities including antibacterial, antitumor, CNS effect, and so on [9]. Tetralone derivatives have attracted wide interest due to their antimicrobial properties. Ampicillin-tetralone derivatives have been shown to overcome drug resistance and demonstrate effective activity against S. aureus [10]. The natural product 4-hydroxy-α-tetralone and its derivatives can reverse the resistance of multi-drug resistant Escherichia coli by inhibiting the ATP-dependent efflux pump [11]. Furthermore, tetralone derivatives have shown antiviral activity [12,13] and antifungal activity [14]. Since tetralone derivatives exhibit excellent antibacterial potential [15], the molecules containing this scaffold are likely to be promising antibacterial drugs in the future.
Antimicrobials have been shown to cross the membrane, subsequently interacting with the target of action [16], a process which poses a significant challenge to many compounds [17]. Study has also shown that amphiphilic and rigid small molecules containing amines with low sphericity were most likely to accumulate to effect concentrations [18]. Aminoguanidine, a group with strongly alkaline and multiple nitrogen atoms, has attracted significant attention in the field of antimicrobial agents [19,20,21,22]. Quinolone-tethered aminoguanidine molecules have demonstrated noteworthy anti-tuberculosis activity [23]. Aminoguanidine derivatives with 1,2,4-triazol moieties exhibited substantial antibacterial activity against both bacteria and Candida albicans [24]. Diphenylurea derivatives bearing aminoguanidine moiety exhibited potent anti-staphylococcal activity [25]. Consequently, aminoguanidine has emerged as a promising antibacterial pharmacophore.
In this study, a series of tetralone indole derivatives (1A–1X, 2A–2J) were first designed and synthesized, including 6-hydroxy-1-tetralone with various substituted benzyl bromides or bromoalkane and aminoguanidine to produce the target compound. Then, their structures were characterized. Antibacterial activity against both Gram-positive and Gram-negative bacteria was assessed. The role of the active molecule 2D in disrupting cell membrane integrity was investigated. A molecular docking study was performed with 2D and S. aureus dihydrofolate reductase (DHFR) to find the potential antibacterial target and elucidate the binding mechanism. Furthermore, the hemolytic activity and cytotoxicity of 2D were also examined. This study provides a class of compounds for the development of novel antibacterial agents.
2. Results and Discussion
2.1. Chemistry
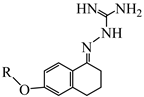
The synthetic route for the preparation of the aminoguanidine tetralone derivatives (1A–1X, 2A–2J) and intermediates is shown in Scheme 1. The key intermediates were prepared from 6-hydroxy-1-tetralone and substituted benzyl bromides or brominated alkanes or brominated alkenes under alkaline conditions. With the key intermediate in hand, all the target derivatives (Table 1) were prepared directly in the presence of aminoguanidine hydrochloride and concentrated hydrochloric acid by stirring in an oil bath at 80 °C. Finally, the structures of the desired compounds were characterized by 1H NMR, 13C NMR and high-resolution mass spectrometry (HR-MS) (Supplementary Materials).
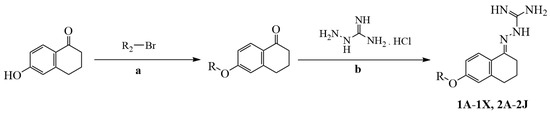
Scheme 1.
The synthesis routes of compounds 1A–1X and 2A–2J. (a) Dry acetonitrile, K2CO3, KI, 80 °C, 0.5–12 h; (b) ethanol absolute, 15 drops of concentrated HCl, 80 °C, 0.5–1 h.

Table 1.
Chemical structures of target compounds.
2.2. Evaluation of In Vitro Antimicrobial Activity
The minimum inhibitory concentrations (MICs) and minimum bactericidal concentrations (MBCs) of the synthesized compounds effective against ESKAPE strains, S. aureus clinical isolates, and MRSA were investigated by the broth dilution method. Levofloxacin is a broad-spectrum antibacterial agent that is widely used in the treatment of ESKAPE infections [26]. Therefore, levofloxacin was selected as the positive control for the preliminary activity screening. DMSO (0.1%) was selected as the negative control.
Initial screening results are presented in Table 2. In general, most of the compounds showed strong activity against ESKAPE strains, with MICs ranging from 0.5 μg/mL to 32 μg/mL. Most of the compounds showed higher antibacterial activity against Gram-positive bacteria, with MICs in the ranging from 0.5 μg/mL to 4 μg/mL. For Gram-negative bacteria, the compounds showed better activity against Escherichia coli and Acinetobacter baumannii in comparison to Klebsiella pneumoniae and Pseudomonas aeruginosa. Compound 2D exhibited the highest level of activity against S. aureus. Compared with the aminoguanidine derivatives synthesized by Ping Yang et al. (MICS. aureus = 2–128 μg/mL) [27], Wolfgang Dohle et al. (MICS. aureus = 1–32 μg/mL) [28] and Xueqian Bai et al. (MICS. aureus = 4–64 μg/mL) [24], respectively, compound 2D (MICS. aureus = 0.5 μg/mL) showed superior activity against S. aureus.

Table 2.
The MICs and MBCs of target compounds against ESKAPE pathogens.
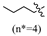
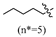
An analysis of the structure–activity relationship reveals the following: In the case of R being a substituted benzyl group, the antibacterial activity of compounds containing one or two halogens (fluorine, chlorine, bromine) or one trifluoromethyl group was similar. In contrast, the activity of compounds containing two trifluoromethyl groups against Gram-positive bacteria decreased by 2–16 times, and the activity against Gram-negative bacteria was lost. In the case of the R being an alkyl group, compounds with more than nine carbon atoms exhibited a loss of all activity against Gram-negative bacteria, while displaying minimal effect on the activity of Gram-positive bacteria. Among all compounds, those comprising medium-chain alkane groups exhibit better activity in comparison to those with halogenated benzyl groups, as well as short- and long-chain alkane compounds. It is speculated that medium-chain compounds possess a more optimal configuration and lipid–water partition coefficient (LogP).
In view of the excellent anti-Staphylococcus aureus activity of the compounds, we further tested the activity of the compounds against four Staphylococcus aureus clinical isolates and two MRSA strains (Table 3). Vancomycin is currently the first-line drug against MRSA infections [29], so it was selected as a positive control. The results showed that the synthesized compounds also showed significant activity against the clinical isolates and the MRSA, with MICs ranging from 1 μg/mL to 32 μg/mL. The activity of 1L, 1M, 1S, 1U, 1X, 2A, 2D–2F, and 2I against MRSA was analogous to that of vancomycin, with MICs ranging from 1 µg/mL to 2 µg/mL. Furthermore, the MBC values of the compounds were detected. The MBC values of most compounds were 1–4 times higher than the MIC values, indicating bactericidal properties of those compounds.

Table 3.
The MICs and MBCs of target compounds against S. aureus clinical isolates and MRSA.
As the most active compound, the bacteriostatic and time-killing ability of 2D was investigated in vitro. 2D was able to suppress the growth of ATCC 29213 and MRSA-2 at concentrations 0.5, 1, 2, 4, and 8 µg/mL. Vancomycin at 0.5 μg/mL and 1 μg/mL failed to completely inhibit the growth of ATCC 29213 and MRSA-2 (Figure 1A,B). The time-kill assay showed that 2D exhibited a concentration-dependent and time-dependent bactericidal response (Figure 1C,D).
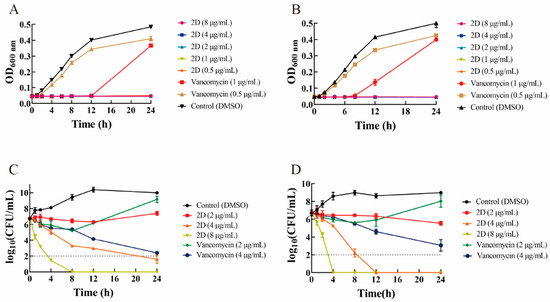
Figure 1.
The result of the growth curve and bactericidal time-kill kinetics assay. (A) Growth curve of ATCC 29213 in the presence of 2D (0.5, 1, 2, 4 and 8 µg/mL) and vancomycin (0.5 and 1 µg/mL). (B) Growth curve of MRSA-2 in the presence of 2D (0.5, 1, 2, 4 and 8 µg/mL) and vancomycin (0.5 and 1 µg/mL). (C) Time-kill curve of ATCC 29213 in the presence of 2D (2, 4 and 8 µg/mL) and vancomycin (2 and 4 µg/mL). (D) Time-kill curve of MRSA-2 in the presence of 2D (2, 4 and 8 µg/mL) and vancomycin (2 and 4 µg/mL).
2.3. 2D Exhibits Potent Anti-Biofilm Properties
The biofilm is a highly structured microbial community that protects bacterial cells from the external environment and resists antibiotics [30]. Therefore, good anti-biofilm activity is an important ability of new antimicrobial agents [31]. The results showed that the biofilm elimination rate of 2D (0.25 μg/mL) was 21.4%, which was significantly superior to that of vancomycin at 0.25 μg/mL (3.63%) (Figure 2A). In the concentration range of 4–32 μg/mL, 2D exhibited a biofilm elimination rate of 91.8% to 93.7%, which was more effective than vancomycin. However, at the test concentration, vancomycin was unable to completely eliminate the biofilm, a finding that is consistent with the existing research [32].
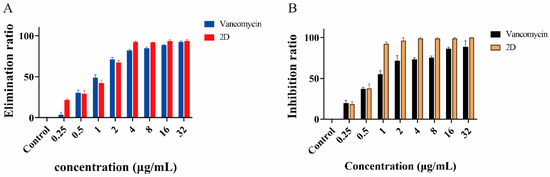
Figure 2.
Elimination and inhibition of biofilm. (A) Elimination rates of biofilm by different concentrations of 2D and vancomycin against MRSA-2. (B) Inhibition rates of biofilm by different concentrations of 2D and vancomycin against MRSA-2.
The ability of 2D and vancomycin to inhibit biofilm formation at 0.25 μg/mL and 0.5 μg/mL (1/4MIC and 1/2MIC) was similar, with an inhibition rate of 18.5% and 37.8% (Figure 2B). 2D at 1 μg/mL (1MIC) reduced biofilm formation by more than 92%. Furthermore, the 2D concentrations of 2–32 μg/mL (2-32MIC) almost completely inhibited the formation of biofilm.
2.4. 2D Destroys the Bacterial Structure
The primary antibacterial mechanism of guanidine involves the destruction of bacterial membranes [33]. Therefore, scanning electron microscopy (SEM) was used to observe the effect of 2D on MRSA cell membranes. As shown in Figure 3, the cell membrane of MRSA-2 was intact in the control group. Following treatment with vancomycin (4 μg/mL) and 2D (2 μg/mL), a small number of bacteria exhibited rupture folds. However, after treatment with 2D at concentrations of 4 and 8 μg/mL, a substantial number of bacteria membranes were broken. These results suggest that 2D can destroy the membrane structure of MRSA-2, and this effect is concentration-dependent.
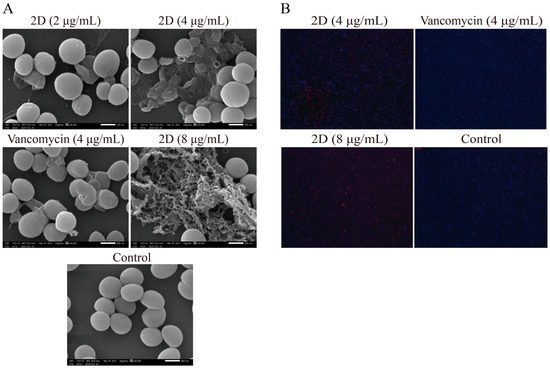
Figure 3.
(A) The results of scanning electron microscopy assay. The strain used was MRSA-2. Scale bar, 500 nm. (B) The results of fluorescence microscopy assay. The strain used was MRSA-2.
2.5. Fluorescence Microscopy
DAPI has been shown to stain living cells blue. PI has the ability to stain dead cells with damaged cell membranes a red color. The results of the fluorescence microscopy analysis showed that the control group exhibited almost no red fluorescence, with only significant blue fluorescence being observed (Figure 3B). The vancomycin (4 µg/mL) group exhibited a very small amount of red fluorescence. The 2D (4 and 8 μg/mL) group exhibited a marked tendency to emit red fluorescence. The findings indicated that 2D exerted a substantial destructive impact on the cell membrane. This finding aligns with the results obtained from the SEM analysis.
2.6. Membrane Permeabilization and Depolarization
Since compound 2D significantly destroyed the membrane structure of bacteria from SEM and fluorescence microscopy, the effect of 2D on bacterial plasma membranes was further studied by fluorescent probes DiSC3(5) and PI. DiSC3(5) localizes in the bacterial membrane depending on an intact membrane potential gradient, where self-quenching occurs [34]. After adding 2D (2, 4, and 8 μg/mL), the fluorescence intensity increased instantaneously (at 10 min), and the final fluorescence intensity was significantly higher than that of the control group (Figure 4A). This finding indicates that 2D has the capacity to depolarize the bacterial cell membrane. The PI enters the bacteria only through the damaged membrane and emits strong fluorescence when combined with DNA [35]. After treatment with 2D (4 and 8 μg/mL), the fluorescence intensity of MRSA-2 was significantly enhanced (Figure 4B). The results showed that 2D could destroy the integrity of the inner membrane of MRSA-2. Concurrently, the effect of 2D on MRSA-2 cell membranes exhibited a concentration-dependent response.
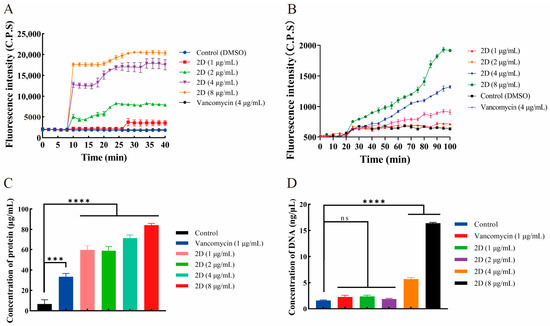
Figure 4.
(A) Cytoplasmic membrane depolarization of 2D using DiSC3(5) assays. (B) The 2D increased the permeability of the inner membrane. (C) Protein leakage caused by the action of 2D on MRSA-2. (D) DNA leakage resulting from the action of 2D on MRSA-2. “ns” is not significant; *** p < 0.001; **** p < 0.0001.
2.7. Leakage of Protein and DNA
The disruption of the bacterial cell membrane can result in the efflux of intracellular substances [36]. As shown in Figure 4C,D, 2D significantly increased the content of extracellular protein and DNA compared with the control group. These results indicated that 2D-induced membrane damage in MRSA-2 can lead to intracellular protein and DNA leakage.
2.8. Docking Analysis
Guanidine has been demonstrated to exhibit a variety of antibacterial mechanisms [37]. As previous studies have shown, aminoguanidine carbazole derivatives [38] and aminoguanidine indole derivatives [39] have strong binding ability with dihydrofolate reductase (DHFR). DHFR is an important antibacterial target [40]. Therefore, the binding capacity of the interaction between 2D and the S. aureus DHFR protein was investigated. The -CDOCKER₋ENERGY of the dihydrofolate reductase inhibitor trimethoprim was determined to be 32.9555. As shown in the docking results, the docked pose of 2D with DHFR had a -CDOCKER₋ENERGY of 33.7945, suggesting that 2D can bind to potential binding sites in the DHFR. Five active site residues (ASN18, LEU20, LEU28, LYS32, and ILE50) were involved in 2D recognition (Figure 5). In DHFR, the ASN18 residue formed a conventional hydrogen bond interaction with the guanidine of the 2D. The side chains of LEU20, LEU28, LYS32, and ILE50 formed alkyl stacking with 2D. The tetralin ring and alkyl moieties enhanced the hydrophobic interaction between 2D and DHFR.
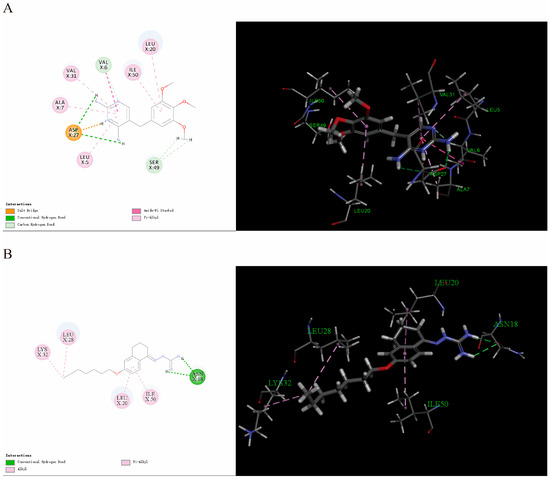
Figure 5.
(A) Interaction of trimethoprim with the S. aureus DHFR protein. (B) Interaction of 2D with the S. aureus DHFR protein.
2.9. Hemolytic Activity and Cytotoxicity
The hemolysis of compound 2D was determined by its ability to lyse sheep red blood cells. The results were expressed as the HC50 value (the concentration at which 50% of red blood cells were lysed). The HC50 value of 2D was 50.65 μg/mL, indicating that the compound exhibited selective toxicity to bacterial cells (SI = 101.30) (Table 4).

Table 4.
The hemolysis and the cytotoxicity of 2D.
In addition, the cytotoxicity of 2D in four mammalian cell lines was also detected (Table 4). The selectivity index (SI) of 2D was determined to be 26.18 for S. aureus ATCC 29213 and the normal human cell line HEK 293-T. In accordance with SI > 10 [41], the results showed that 2D was less toxic to mammalian cells.
2.10. In Vivo Efficacy
As 2D has exhibited potent antibacterial activity in vitro, its antibacterial effect in vivo was assessed by a mouse skin abscess model. After a 48 h period of infection, the dorsal skin of mice in both the model group and the solvent group displayed significant swelling, accompanied by an increase in the volume of subcutaneous abscesses. The abscesses in the vancomycin group and the 2D (5 mg/kg) group exhibited a reduced volume compared to the model group and the solvent group, although complete resolution was not observed. The dorsal skin of mice treated with 2D (10 mg/kg) displayed a smooth appearance and was devoid of any discernible abscesses. 2D (5 and 10 mg/kg) exhibited superiority over vancomycin in terms of reducing the volume of skin abscesses (Figure 6A,D). The results of the histopathological examination of the skin indicated that the mice in the model group developed a suppurative lesion with a distinct boundary with the surrounding tissues, primarily comprising neutrophils and lymphocytes, which was characterized as suppurative dermatitis. A minimal degree of inflammatory cell infiltration was observed beneath the skin of mice treated with 2D (10 mg/kg), with the presence of almost no purulent foci noted. The purulent lesions at the infection site of mice in the vancomycin (5 mg/kg) and 2D (5 mg/kg) groups were significantly smaller (Figure 6B). The results of the tissue bacterial load analysis showed that the bacterial load of the skin at the infected site was significantly reduced after vancomycin and 2D treatment, compared with the model group and the solvent group (Figure 6C,E,F). The results of the histopathological examination of the major organs showed that the major organs of the model group and the solvent group exhibited significant pathological alterations, in comparison with the blank control group. These alterations were characterized by congestion of the central vein of the liver, indistinct boundaries of the marginal area of the spleen, diffuse necrosis of parenchymal cells, glomerular degeneration and atrophy, and local hemorrhage. After the vancomycin and 2D treatment, no discernible lesions were observed in the major organs (Figure 7).
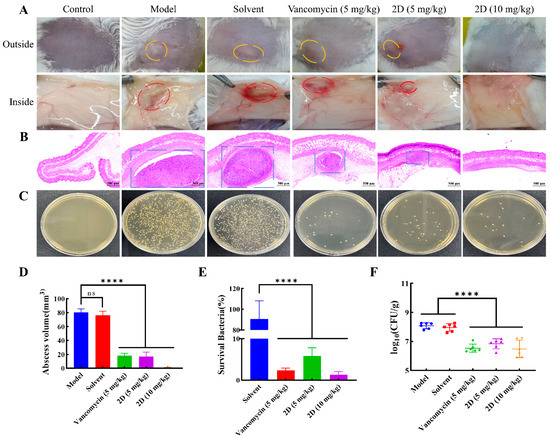
Figure 6.
2D can treat MRSA infection in mice. (A) Photographs of MRSA-2 infection sites with various treatments after infection. Skin abscesses were marked by the orange circles (outside) and red circles (inside). (B) Histological photographs of infected skin that received various treatments. Suppurative lesions were marked by the blue sqaures. Scale bar, 500 μm. (C) Images of MRSA-2 colonies cultured from the homogenate of infected skin after appropriate dilution. (D) Size of abscess at MRSA-2 infection site under different treatments. (E) Bacterial survival in the skin of MRSA-2-infected mice of each group treated with 2D and vancomycin. (F) The tissue bacteria load of MRSA-2-infected mice skin in each group treated with 2D (5 and 10 mg/kg) and vancomycin (5 mg/kg). The data are presented as the means ± SD from six independent experiments; “ns” is not significant; **** p < 0.0001.
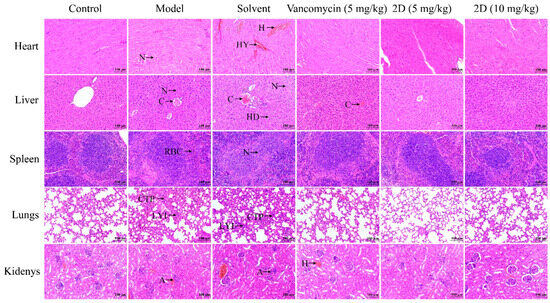
Figure 7.
Histopathological analysis of different tissues using hematoxylin-eosin (H&E) staining. A: atrophy; C: congestion; CTP: connective tissue proliferation; H: hemorrhage; HD: hepatocyte degeneration; HY: hyperemia; LYI: lymphocyte infiltration; N: necrosis; RBC: red blood cell. Scale bar, 100 μm.
In general, the aminoguanidine tetralone derivative 2D exhibited a favorable therapeutic effect in a mouse model of MRSA-infected skin abscesses, suggesting its potential as a treatment for MRSA infection.
3. Materials and Methods
3.1. Chemistry
3.1.1. General
All reagents were commercially available and were used without further purification. The reaction process was monitored by Thin Layer Chromatography (TLC). The 1H and 13C NMR spectra were recorded on an Agilent DD2-600MHz NMR spectrometer at room temperature, operating at 600 MHz for 1H NMR and 151 MHz for 13C NMR by using DMSO-d6 as solvent. The mass spectra were obtained at Agilent 6530 QTOF.
3.1.2. Synthesis of Intermediates
A quantity of 1 mmol (163 mg) of 6-hydroxy-1-tetralone was dissolved in 5 mL of dry acetonitrile. Subsequently, 4 mmol (554 mg) of anhydrous potassium carbonate and 0.1 mmol (16.6 mg) of potassium iodide were added to the solution. The mixture was stirred in an oil bath at 80 °C for 10 min. Then, 1 mmol of substituted benzyl bromide or bromoalkane or bromoalkene was added, and the reaction was stirred at 80 °C for 0.5–12 h. After the completion of the reaction, 50 mL of water was added, filtered or extracted with dichloromethane (15 mL × 3, the dichloromethane layer was washed twice with saturated salt water, anhydrous sodium sulfate was used to remove water, and dichloromethane was removed by vacuum distillation). The obtained product was used for the next reactions without further treatment after drying.
3.1.3. General Procedure for the Synthesis of Compounds 1A–1X and 2A–2J
The intermediate (1 mmol) was meticulously suspended in 5 mL of anhydrous ethanol, and 15 drops of concentrated hydrochloric acid were added to the suspension. Subsequently, aminoguanidine hydrochloride (1.2 mmol, 132.7 mg) was added to the mixture and stirred at 80 °C for 0.5–1 h. After the completion of the reaction, the solution was cooled to room temperature. In instances where a substantial quantity of solid precipitation was present, the collected solid was subjected to three cycles of washing with anhydrous ethanol, followed by three additional cycles of washing with water. In the absence of solid precipitation, a small amount of ethyl acetate was added to the reaction solution to precipitate a large amount of solid. The precipitated solids were subsequently collected and washed thrice with ethyl acetate and water, respectively. After solids were dry, the target compounds 1A–1X and 2A–2J were obtained.
The LogP of the compounds was predicted by ADMETlab 3.0 (https://admetlab3.scbdd.com./server/evaluationCal (accessed on 25 April 2025)). The spectral data of the novel compounds are listed below.
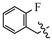
(E)-2-(6-((2-fluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1A). Yield: 56.0%, 0.177 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.30–8.26 (m, 1H), 7.95–7.67 (br s, 3H), 7.56 (td, J = 7.5, 1.4 Hz, 1H), 7.45–7.40 (m, 1H), 7.28–7.21 (m, 2H), 6.90–6.86 (m, 2H), 5.17 (s, 2H), 2.74 (t, J = 6.0 Hz, 1H), 2.66 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 160.39 (d, JCF = 246.3 Hz), 159.19, 155.96, 151.12, 142.11, 130.76 (d, JCF = 4.0 Hz), 130.47 (d, JCF = 8.2 Hz), 127.39, 124.53 (d, JCF = 3.4 Hz), 124.41, 123.60 (d, JCF = 14.6 Hz), 115.39 (d, JCF = 20.9 Hz), 113.61, 113.25, 63.54 (d, JCF = 3.5 Hz), 28.99, 26.43, 21.18; HRMS (ESI+): m/z calcd for C18H19FN4O [M + H]+ 327.1615, found 327.1628.
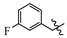
(E)-2-(6-((3-fluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1B). Yield: 89.3%, 0.263 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.28 (d, J = 8.7 Hz, 1H), 7.94–7.64 (br s, 3H), 7.44 (td, J = 8.0, 6.0 Hz, 1H), 7.30–7.26 (m, 2H), 7.18–7.14 (m, 1H), 6.89–6.85 (m, 2H), 5.17 (s, 2H), 2.73 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 162.18 (d, JCF = 243.6 Hz), 159.14, 155.97, 151.12, 142.08, 139.84 (d, JCF = 7.5 Hz), 130.48 (d, JCF = 8.6 Hz), 127.39, 124.39, 123.52 (d, JCF = 2.8 Hz), 114.60 (d, JCF = 21.1 Hz), 114.22 (d, JCF = 21.6 Hz), 113.70, 113.42, 68.33 (d, JCF = 2.1 Hz), 28.99, 26.43, 21.18; HRMS (ESI+): m/z calcd for C18H19FN4O [M + H]+ 327.1615, found 327.1625.
(E)-2-(6-((4-fluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1C). Yield: 67.5%, 0.207 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.29–8.26 (m, 1H), 7.95–7.66 (br s, 3H), 7.52–7.48 (m, 2H), 7.24–7.19 (m, 2H), 6.86 (t, J = 7.9 Hz, 2H), 5.12 (s, 2H), 2.73 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H), 13C NMR (151 MHz, DMSO-d6) δ 161.77 (d, JCF = 243.9 Hz), 159.28, 155.97, 151.15, 142.06, 133.09 (d, JCF = 2.9 Hz), 129.98 (d, JCF = 8.2 Hz, 2C), 127.37, 124.27, 115.25 (d, JCF = 21.4 Hz, 2C), 113.72, 113.37, 68.49, 29.00, 26.44, 21.19; HRMS (ESI+): m/z calcd for C18H19FN4O [M + H]+ 327.1615, found 327.1624.
(E)-2-(6-((2,4-difluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1D). Yield: 71.4%, 0.231 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.30–8.27 (m, 1H), 8.05–7.67 (br s, 3H), 7.63 (td, J = 8.6, 6.6 Hz, 1H), 7.30 (ddd, J = 10.3, 9.3, 2.6 Hz, 1H), 7.17–7.10 (m, 1H), 6.88 (t, J = 7.6 Hz, 2H), 5.14 (s, 2H), 2.74 (t, J = 6.0 Hz, 2H), 2.66 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 162.32 (dd, JCF = 254.6, 12.5 Hz), 160.68 (dd, JCF = 256.7, 12.4 Hz), 159.12, 155.97, 151.12, 142.13, 132.23 (dd, JCF = 9.9, 5.6 Hz), 127.41, 124.47, 120.11 (dd, JCF = 14.9, 3.5 Hz), 113.63, 113.27, 111.61 (dd, JCF = 21.3, 3.5 Hz), 104.06 (t, JCF = 25.7 Hz), 63.11 (d, JCF = 3.0 Hz), 28.99, 26.43, 21.18; HRMS (ESI+): m/z calcd for C18H18F2N4O [M + H]+ 345.1521, found 345.1526.
(E)-2-(6-((2,5-difluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1E). Yield: 80.2%, 0.254 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.31-8.27 (m, 1H), 7.97–7.61 (br s, 3H), 7.43 (ddd, J = 8.8, 5.6, 3.2 Hz, 1H), 7.32 (td, J = 9.2, 4.5 Hz, 1H), 7.27 (td, J = 8.8, 8.3, 4.5 Hz, 1H), 6.92–6.88 (m, 2H), 5.16 (s, 2H), 2.74 (t, J = 6.0 Hz, 2H), 2.66 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 158.95, 158.05 (dd, JCF = 240.4, 2.1 Hz), 156.35 (dd, JCF = 242.3, 2.4 Hz), 155.96, 151.08, 142.14, 127.42, 125.68 (dd, JCF = 17.4, 8.1 Hz), 124.58, 117.00 (dd, JCF = 24.2, 9.0 Hz), 116.70 (dd, JCF = 24.7, 4.4 Hz), 116.61 (dd, JCF = 24.2, 8.7 Hz), 113.63, 113.31, 63.06 (d, JCF = 2.5 Hz), 28.98, 26.43, 21.17; HRMS (ESI+): m/z calcd for C18H18F2N4O [M + H]+ 345.1521, found 345.1530.
(E)-2-(6-((2,6-difluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1F). Yield: 50.9%, 0.143 g; 1H NMR (600 MHz, DMSO-d6) δ 11.17 (s, 1H), 8.30 (d, J = 8.6 Hz, 1H), 8.10–7.60 (br s, 3H), 7.53 (tt, J = 8.4, 6.6 Hz, 1H), 7.18 (t, J = 8.0 Hz, 2H), 6.96–6.80 (m, 2H), 5.14 (s, 2H), 2.74 (t, J = 6.0 Hz, 2H), 2.66 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 161.19 (d, JCF = 256.8 Hz), 161.19 (d, JCF = 241.6 Hz), 159.09, 156.01, 151.07, 142.16, 131.81 (t, JCF= 10.4 Hz), 127.45, 124.63, 113.47, 113.22, 112.06 (t, JCF = 19.4 Hz), 111.88 (d, JCF = 4.6 Hz), 111.74 (d, JCF = 4.5 Hz), 57.61 (t, JCF = 3.6 Hz), 29.00, 26.46, 21.17. HRMS (ESI+): m/z calcd for C18H18F2N4O [M + H]+ 345.1521, found 345.1526.
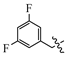
(E)-2-(6-((3,5-difluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1G). Yield: 86.4%, 0.257 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.28 (d, J = 8.8 Hz, 1H), 8.10–7.50 (br s, 3H), 7.23–7.15 (m, 3H), 6.92–6.83 (m, 2H), 5.18 (s, 2H), 2.73 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 162.45 (d, JCF = 246.4 Hz), 162.36 (d, JCF = 246.6 Hz), 158.90, 155.97, 151.09, 142.11, 141.64 (t, JCF = 9.3 Hz), 127.43, 124.56, 113.68, 113.46, 110.48 (d, JCF = 5.2 Hz), 110.34 (d, JCF = 5.2 Hz), 103.19 (t, JCF = 25.7 Hz), 67.78, 28.98, 26.41, 21.16; HRMS (ESI+): m/z calcd for C18H18F2N4O [M + H]+ 345.1521, found 345.1527.
(E)-2-(6-((3,4-difluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1H). Yield: 80.0%, 0.262 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.28 (d, J = 8.7 Hz, 1H), 7.95–7.64 (br s, 3H), 7.53 (ddd, J = 11.6, 7.9, 2.2 Hz, 1H), 7.45 (dt, J = 10.8, 8.4 Hz, 1H), 7.34–7.29 (m, 1H), 6.93–6.82 (m, 2H), 5.13 (s, 2H), 2.73 (t, J = 5.9 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.07, 155.97, 151.12, 149.34 (dd, JCF = 245.7, 12.7 Hz), 149.00 (dd, JCF = 245.6, 12.6 Hz), 142.09, 134.70 (dd, JCF = 5.8, 3.5 Hz), 127.40, 124.60 (dd, JCF = 6.8, 3.4 Hz), 124.44, 117.55 (d, JCF = 17.2 Hz), 116.80 (d, JCF = 17.6 Hz), 113.71, 113.41, 67.88, 28.99, 26.43, 21.18; HRMS (ESI+): m/z calcd for C18H18F2N4O [M + H]+ 345.1521, found 345.1526.
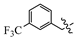
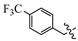
(E)-2-(6-((3-(trifluoromethyl)benzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1I). Yield: 72.6%, 0.251 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.31-8.26 (m, 1H), 8.01–7.79 (br s, 3H), 7.82 (d, J = 2.0 Hz, 1H), 7.77 (d, J = 8.0, 1.7 Hz, 1H), 7.70 (dd, J = 8.0, 1.7 Hz, 1H), 7.64 (t, J = 7.7 Hz, 1H), 6.90 (t, J = 8.0 Hz, 2H), 5.25 (s, 2H), 2.74 (t, J = 6.0 Hz, 2H), 2.66 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.11, 155.97, 151.11, 142.10, 138.44, 131.67, 129.59, 129.18 (d, JCF = 31.7 Hz), 127.42, 124.58 (q, JCF = 3.8 Hz), 124.47, 124.17 (d, JCF = 272.3 Hz), 124.03 (q, JCF = 3.8 Hz), 113.66, 113.47, 68.30, 28.99, 26.42, 21.18; HRMS (ESI+): m/z calcd for C19H19F3N4O [M + H]+ 377.1583, found 377.1587.
(E)-2-(6-((4-(trifluoromethyl)benzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1J). Yield: 60.8%, 0.205 g; 1H NMR (600 MHz, DMSO-d6) δ 8.21 (d, J = 8.5 Hz, 1H), 7.76 (d, J = 8.0 Hz, 2H), 7.67 (d, J = 8.0 Hz, 2H), 7.50–6.85 (br s, 4H), 6.87–6.81 (m, 2H), 5.25 (s, 2H), 2.71 (t, J = 5.9 Hz, 2H), 2.66 (t, J = 6.5 Hz, 2H), 1.80 (p, J = 6.2 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 158.45, 157.07, 149.89, 141.93, 141.33, 128.30 (d, JCF = 31.7 Hz), 127.98 (2C), 126.75, 125.73, 125.32 (q, JCF = 3.9 Hz, 2C), 124.23 (d, JCF = 271.9 Hz), 113.51, 113.44, 68.24, 29.31, 26.31, 21.47; HRMS (ESI+): m/z calcd for C19H19F3N4O [M + H]+ 377.1583, found 377.1587.
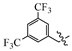
(E)-2-(6-((3,5-bis(trifluoromethyl)benzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1K). Yield: 82.7%, 0.332 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.30 (d, J = 8.6 Hz, 1H), 8.18 (d, J = 1.8 Hz, 2H), 8.09 (s, 1H), 7.62–7.97 (br s, 3H), 6.95–6.90 (m, 2H), 5.35 (s, 2H), 2.75 (t, J = 5.9 Hz, 2H), 2.66 (t, J = 6.5 Hz, 2H), 1.85 (p, J = 6.4 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 158.85, 155.96, 151.05, 142.13, 140.53, 130.35 (q, JCF = 32.9 Hz, 2C), 128.24 (d, JCF = 3.4 Hz, 2C), 127.47, 124.69, 123.28 (q, JCF = 272.8 Hz, 2C), 121.68–121.52 (m), 113.60, 113.52, 67.58, 28.98, 26.41, 21.17; HRMS (ESI+): m/z calcd for C20H18F6N4O [M + H]+ 445.1457, found 445.1458.
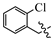
(E)-2-(6-((2-chlorobenzyl)oxy)-3,4-dihydro-1(2H)-naphthalenylidene)hydrazine-1-carboxamide(1L); Yield: 87.6%, 0.300 g; 1H NMR (600 MHz, DMSO-d6) δ 11.06 (s, 1H), 8.26 (d, J = 8.7 Hz, 1H), 8.1–7.6 (br s, 3H), 7.60–7.56 (m, 1H), 7.52–7.47 (m, 1H), 7.41–7.34 (m, 2H), 6.88–6.84 (m, 2H), 5.17 (s, 2H), 2.75–2.70 (m, 2H), 2.63 (t, J = 6.5 Hz, 2H), 1.85–1.79 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.65, 156.35, 151.59, 142.60, 134.54, 133.14, 130.74, 130.42, 129.85, 127.86, 127.83, 124.92, 114.10, 113.69, 67.31, 29.43, 26.84, 21.62; HRMS (ESI+): m/z calcd for C18H19ClN4O [M + H]+ 343.1320, found 343.1332.
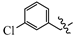
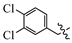
(E)-2-(6-((3-chlorobenzyl)oxy)-3,4-dihydro-1(2H)-naphthalenylidene)hydrazine-1-carboxamide(1M). Yield: 82.2%, 0.282 g; 1H NMR (600 MHz, DMSO-d6) δ 11.05 (s, 1H), 8.25 (d, J = 8.6 Hz, 1H), 8.13–7.52 (br s, 3H), 7.50 (s, 1H), 7.42–7.36 (m, 3H), 6.87–6.83 (m, 2H), 5.14 (s, 2H), 2.73–2.69 (m, 2H), 2.62 (t, J = 6.5 Hz, 2H), 1.85–1.78 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.55, 156.35, 151.58, 142.53, 139.93, 133.54, 130.82, 128.23, 127.83, 127.75, 126.63, 124.83, 114.13, 113.87, 68.69, 29.43, 26.82, 21.62. HRMS (ESI+): m/z calcd for C18H19ClN4O [M + H]+ 343.1320, found 343.1330.
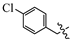
(E)-2-(6-((4-chlorobenzyl)oxy)-3,4-dihydro-1(2H)-naphthalenylidene)hydrazine-1-carboxamide(1N). Yield: 57.9%, 0.198 g; 1H NMR (600 MHz, DMSO-d6) δ 11.05 (s, 1H), 8.24 (d, J = 8.8 Hz, 1H), 8.11–7.50 (br s, 3H), 7.47–7.42 (m, 4H), 6.85–6.82 (m, 2H), 5.12 (s, 2H), 2.73–2.68 (m, 2H), 2.62 (t, J = 6.5 Hz, 2H), 1.85-1.78 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.63, 156.34, 151.59, 142.52, 136.38, 132.88, 129.95 (2C), 128.88 (2C), 127.81, 124.77, 114.16, 113.85, 68.79, 29.44, 26.82, 21.62; HRMS (ESI+): m/z calcd for C18H19ClN4O [M + H]+ 343.1320, found 343.1328.
(E)-2-(6-((2,4-dichlorobenzyl)oxy)-3,4-dihydro-1(2H)-naphthalenylidene)hydrazine-1-carboxamide(1O). Yield: 86.1%, 0.325 g; 1H NMR (600 MHz, DMSO-d6) δ 11.05 (s, 1H), 8.28–8.25 (m, 1H), 8.07–7.68 (br s, 3H), 7.67 (d, J = 2.1 Hz, 1H), 7.60 (d, J = 8.3 Hz, 1H), 7.46 (dd, J = 8.3, 2.1 Hz, 1H), 6.87–6.84 (m, 2H), 5.15 (s, 2H), 2.75–2.70 (m, 2H), 2.63 (t, J = 6.5 Hz, 2H), 1.85–1.79 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.46, 156.34, 151.56, 142.61, 134.10, 134.05, 133.79, 131.92, 129.38, 128.02, 127.87, 125.04, 114.10, 113.73, 66.72, 29.42, 26.82, 21.60; HRMS (ESI+): m/z calcd for C18H18Cl2N4O [M + H]+ 377.0930, found 377.0935.
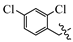
(E)-2-(6-((3,4-dichlorobenzyl)oxy)-3,4-dihydro-1(2H)-naphthalenylidene)hydrazine-1-carboxamide(1P). Yield: 80.3%, 0.303 g; 1H NMR (600 MHz, DMSO-d6) δ 11.04 (s, 1H), 8.25 (d, J = 8.7 Hz, 1H), 8.12–7.60 (br s,3H), 7.70 (d, J = 1.8 Hz, 1H), 7.64 (d, J = 8.3 Hz, 1H), 7.42 (dd, J = 8.3, 1.9 Hz, 1H), 6.85 (dt, J = 5.6, 2.4 Hz, 2H), 5.14 (s, 2H), 2.74–2.68 (m, 2H), 2.62 (t, J = 6.5 Hz, 2H), 1.85–1.78 (m, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.41, 156.33, 151.55, 142.55, 138.60, 131.53, 131.14, 130.83, 129.91, 128.27, 127.85, 124.92, 114.13, 113.89, 68.05, 29.42, 26.81, 21.60; HRMS (ESI+): m/z calcd for C18H18Cl2N4O [M + H]+ 377.0930, found 377.0937.
(E)-2-(6-((2-bromobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1Q). Yield: 80.2%, 0.312 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.30–8.28 (m, 1H), 8.08–7.72 (br s, 3H), 7.68 (dd, J = 8.1, 1.2 Hz, 1H), 7.59 (dd, J = 7.6, 1.7 Hz, 1H), 7.43 (td, J = 7.5, 1.2 Hz, 1H), 7.32 (td, J = 7.7, 1.8 Hz, 1H), 6.87 (t, J = 7.5 Hz, 2H), 5.15 (s, 2H), 2.75 (t, J = 6.0 Hz, 2H), 2.66 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.18, 155.96, 151.12, 142.14, 135.68, 132.65, 130.41, 130.22, 127.93, 127.42, 124.51, 122.95, 113.64, 113.25, 69.07, 28.98, 26.41, 21.17; HRMS (ESI+): m/z calcd for C18H19BrN4O [M + H]+ 387.0815, found 387.0810.
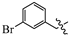
(E)-2-(6-((3-bromobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1R). Yield: 71.5%, 0.255 g; 1H NMR (600 MHz, DMSO-d6) δ 11.12 (s, 1H), 8.28 (d, J = 8.6 Hz, 1H), 8.00–7.69 (br s, 3H), 7.66 (t, J = 1.8 Hz, 1H), 7.53 (ddd, J = 8.0, 2.0, 1.0 Hz, 1H), 7.46 (dt, J = 7.6, 1.2 Hz, 1H), 7.36 (t, J = 7.8 Hz, 1H), 6.90–6.83 (m, 2H), 5.15 (s, 2H), 2.73 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.4 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.11, 155.95, 151.12, 142.08, 139.73, 130.69, 130.67, 130.19, 127.40, 126.58, 124.40, 121.69, 113.68, 113.43, 68.19, 28.99, 26.41, 21.17; HRMS (ESI+): m/z calcd for C18H19BrN4O [M + H]+ 387.0815, found 387.0806.
(E)-2-(6-((4-bromobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1S). Yield: 95.6%, 0.342 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.27 (d, J = 8.5 Hz, 1H), 7.98–7.68 (br s, 3H), 7.63–7.54 (m, 2H), 7.44–7.37 (m, 2H), 6.90–6.82 (m, 2H), 5.12 (s, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.16, 155.96, 151.13, 142.07, 136.36, 131.36(2C), 129.80(2C), 127.38, 124.35, 120.97, 113.71, 113.42, 68.37, 28.99, 26.42, 21.17; HRMS(ESI+): m/z calcd for C18H19BrN4O [M + H]+ 387.0815, found 387.0811.
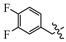
(E)-2-(6-((2-chloro-4-fluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1T). Yield: 99.8%, 0.360 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.33–8.23 (m, 1H), 8.03–7.70 (br s, 3H), 7.66 (dd, J = 8.7, 6.2 Hz, 1H), 7.52 (dd, J = 8.8, 2.6 Hz, 1H), 7.28 (td, J = 8.5, 2.6 Hz, 1H), 6.88 (t, J = 7.8 Hz, 2H), 5.15 (s, 2H), 2.74 (t, J = 6.0 Hz, 2H), 2.66 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 161.76 (d, JCF = 248.4 Hz), 159.14, 155.97, 151.11, 142.15, 133.83 (d, JCF = 10.6 Hz), 132.05 (d, JCF = 9.2 Hz), 130.61 (d, JCF = 3.4 Hz), 127.42, 124.53, 116.82 (d, JCF = 25.3 Hz), 114.53 (d, JCF = 21.0 Hz), 113.66, 113.25, 66.35, 28.99, 26.43, 21.17; HRMS (ESI+): m/z calcd for C18H18ClFN4O [M + H]+ 361.1225, found 361.1221.
(E)-2-(6-((3-chloro-4-fluorobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1U). Yield:70.7%, 0.255 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.28 (d, J = 8.7 Hz, 1H), 8.00–7.71 (br s, 3H), 7.68 (dd, J = 7.2, 2.1 Hz, 1H), 7.50–7.40 (m, 2H), 6.90–6.84 (m, 2H), 5.13 (s, 2H), 2.73 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.04, 156.76 (d, JCF = 246.7 Hz), 155.97, 151.09, 142.08, 134.87 (d, JCF = 3.8 Hz), 129.82, 128.47 (d, JCF = 7.5 Hz), 127.40, 124.45, 119.43 (d, JCF = 17.9 Hz), 116.95 (d, JCF = 20.9 Hz), 113.69, 113.42, 67.74, 28.99, 26.42, 21.18; HRMS(ESI+): m/z calcd for C18H18ClFN4O [M + H]+ 361.1225, found 361.1221.
(E)-2-(6-((4-cyanobenzyl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1V). Yield: 90.6%, 0.302 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.28 (d, J = 8.6 Hz, 1H), 8.05–7.70 (br s, 3H), 7.88–7.84 (m, 2H), 7.66–7.59 (m, 2H), 6.90–6.84 (m, 2H), 5.26 (s, 2H), 2.73 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 158.97, 155.97, 151.08, 142.72, 142.11, 132.42(2C), 128.06(2C), 127.43, 124.53, 118.73, 113.68, 113.45, 110.50, 68.22, 28.98, 26.41, 21.16; HRMS (ESI+): m/z calcd for C19H19N5O [M + H]+ 334.1662, found 334.1659.
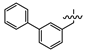
(E)-2-(6-([1,1’-biphenyl]-3-ylmethoxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1W). Yield: 96.2%, 0.370 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.28 (d, J = 8.5 Hz, 1H), 8.00–7.75 (br s, 3H), 7.75 (s, 1H), 7.68–7.65 (m, 2H), 7.62 (dt, J = 7.6, 1.6 Hz, 1H), 7.52–7.43 (m, 4H), 7.39–7.36 (m, 1H), 6.93–6.88 (m, 2H), 5.22 (s, 2H), 2.73 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.84 (p, J = 6.3 Hz, 2H); 13C NMR (151 MHz, DMSO-d6) δ 159.39, 155.96, 151.17, 142.07, 140.34, 139.90, 137.61, 129.11, 128.97(2C), 127.57, 127.38, 126.77, 126.71(2C), 126.23, 126.06, 124.26, 113.73, 113.46, 69.17, 29.01, 26.43, 21.19; HRMS (ESI+): m/z calcd for C24H24N4O [M + H]+ 385.2022, found 385.2016.
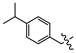
(E)-2-(6-(4-isopropylphenoxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(1X). Yield: 83.6%, 0.271 g; 1H NMR (600 MHz, DMSO-d6) δ 8.07 (d, J = 8.6 Hz, 1H), 7.35 (d, J = 7.6 Hz, 2H), 7.25 (d, J = 7.7 Hz, 2H), 6.81–6.70 (m, 2H), 5.78 (s, 2H), 5.32 (s, 2H), 5.03 (s, 2H), 2.88 (p, J = 6.9 Hz, 1H), 2.68 (dt, J = 13.4, 6.2 Hz, 4H), 1.73 (p, J = 6.2 Hz, 2H), 1.20 (d, J = 6.9 Hz, 6H); 13C NMR (151 MHz, DMSO-d6) δ 159.03, 157.71, 148.00, 147.76, 139.81, 134.56, 127.84(2C), 127.69, 126.29(2C), 125.41, 113.30, 113.21, 68.97, 33.18, 29.92, 26.25, 23.87(2C), 22.07; HRMS (ESI+): m/z calcd for C21H26N4O [M + H]+ 351.2179, found 351.2172.
(E)-2-(6-butoxy-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2A). Yield: 89.2%, 0.245 g; 1H NMR (600 MHz, DMSO-d6) δ 11.14 (s, 1H), 8.25 (d, J = 8.8 Hz, 1H), 7.95–7.63 (br s, 3H), 6.80–6.73 (m, 2H), 3.98 (t, J = 6.5 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H), 1.71–1.64 (m, 2H), 1.46–1.38 (m, 2H), 0.92 (t, J = 7.4 Hz, 3H); 13C NMR (151 MHz, DMSO-d6) δ 159.77, 155.98, 151.25, 142.06, 127.32, 123.86, 113.50, 112.80, 67.15, 30.70, 29.01, 26.47, 21.21, 18.69, 13.67; HRMS (ESI+): m/z calcd for C15H22N4O [M + H]+ 275.1866, found 275.1864.
(E)-2-(6-(pentyloxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2B). Yield: 78.7%, 0.267 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.24 (d, J = 8.8 Hz, 1H), 7.95–7.64 (br s, 3H), 6.80–6.71 (m, 2H), 3.97 (t, J = 6.5 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H), 1.74–1.66 (m, 2H), 1.42–1.30 (m, 4H), 0.89 (t, J = 7.1 Hz, 3H); 13C NMR (151 MHz, DMSO-d6) δ 159.76, 155.96, 151.25, 142.06, 127.32, 123.85, 113.48, 112.80, 67.44, 29.00, 28.32, 27.67, 26.46, 21.86, 21.21, 13.89; HRMS (ESI+): m/z calcd for C16H24N4O [M + H]+ 289.2022, found 289.2014.
(E)-2-(6-(hexyloxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2C). Yield: 83.7%, 0.253 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.24 (d, J = 8.7 Hz, 1H), 7.96–7.64 (br s, 3H), 6.79–6.72 (m, 2H), 3.97 (t, J = 6.5 Hz, 2H), 2.72 (t, J = 5.9 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H), 1.72–1.66 (m, 2H), 1.43–1.36 (m, 2H), 1.29 (h, J = 3.8 Hz, 4H), 0.91–0.82 (m, 3H); 13C NMR (151 MHz, DMSO-d6) δ 159.76, 155.96, 151.25, 142.06, 127.31, 123.85, 113.49, 112.80, 67.45, 30.97, 29.00, 28.60, 26.46, 25.15, 22.05, 21.21, 13.89; HRMS (ESI+): m/z calcd for C17H26N4O [M + H]+ 303.2179, found 303.2176.
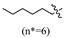
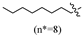
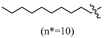
(E)-2-(6-(heptyloxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2D). Yield: 88.9%, 0.281 g; 1H NMR (600 MHz, DMSO-d6) δ 11.11 (s, 1H), 8.24 (d, J = 8.8 Hz, 1H), 7.94–7.62 (br s, 3H), 6.78–6.72 (m, 2H), 3.97 (t, J = 6.5 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.64 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H), 1.69 (p, J = 6.7 Hz, 2H), 1.43–1.35 (m, 2H), 1.34–1.20 (m, 6H), 0.86 (t, J = 6.9 Hz, 3H); 13C NMR (151 MHz, DMSO-d6) δ 159.77, 155.95, 151.27, 142.07, 127.32, 123.85, 113.50, 112.80, 67.45, 31.23, 29.01, 28.64, 28.43, 26.45, 25.46, 22.04, 21.22, 13.93; HRMS (ESI+): m/z calcd for C18H28N4O [M + H]+ 317.2335, found 317.2330.
(E)-2-(6-(octyloxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2E). Yield:75.0%, 0.248 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.24 (d, J = 8.8 Hz, 1H), 7.95–7.62 (br s, 3H), 6.78–6.72 (m, 2H), 3.97 (t, J = 6.5 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H), 1.69 (p, J = 6.7 Hz, 2H), 1.42–1.35 (m, 2H), 1.33–1.19 (m, 8H), 0.88–0.82 (m, 3H); 13C NMR (151 MHz, DMSO-d6) δ 159.76, 155.96, 151.24, 142.04, 127.31, 123.84, 113.49, 112.79, 67.44, 31.23, 29.01, 28.72, 28.65, 28.63, 26.46, 25.49, 22.07, 21.21, 13.93; HRMS (ESI+): m/z calcd for C19H30N4O [M + H]+ 331.2492, found 331.2483.
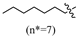
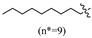
(E)-2-(6-(nonyloxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2F). Yield: 30.0%, 0.103 g; 1H NMR (600 MHz, DMSO-d6) δ 11.11 (s, 1H), 8.24 (d, J = 8.8 Hz, 1H), 7.94–7.60 (br s, 3H), 6.77 (dd, J = 8.8, 2.6 Hz, 1H), 6.74 (d, J = 2.6 Hz, 1H), 3.97 (t, J = 6.5 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.64 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.2 Hz, 2H), 1.69 (p, J = 6.7 Hz, 2H), 1.42–1.35 (m, 2H), 1.33–1.19 (m, 10H), 0.87–0.82 (m, 3H); 13C NMR (151 MHz, DMSO-d6) δ 159.77, 155.95, 151.25, 142.05, 127.32, 123.85, 113.49, 112.79, 67.44, 31.27, 29.01, 28.96, 28.76, 28.65, 28.63, 26.46, 25.48, 22.09, 21.22, 13.93; HRMS (ESI+): m/z calcd for C20H32N4O [M + H]+ 345.2648, found 345.2639.
(E)-2-(6-(decyloxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2G). Yield: 82.7%, 0.296 g; 1H NMR (600 MHz, DMSO-d6) δ 11.10 (s, 1H), 8.24 (d, J = 8.7 Hz, 1H), 7.94–7.63 (br s, 3H), 6.78–6.72 (m, 2H), 3.97 (t, J = 6.5 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.64 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H), 1.69 (p, J = 6.7 Hz, 2H), 1.42–1.35 (m, 2H), 1.33–1.19 (m, 12H), 0.84 (t, 3H); 13C NMR (151 MHz, DMSO-d6) δ 159.77, 155.95, 151.25, 142.05, 127.32, 123.85, 113.49, 112.79, 67.44, 31.29, 29.02, 29.00, 28.95, 28.75, 28.69, 28.63, 26.45, 25.48, 22.09, 21.22, 13.93; HRMS (ESI+): m/z calcd for C21H34N4O [M + H]+ 359.2805, found 359.2791.
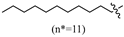
(E)-2-(6-(undecyloxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2H). Yield: 71.5%, 0.266 g; 1H NMR (600 MHz, DMSO-d6) δ 11.11 (s, 1H), 8.24 (d, J = 8.8 Hz, 1H), 7.95–7.63 (br s, 3H), 6.79–6.71 (m, 2H), 3.97 (t, J = 6.5 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.64 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.3 Hz, 2H), 1.69 (p, J = 6.7 Hz, 2H), 1.38 (td, J = 9.3, 8.5, 4.8 Hz, 2H), 1.33–1.18 (m, 14H), 0.84 (t, J = 7.0 Hz, 3H); 13C NMR (151 MHz, DMSO-d6) δ 159.77, 155.95, 151.24, 142.04, 127.32, 123.85, 113.49, 112.79, 67.44, 31.30, 29.02, 28.99 (3C), 28.75, 28.71, 28.63, 26.45, 25.48, 22.09, 21.22, 13.93; HRMS (ESI+): m/z calcd for C22H36N4O [M + H]+ 373.2961, found 373.2946.
(E)-2-(6-((3-methylbut-2-en-1-yl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2I). Yield: 22.8%, 0.065 g; 1H NMR (600 MHz, DMSO-d6) δ 11.13 (s, 1H), 8.24 (d, J = 8.6 Hz, 1H), 7.93–7.63 (br s, 3H), 6.80–6.75 (m, 2H), 5.42 (ddt, J = 6.7, 5.2, 1.4 Hz, 1H), 4.54 (d, J = 6.7 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.65 (t, J = 6.5 Hz, 2H), 1.83 (p, J = 6.4 Hz, 2H), 1.72 (dd, J = 19.9, 1.4 Hz, 6H); 13C NMR (151 MHz, DMSO-d6) δ 159.54, 155.96, 151.25, 142.02, 137.27, 127.28, 123.88, 119.74, 113.66, 113.01, 64.32, 29.03, 26.45, 25.41, 21.22, 18.01; HRMS (ESI+): m/z calcd for C16H22N4O [M + H]+ 287.1866, found 287.1859.
2-((E)-6-(((E)-3,7-dimethylocta-2,6-dien-1-yl)oxy)-3,4-dihydronaphthalen-1(2H)-ylidene)hydrazine-1-carboximidamide(2J). Yield: 86.0%, 0.305 g; 1H NMR (600 MHz, DMSO-d6) δ 8.05 (d, J = 8.7 Hz, 1H), 6.70 (dd, J = 8.7, 2.7 Hz, 1H), 6.66 (d, J = 2.7 Hz, 1H), 6.10–5.60 (br s, 2H), 5.56–5.30 (m, 3H), 5.07 (t, J = 6.9 Hz, 1H), 4.52 (d, J = 6.5 Hz, 2H), 2.68 (dt, J = 12.1, 6.1 Hz, 4H), 2.08 (q, J = 7.2 Hz, 2H), 2.03 (dd, J = 8.8, 6.2 Hz, 2H), 1.72 (d, J = 22.6 Hz, 5H), 1.63 (s, 3H), 1.56 (s, 3H); 13C NMR (151 MHz, DMSO-d6) δ 158.89, 157.83, 147.99, 139.94, 139.84, 131.00, 127.30, 125.43, 123.78, 119.88, 113.23, 113.03, 64.21, 38.94, 29.92, 26.26, 25.82, 25.46, 22.06, 17.55, 16.33; HRMS (ESI+): m/z calcd for C21H30N4O [M + H]+ 355.2492, found 355.2479.
3.2. Antibacterial Evaluation
3.2.1. MIC and MBC Testing
The minimum inhibitory concentration (MIC) of all synthesized compounds was tested by the broth microdilution method [42]. In brief, compounds dissolved in DMSO were diluted twice continuously in a 96-well plate with MH broth. The ESKAPE pathogens (E. coli ATCC 25922, S. aureus ATCC 29213, K. pneumoniae ATCC 700603, A. baumannii ATCC 19606, P. aeruginosa ATCC 27853, and E. faecium ATCC 35667), clinical S. aureus isolates (LMY45, LMY46, LMY47, and LMY48) and methicillin-resistant Staphylococcus aureus (MRSA-1 and MRSA-2) were cultured in MH broth to logarithmic growth phase and then diluted to 1 × 106 CFU/mL in MH broth. Subsequently, the diluted bacterial solution was mixed with the diluted compound solution in a 96-well plate and incubated at 37 °C for a duration of 18 h. The MIC of the compounds was recorded as the lowest concentration at which no visible growth was observed. Subsequently, the MBC values were also determined (only for the compounds with MIC values of ≤32 µg/mL). The levofloxacin or vancomycin was used as a positive control and DMSO (0.1%) was used as a negative control. Each concentration was replicated thrice.
3.2.2. Growth Curve and Bactericidal Time-Kill Kinetics Assay
The growth inhibition and time-dependent killing abilities of 2D were evaluated against S. aureus (ATCC 29213) and MRSA-2. After culturing the strains in MH broth until the logarithmic growth phase, they were diluted with MH broth to 1 × 106 CFU/mL. Subsequently, different final concentrations of 2D (0.5, 1, 2, 4, and 8 µg/mL) and vancomycin (0.5 and 1 µg/mL) were added to a diluted bacterial suspension and incubated at 37 °C, respectively. OD600 values were measured at predetermined time intervals (0, 1, 2, 4, 6, 8, 10, 12, and 24 h).
S. aureus (ATCC 29213) and MRSA-2 were cultivated in MH broth until the logarithmic growth phase and then diluted to 1 × 107 CFU/mL, respectively. Subsequently, different final concentrations of 2D (2, 4, and 8 µg/mL) and vancomycin (2 and 4 µg/mL) were added to a diluted bacterial suspension and incubated at 37 °C. After a specified time interval (0, 1, 2, 4, 8, 12, and 24 h), the culture suspension was serially diluted in 0.9% saline over a concentration range from 10−1 to 10−9. A volume of 100 μL of the obtained diluent and culture solution was dispensed onto a sterile MH agar plate. The plate was subsequently cultured at 37 °C for a duration of 24 h. The total number of colonies was enumerated and expressed as log10(CFU/mL).
3.2.3. Biofilm Formation Inhibition and Elimination Assay
The effect of 2D on the biofilm formation of MRSA was evaluated. MRSA-2 was cultured in MH broth for a period of 3–5 h, after which it was diluted to a density of 1 × 106 CFU/mL with MH broth. In 96-well plates, different concentrations of 2D and vancomycin were mixed with bacterial suspension and subsequently incubated at 37 °C for 24 h. After the removal of the suspended bacteria, the well plates were washed thrice with PBS. Then, 150 μL of methanol was added and fixed for 30 min. Subsequent to air-drying, 150 μL of a 0.1% crystal violet aqueous solution was added. Following a 15 min staining period, the excess crystal violet was removed by rinsing with flowing water. Finally, 150 μL of anhydrous ethanol was added and the mixture was incubated for 20 min. The optical density at 575 nm was detected.
The ability of 2D to eliminate biofilm was evaluated. MRSA-2 (1 × 106 CFU/mL) was incubated in 96-well plates for 24 h until biofilm formation. The planktonic bacteria were discarded and washed thrice with PBS. Subsequently, various concentrations of 2D and vancomycin were added and cultivated at 37 °C for 24 h. According to the above method, the OD575 nm value was detected after the following steps: PBS washing, methanol fixation, crystal violet staining, and ethanol incubation.
3.3. Mode of Action Study
3.3.1. Scanning Electron Microscopy (SEM) Analysis
MRSA-2 was cultivated to reach the logarithmic growth phase, washed thrice with PBS (pH = 7.2–7.4), and then resuspended to a bacterial concentration of 1×108 CFU/mL. The bacterial suspension was treated with 2D (2, 4, and 8 μg/mL) and vancomycin (4 μg/mL), followed by incubation at 37 °C for 3 h. Subsequently, the bacterial precipitate was collected via centrifugation and washed. The bacterial precipitate was incubated in 2.5% glutaraldehyde fixative at room temperature for 2 h, and then transferred to 4 °C overnight in the dark. Then, the samples were processed according to the following steps: washed with PBS, fixed with 1% osmium tetraoxide, and dehydrated with ethanol. After spraying with gold, the bacteria were observed by electron microscope.
3.3.2. Membrane Depolarization Assay
MRSA-2 was cultivated in MH broth, with a final concentration of 1 × 108 CFU/mL. After centrifugation, the supernatant was discarded. The bacterial precipitate was washed thrice and resuspended with PBS. Subsequently, the bacterial resuspension (180 μL) and the DiSC5 (10 μL, 3 μM) were incubated in the dark for 30 min. The fluorescence intensity was measured at 2 min intervals for a period of 10 min. After the fluorescence intensity was stabilized, 10 μL of 2D (final concentrations of 1, 2, 4, and 8 μg/mL) was added, respectively. The fluorescence intensity was measured at 2 min intervals for a total duration of 30 min. The excitation/emission wavelength was 622/670 nm.
3.3.3. Fluorescence Microscopy Assay
MRSA-2 was cultivated to the logarithmic growth phase, then washed with PBS and resuspended in PBS until the concentration of bacterial suspension reached 1 × 108 CFU/mL. The compound 2D (final concentration of 4 and 8 μg/mL) and vancomycin (4 μg/mL) were mixed with the bacterial suspension and incubated at 37 °C for 2 h. Thereafter, the samples were incubated with DAPI (10 μM) and PI (10 μM) in the dark for 15 min. The bacterial cells were then subjected to observation under a fluorescence microscope. DMSO served as the negative controls.
3.3.4. Inner Membrane Permeabilization Assay
MRSA-2 bacterial precipitates in the logarithmic growth phase were collected by centrifugation, washed, and resuspended in PBS to a density of 1 × 106 CFU/mL. PI (10 μM, 40 μL) was added to a black 96-well plate along with 150 μL of bacterial suspension. After an incubation period of 30 min, fluorescence was detected at excitation wavelengths of 535 nm and emission wavelengths of 617 nm at 5 min intervals until a stable state was reached. Various concentrations of 2D and vancomycin were then added and the fluorescence intensity was immediately detected every 5 min.
3.3.5. Bacterial DNA and Protein Leakage Experiment
MRSA-2 was cultured to logarithmic growth phase. The bacterial precipitate was collected, washed twice with PBS (pH = 7.2–7.4), and resuspended in PBS to 1 × 107 CFU/mL. 2D (1, 2, 4, and 8 μg/mL) and vancomycin (1 μg/mL) were subsequently added and incubated for 4 h at 37 °C. After centrifugation of 1 mL of the suspension, the supernatant was collected. The protein level was measured by BCA protein concentration kits (Servicebio, Hubei, China), and the DNA level was detected by microspectrophotometer.
3.3.6. Predicted Binding Mode of 2D in DHFR
Preliminary docking was performed to evaluate whether binding to DHFR might account for the bactericidal effect of the compound 2D. All docking studies were carried out using Discovery Studio 2019 Client (Accelrys, San Diego, CA, USA). The crystal structure data were obtained from the protein data bank (https://www.rcsb.org/structure/6P9Z (accessed on 6 May 2025)). Enzyme structures were checked for missing atoms, bonds, and contacts. Hydrogen atoms were added to the enzyme structure. Water molecules and bound ligands were manually deleted. The structures of compounds were sketched in 2D (2 Dimensions) and converted into 3D (3 Dimensions) using the DS molecule editor. Automated docking studies were carried out to investigate the binding mode of 2D utilizing DS-CDOCKER protocol. The pose with the top-CDOCKER_ENERGY was chosen for analyzing the binding features of 2D with DHFR. Dihydrofolate reductase inhibitor trimethoprim was used as a control.
3.4. Safety Assay
The sterile defibrillated sheep blood cells were centrifuged (5000 rpm, 5 min) and then washed three times with 0.9% saline. Subsequently, 1 μL of different concentrations of 2D (0.5–128 μg/mL) was mixed with 999 μL of red blood cell suspension. After incubation at 37 °C for 1 h, the mixed solution was centrifuged. The absorbance of the supernatant at 570 nm was measured. Triton X-100 (0.1%) was used as a positive control, and DMSO was used as a negative control. The hemolysis rate was calculated according to the following formula:
Hemolysis rate (%) = (ODTreatment − ODNegative)/(ODPositive − ODNegative) × 100%
The CCK-8 method was used to evaluate the toxicity of 2D to four cell lines (lung cancer cell line (A549), human liver cell line (HepG2), human embryonic kidney 293T cells (HEK 293-T), and human colon adenocarcinoma cell line (Caco-2)). Cells with a density of 1 × 105 cells/well were added to 96-well plates and then incubated for 24 h at 5% CO2 and 37 °C. Subsequently, after treatment with different concentrations of 2D for 24 h, the CCK-8 kit was added according to the prescribed procedure. Following an incubation at 37 °C for 0.5 h, the absorbances at 450 nm were detected. The hemolysis rate was calculated according to the following formula:
Cytotoxic (%) = (Atest/Acontrol) × 100%
3.5. In Vivo Infection Model
The MRSA-induced mouse skin abscess model was used to assess the in vivo antibacterial effects of 2D. Female KM mice, aged 4–6 weeks and weighing 20 ± 2 g, were purchased from the Laboratory Animal Center, Lanzhou Institute of Veterinary Medicine, Chinese Academy of Agricultural Sciences. The animal experiments were approved by the Ethics Committee of Research Involving Animals of the Lanzhou Institute of Husbandry and Pharmaceutical Science of CAAS (Approval number: 2025-03) and performed in accordance with the guidelines of the Ethics Committee for Animal Experiments. Prior to the formal experiment, all mice were acclimated for 5 days. The mice were kept in a pathogen-free environment with a 12 h light/12 h dark cycle, 50% ± 10% humidity, and a temperature of 24 °C ± 2 °C. After removing the dorsal hair, the mice were randomly divided into six groups, with six mice in each group: a control group with no MRSA infection and no treatment, a model group with MRSA infection only, a solvent group with MRSA infection and solvent (75% normal saline, 10% DMSO, 5% Tween-80, 10% PEG 300), a vancomycin group with 5 mg/kg doses, and a 2D group with 5 mg/kg and 10 mg/kg doses. The depilation method employed was as follows: depilatory paste was applied to the designated area for 10 min, after which it was thoroughly washed with 0.9% saline. The mice were semi-anesthetized by intraperitoneal injection with sodium Ulatan (TCI, 20%, 100 μL). Subcutaneous abscesses were induced by injecting 60 μL of MRSA-2 bacterial suspension (1 × 108 CFU/mL) into the back of anesthetized mice, followed by an injection of 60 μL of 2D, vancomycin, or solvent 2 h postinfection. After a 24 h interval, the administration was repeated, and 24 h after the last administration, the volume of abscess was measured and calculated. Then, the skin of the infected site and main organs were collected for tissue bacterial load and pathological detection.
3.6. Statistical Analysis
The results of all experiments are presented as the mean ± SD. Statistical significance was analyzed by GraphPad Prism 9 software (GraphPad Software, San Diego, CA, USA) with unpaired Student’s t-test or non-parametric one-way ANOVA. Significant values are represented by an asterisk: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
4. Conclusions
A total of 34 novel aminoguanidine indole derivatives were designed, synthesized, and identified. The synthesized compounds showed better activity against Gram-positive bacteria than Gram-negative bacteria. Compounds 1L, 1M, 1S, 1U, 1X, 2A, 2D–2F, and 2I demonstrated strong antibacterial activity against S. aureus, including MRSA, with MIC values ranging from 0.5 to 4 μg/mL. Compound 2D, with the most antibacterial activity, demonstrated a favorable anti-MRSA effect in vivo, similar to that of vancomycin. The main antibacterial mechanisms of 2D may be related to the disruption of the membrane integrity and inhibition of dihydrofolate reductase. Furthermore, the hemolytic activity and cytotoxicity of 2D were acceptable. Therefore, compound 2D, as an aminoguanidine tetralone derivative, may be a promising candidate for a novel broad-spectrum antibacterial agent.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms26135980/s1.
Author Contributions
X.-W.L. and J.-Y.L. conceived of and proposed the idea, and designed the study. Q.-J.Z. and Y.-X.L. synthesized and identified the compounds. Q.-J.Z. and Y.-X.L. evaluated the biological activities. Q.-J.Z., Y.-X.L., W.-B.G., L.-X.B., X.X. and Y.-J.Y. performed the acquisition and analysis of data. Y.-X.L., Q.-J.Z., X.-W.L. and J.-Y.L. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from the National Key R&D Program of China (2021YFD1800900), and Science-Technology Innovation Engineering of CAAS (25-LZIHPS-02).
Institutional Review Board Statement
Animal experiments were approved by the guidelines of the Animal Experimental Ethical Com-mittee of Lanzhou Institute of Husbandry and Pharmaceutical Science of CAAS (2025–03).
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusion of this study will be made available by the authors.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
MRSA, methicillin-resistant Staphylococcus aureus; HRMS, high-resolution mass spectrometry; NMR, nuclear magnetic resonance; MH, Mueller−Hinton; MIC, minimum inhibitory concentration; DMSO, dimethyl sulfoxide; CFU, colony forming units; PBS, phosphate-buffered saline; DAPI, 4′,6-diamidino-2-phenylindole; PI, propidium iodide; DiSC3(5), 3,3′-dipropylthiadicarbocyanine iodide; CCK-8, cell counting kit-8; H&E, hematoxylin and eosin; RBCs, red blood cells; SEM, scanning electron microscopy; SI, selectivity index; TLC, thin-layer chromatography.
References
- Okeke, I.N.; de Kraker, M.E.A.; Van Boeckel, T.P.; Kumar, C.K.; Schmitt, H.; Gales, A.C.; Bertagnolio, S.; Sharland, M.; Laxminarayan, R. The Scope of the Antimicrobial Resistance Challenge. Lancet 2024, 403, 2426–2438. [Google Scholar] [CrossRef]
- Venkateswaran, P.; Vasudevan, S.; David, H.; Shaktivel, A.; Shanmugam, K.; Neelakantan, P.; Solomon, A.P. Revisiting ESKAPE Pathogens: Virulence, Resistance, and Combating Strategies Focusing on Quorum Sensing. Front. Cell. Infect. Microbiol. 2023, 13, 1159798. [Google Scholar] [CrossRef]
- Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. Available online: https://www.who.int/publications/i/item/WHO-EMP-IAU-2017.12 (accessed on 24 October 2024).
- WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 1 November 2024).
- Ahmad-Mansour, N.; Loubet, P.; Pouget, C.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.-P.; Molle, V. Staphylococcus aureus Toxins: An Update on Their Pathogenic Properties and Potential Treatments. Toxins 2021, 13, 677. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-Resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Jin, Y.; Chen, P.; Ge, Q.; Dong, X.; Chen, Y.; Jiang, M.; Xiao, Y. Reshaping the Battlefield: A Decade of Clonal Wars among Staphylococcus aureus in China. Drug Resist. Updates 2025, 78, 101178. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-H.; Tang, H.-J.; Chen, C.-C.; Chen, C.-J. Clinical Characteristics and Genomic Changes of Recurrent Methicillin-Resistant Staphylococcus aureus Bacteremia. J. Microbiol. Immunol. Infect. 2024, 58, 251–257. [Google Scholar] [CrossRef]
- Sheng, K.; Song, Y.; Lei, F.; Zhao, W.; Fan, L.; Wu, L.; Liu, Y.; Wu, S.; Zhang, Y. Research Progress in Pharmacological Activities and Structure-Activity Relationships of Tetralone Scaffolds as Pharmacophore and Fluorescent Skeleton. Eur. J. Med. Chem. 2022, 227, 113964. [Google Scholar] [CrossRef]
- Chakraborty, S.; Baindara, P.; Mondal, S.K.; Roy, D.; Mandal, S.M. Synthesis of a Tetralone Derivative of Ampicillin to Control Ampicillin-Resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2024, 714, 149974. [Google Scholar] [CrossRef]
- Dwivedi, G.R.; Upadhyay, H.C.; Yadav, D.K.; Singh, V.; Srivastava, S.K.; Khan, F.; Darmwal, N.S.; Darokar, M.P. 4-Hydroxy-α-Tetralone and Its Derivative as Drug Resistance Reversal Agents in Multi Drug Resistant Escherichia coli. Chem. Biol. Drug Des. 2014, 83, 482–492. [Google Scholar] [CrossRef]
- Manvar, D.; Fernandes, T.d.A.; Domingos, J.L.O.; Baljinnyam, E.; Basu, A.; Junior, E.F.T.; Costa, P.R.R.; Kaushik-Basu, N. Synthesis and Biological Evaluation of α-Aryl-α-Tetralone Derivatives as Hepatitis C Virus Inhibitors. Eur. J. Med. Chem. 2015, 93, 51–54. [Google Scholar] [CrossRef]
- Fernandes, T.d.A.; Manvar, D.; Domingos, J.L.O.; Basu, A.; Nichols, D.B.; Kaushik-Basu, N.; Costa, P.R.R. 5-Carba-Pterocarpens: A New Scaffold with Anti-HCV Activity. Eur. J. Med. Chem. 2016, 112, 33–38. [Google Scholar] [CrossRef]
- Meddeb, A.; Thebti, A.; Elleuch, H.; Ayari, S.; Bouslama, L.; Ouzari, H.-I. Regioselective Oxidation of Tetrahydronaphthalenes to α-Tetralone Derivatives Using DDQ as Oxidizing Agent: Synthesis and Evaluation of Antibacterial and Antifungal Activities. ACS Omega 2024, 9, 39344–39352. [Google Scholar] [CrossRef] [PubMed]
- Gauni, B.; Mehariya, K.; Shah, A.; Duggirala, S.M. Tetralone Scaffolds and Their Potential Therapeutic Applications. Lett. Drug Des. Discov. 2020, 18, 222–238. [Google Scholar] [CrossRef]
- Castro, R.A.D.; Borrell, S.; Gagneux, S. The Within-Host Evolution of Antimicrobial Resistance in Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2021, 45, fuaa071. [Google Scholar] [CrossRef] [PubMed]
- Saxena, D.; Maitra, R.; Bormon, R.; Czekanska, M.; Meiers, J.; Titz, A.; Verma, S.; Chopra, S. Tackling the Outer Membrane: Facilitating Compound Entry Into Gram-Negative Bacterial Pathogens. npj Antimicrob. Resist. 2023, 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Perlmutter, S.J.; Geddes, E.J.; Drown, B.S.; Motika, S.E.; Lee, M.R.; Hergenrother, P.J. Compound Uptake into E. coli Can Be Facilitated by N-Alkyl Guanidiniums and Pyridiniums. ACS Infect. Dis. 2021, 7, 162–173. [Google Scholar] [CrossRef]
- Mohammad, H.; Younis, W.; Chen, L.; Peters, C.E.; Pogliano, J.; Pogliano, K.; Cooper, B.; Zhang, J.; Mayhoub, A.; Oldfield, E.; et al. Phenylthiazole Antibacterial Agents Targeting Cell Wall Synthesis Exhibit Potent Activity In Vitro and In Vivo against Vancomycin-Resistant Enterococci. J. Med. Chem. 2017, 60, 2425–2438. [Google Scholar] [CrossRef]
- Dantas, N.; de Aquino, T.M.; de Araújo-Júnior, J.X.; da Silva-Júnior, E.; Gomes, E.A.; Gomes, A.A.S.; Siqueira-Júnior, J.P.; Mendonça Junior, F.J.B. Aminoguanidine Hydrazones (AGH’s) as Modulators of Norfloxacin Resistance in Staphylococcus aureus that Overexpress NorA Efflux Pump. Chem. Biol. Interact. 2018, 280, 8–14. [Google Scholar] [CrossRef]
- Huang, Y.; Hu, H.; Yan, R.; Lin, L.; Song, M.; Yao, X. Synthesis and Evaluation of Antimicrobial and Anticancer Activities of 3-Phenyl-1-Phenylsulfonyl Pyrazoles Containing an Aminoguanidine Moiety. Arch. Pharm. 2021, 354, 2000165. [Google Scholar] [CrossRef]
- Deng, X.; Song, M. Synthesis, Antibacterial and Anticancer Activity, and Docking Study of Aminoguanidines Containing an Alkynyl Moiety. J. Enzyme Inhib. Med. Chem. 2019, 35, 354–364. [Google Scholar] [CrossRef]
- Angula, K.T.; Legoabe, L.J.; Jordaan, A.; Warner, D.F.; Beteck, R.M. Investigation of Quinolone-Tethered Aminoguanidine as Novel Antibacterial Agents. Arch. Pharm. 2022, 355, 2200172. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Wang, J.; Jiao, F.; Zhang, H.; Zhang, T. Synthesis and Biological Evaluation of Novel Aminoguanidine Derivatives as Potential Antibacterial Agents. Sci. Rep. 2024, 14, 26896. [Google Scholar] [CrossRef]
- Elsebaie, M.M.; Nour El-Din, H.T.; Abutaleb, N.S.; Abuelkhir, A.A.; Liang, H.-W.; Attia, A.S.; Seleem, M.N.; Mayhoub, A.S. Exploring the Structure-Activity Relationships of Diphenylurea as an Antibacterial Scaffold Active Against Methicillin- and Vancomycin-Resistant Staphylococcus aureus. Eur. J. Med. Chem. 2022, 234, 114204. [Google Scholar] [CrossRef]
- Noreddin, A.M.; Elkhatib, W.F. Levofloxacin in the Treatment of Community-Acquired Pneumonia. Expert Rev. Anti Infect. Ther. 2010, 8, 505–514. [Google Scholar] [CrossRef]
- Yang, P.; Liu, H.-Z.; Wang, Y.-S.; Qi, H.; Wang, L.-L.; Wang, B.-B.; Xie, X.-B. Synthesis and Structure–Activity Relationship of Novel Thiazole Aminoguanidines Against MRSA and Escherichia coli. RSC Med. Chem. 2025, 15, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Dohle, W.; Su, X.; Nigam, Y.; Dudley, E.; Potter, B.V.L. Synthesis and In Vitro Antimicrobial SAR of Benzyl and Phenyl Guanidine and Aminoguanidine Hydrazone Derivatives. Molecules 2022, 28, 5. [Google Scholar] [CrossRef] [PubMed]
- García Aragonés, L.; Blanch Sancho, J.J.; Segura Luque, J.C.; Mateos Rodriguez, F.; Martínez Alfaro, E.; Solís García del Pozo, J. What Do Beta-Lactams Add to Vancomycin or Daptomycin in the Treatment of Patients with Methicillin-Resistant Staphylococcus aureus Bacteraemia? A Review. Postgrad. Med. J. 2022, 98, 48–56. [Google Scholar] [CrossRef]
- Guo, H.; Tong, Y.; Cheng, J.; Abbas, Z.; Li, Z.; Wang, J.; Zhou, Y.; Si, D.; Zhang, R. Biofilm and Small Colony Variants—An Update on Staphylococcus aureus Strategies toward Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1241. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Wozniak, D.J.; Stoodley, P.; Hall-Stoodley, L. Prevention and Treatment of Staphylococcus aureus Biofilms. Expert Rev. Anti Infect. Ther. 2015, 13, 1499–1516. [Google Scholar] [CrossRef]
- Post, V.; Wahl, P.; Richards, R.G.; Moriarty, T.F. Vancomycin Displays Time-Dependent Eradication of Mature Staphylococcus aureus Biofilms. J. Orthop. Res. 2017, 35, 381–388. [Google Scholar] [CrossRef]
- Gomes, A.R.; Varela, C.L.; Pires, A.S.; Tavares-da-Silva, E.J.; Roleira, F.M.F. Synthetic and Natural Guanidine Derivatives as Antitumor and Antimicrobial Agents: A Review. Bioorg. Chem. 2023, 138, 106600. [Google Scholar] [CrossRef]
- Weiss, A.; Delavenne, E.; Matias, C.; Lagler, H.; Simon, D.; Li, P.; Hansen, J.U.; dos Santos, T.P.; Jana, B.; Priemel, P.; et al. Topical Niclosamide (ATx201) Reduces Staphylococcus aureus Colonization and Increases Shannon Diversity of the Skin Microbiome in Atopic Dermatitis Patients in a Randomized, Double-blind, Placebo-controlled Phase 2 Trial. Clin. Transl. Med. 2022, 12, e790. [Google Scholar] [CrossRef]
- Zhang, E.; Bai, P.-Y.; Cui, D.-Y.; Chu, W.-C.; Hua, Y.-G.; Liu, Q.; Yin, H.-Y.; Zhang, Y.-J.; Qin, S.; Liu, H.-M. Synthesis and Bioactivities Study of New Antibacterial Peptide Mimics: The Dialkyl Cationic Amphiphiles. Eur. J. Med. Chem. 2018, 143, 1489–1509. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Zhang, F.; Yao, J.; Zhu, Y.; Zhu, N.; Zhang, Y.; Liu, H.; Gou, S.; Ni, J. Antimicrobial Peptides with Symmetric Structures Against Multidrug-Resistant Bacteria While Alleviating Antimicrobial Resistance. Biochem. Pharmacol. 2021, 186, 114470. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Semenya, D.; Castagnolo, D. Antimicrobial Drugs Bearing Guanidine Moieties: A Review. Eur. J. Med. Chem. 2021, 216, 113293. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.-J.; Li, M.-Y.; Jin, X.-J.; Zheng, C.-J.; Piao, H.-R. Design, Synthesis and Evaluation of Carbazole Derivatives as Potential Antimicrobial Agents. J. Enzyme Inhib. Med. Chem. 2021, 36, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-X.; Geng, X.; Tao, Q.; Hao, R.-C.; Yang, Y.-J.; Liu, X.-W.; Li, J.-Y. Synthesis, Antimicrobial Activities, and Model of Action of Indolyl Derivatives Containing Amino-Guanidinium Moieties. Molecules 2025, 30, 887. [Google Scholar] [CrossRef]
- Srinivasan, B.; Tonddast-Navaei, S.; Roy, A.; Zhou, H.; Skolnick, J. Chemical Space of Escherichia coli Dihydrofolate Reductase Inhibitors: New Approaches for Discovering Novel Drugs for Old Bugs. Med. Res. Rev. 2019, 39, 684–705. [Google Scholar] [CrossRef]
- Wang, B.; Pachaiyappan, B.; Gruber, J.D.; Schmidt, M.G.; Zhang, Y.-M.; Woster, P.M. Antibacterial Diamines Targeting Bacterial Membranes. J. Med. Chem. 2016, 59, 3140–3151. [Google Scholar] [CrossRef]
- Kadeřábková, N.; Mahmood, A.J.S.; Mavridou, D.A.I. Antibiotic Susceptibility Testing Using Minimum Inhibitory Concentration (MIC) Assays. npj Antimicrob. Resist. 2024, 2, 37. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).