Comparison Bioinformatic Analysis of Extracellular Vesicles-Related Genes and MicroRNAs in Breast Cancer
Abstract
1. Introduction
2. Results
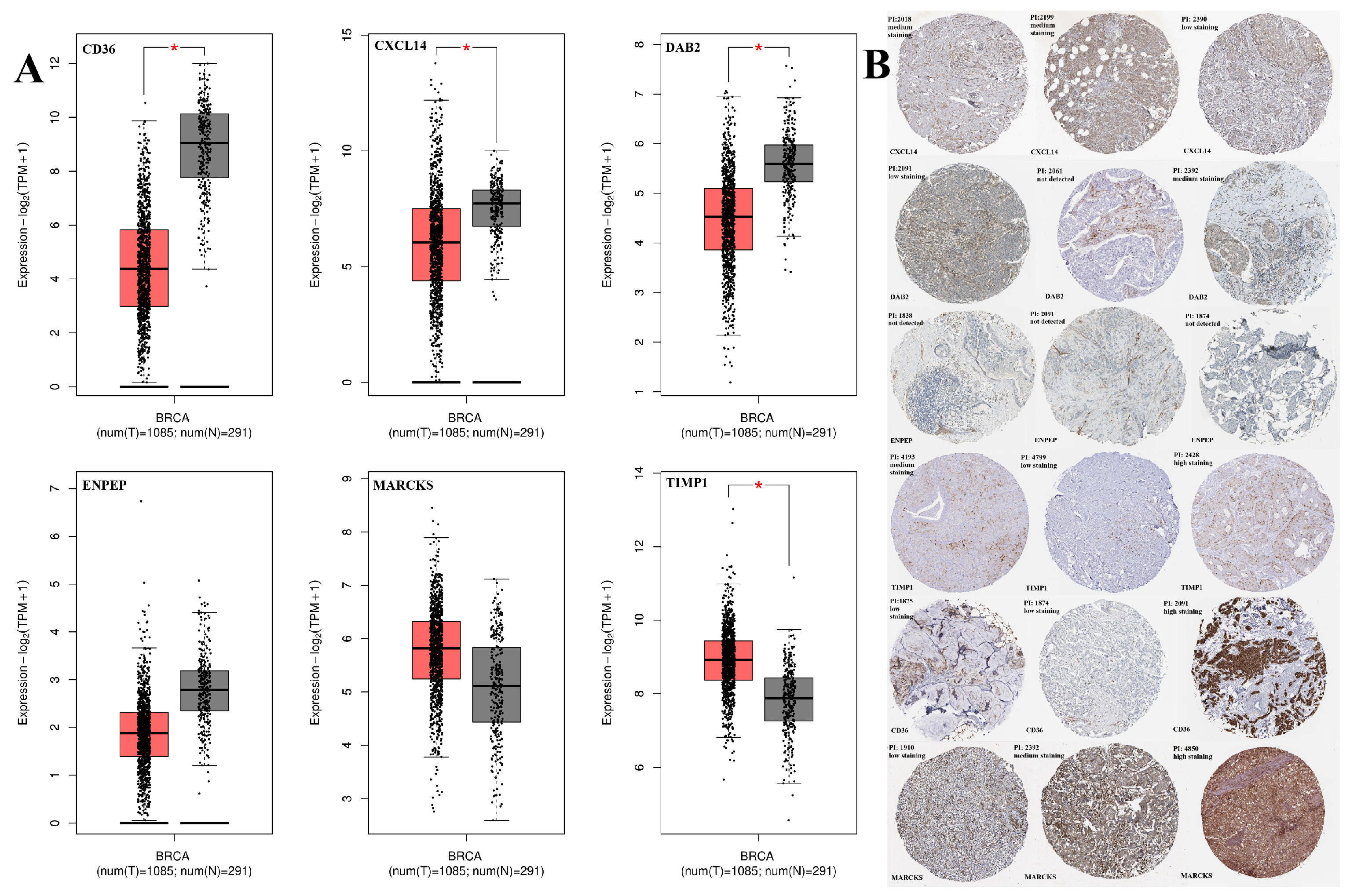
2.1. Expression Profile Results
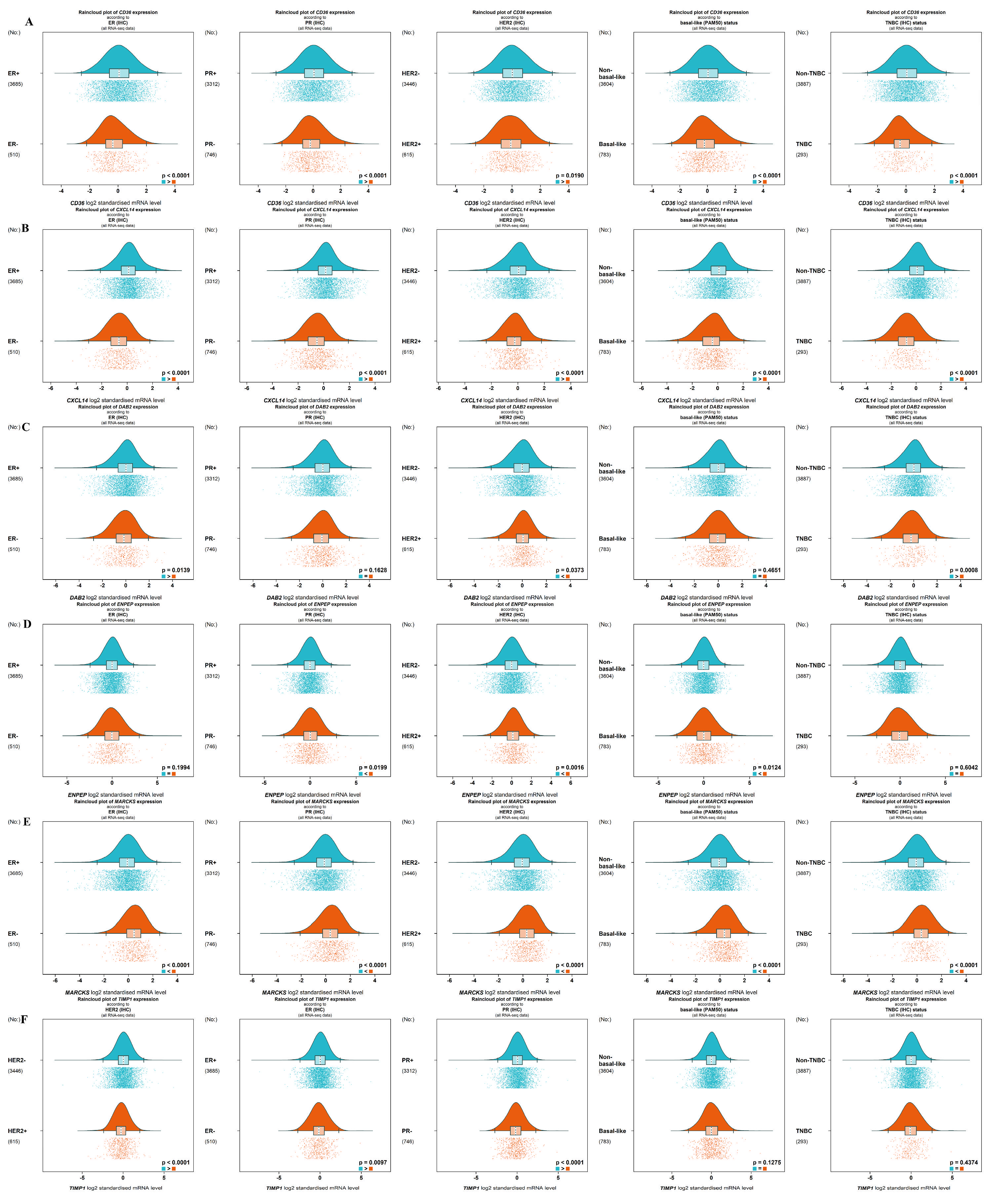
2.2. Gene Correlation and Targeted Expression Analysis Results
2.3. The Promotor Methylation Analysis
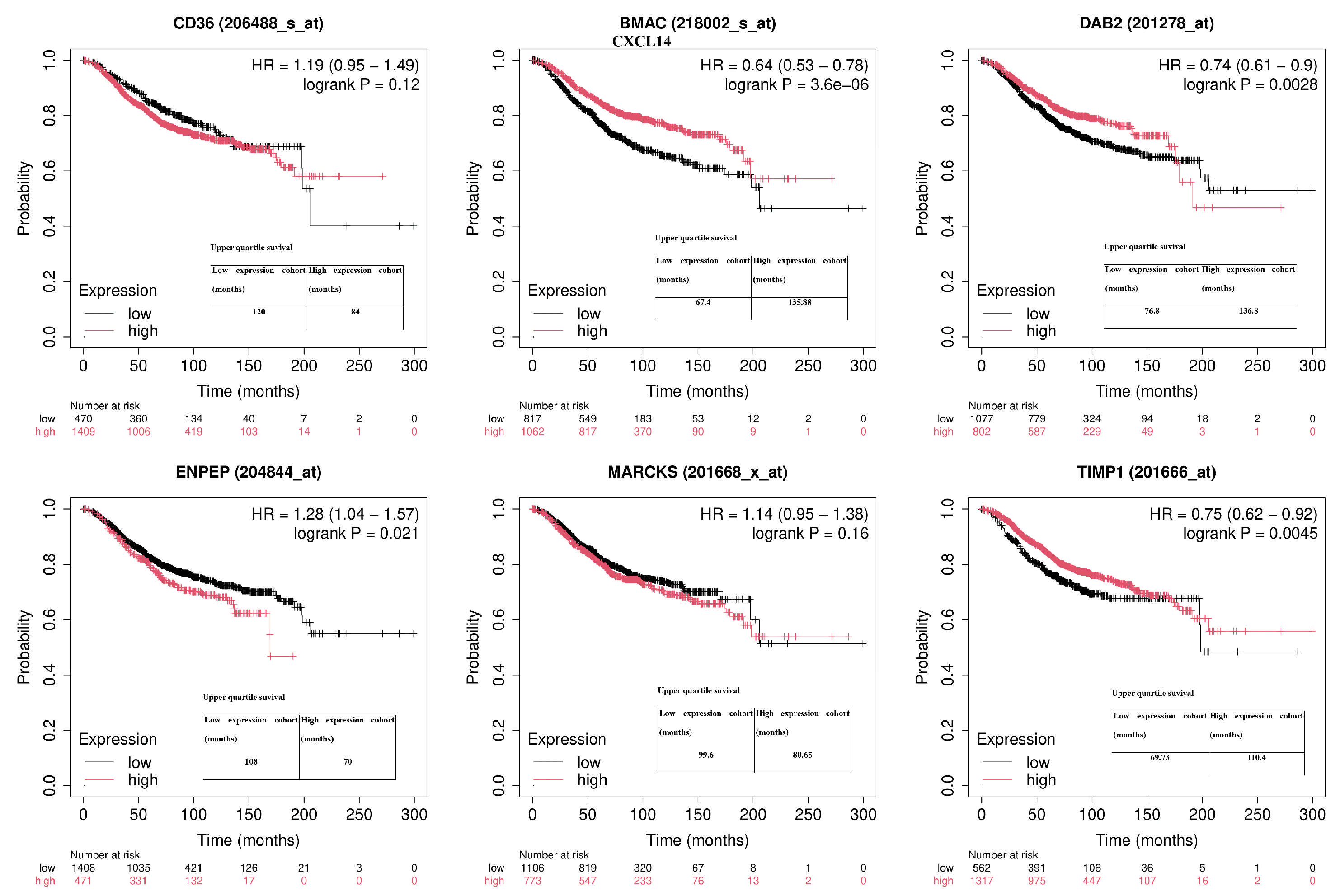
2.4. The Overall Survival Status
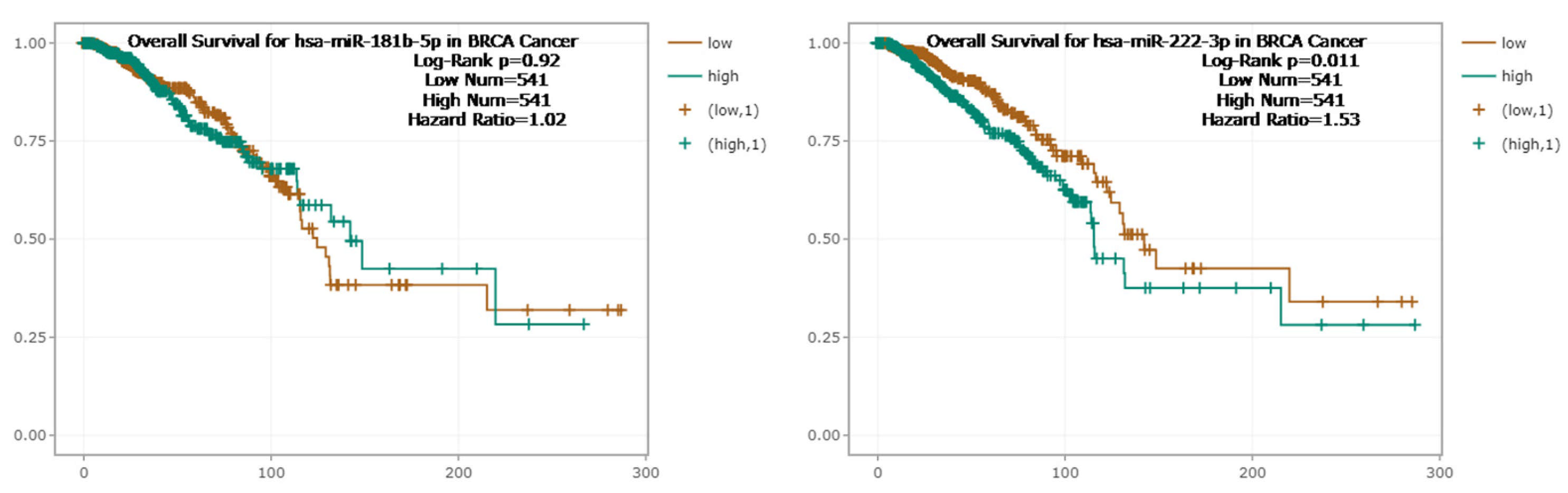
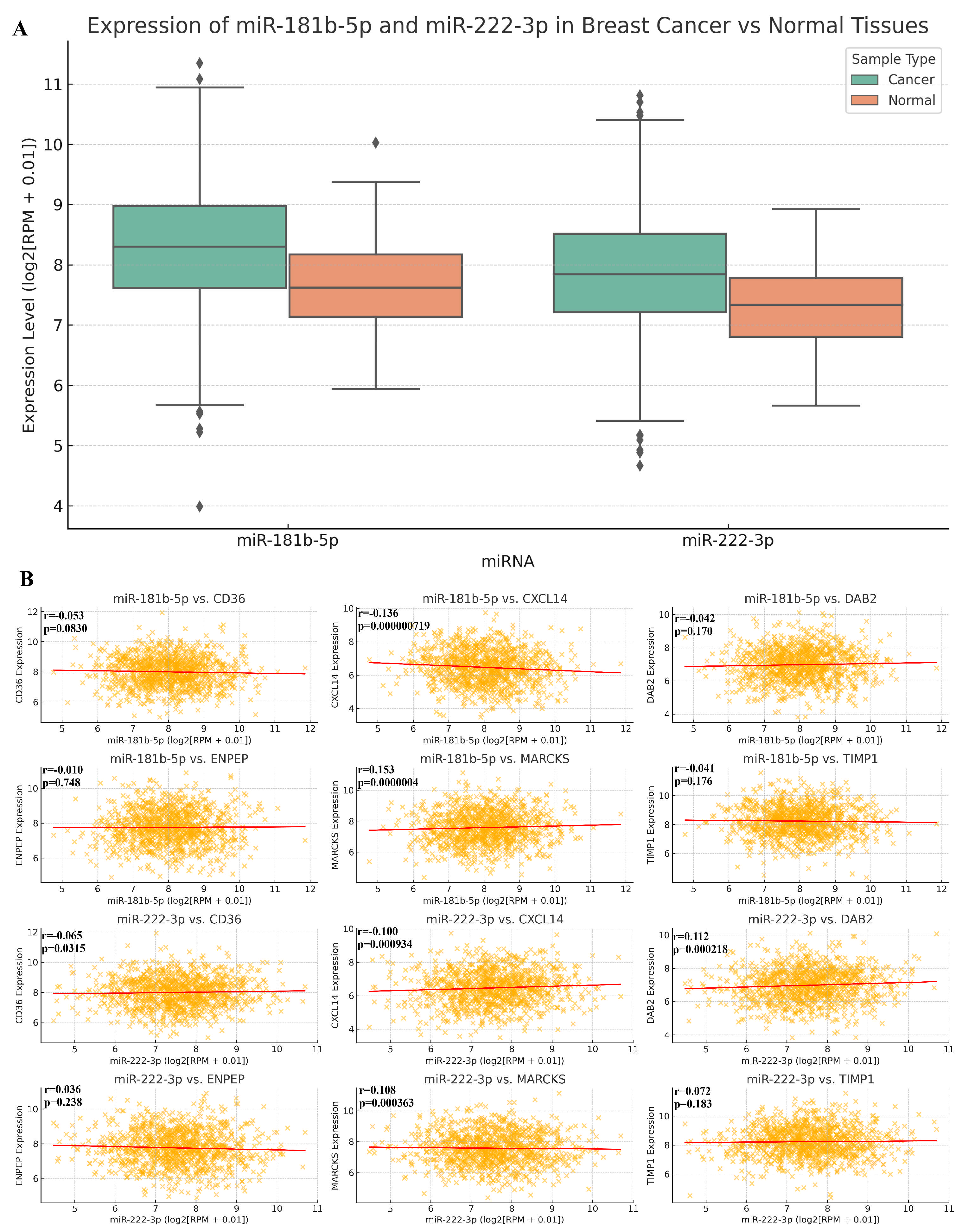
2.5. The Relationship Between MicroRNA vs. BC and MicroRNA vs. RNA
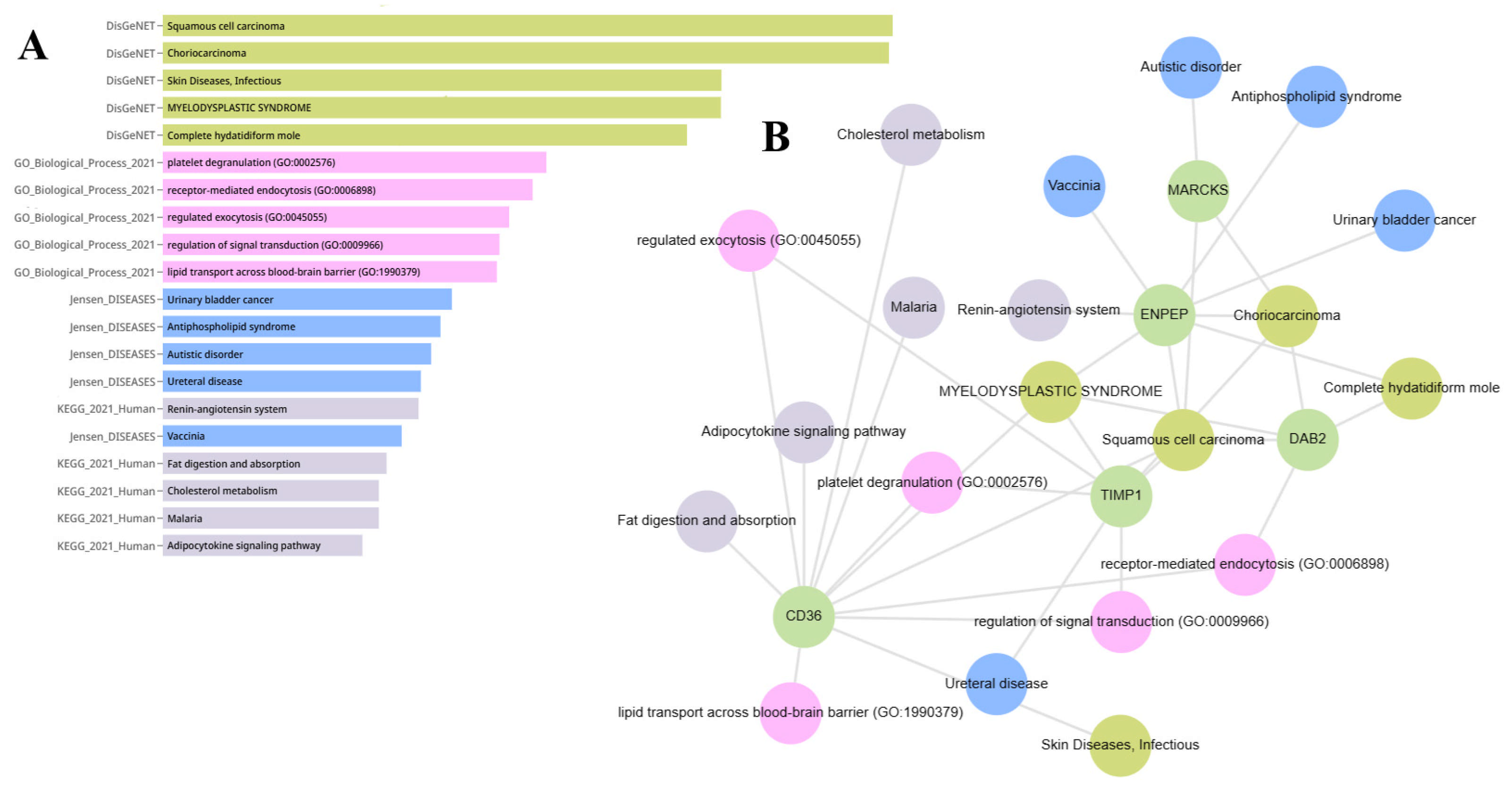
2.6. Gene Set Enrichment Analysis of ENPEP, TIMP1, CD36, MARCKS, DAB2, and CXCL14
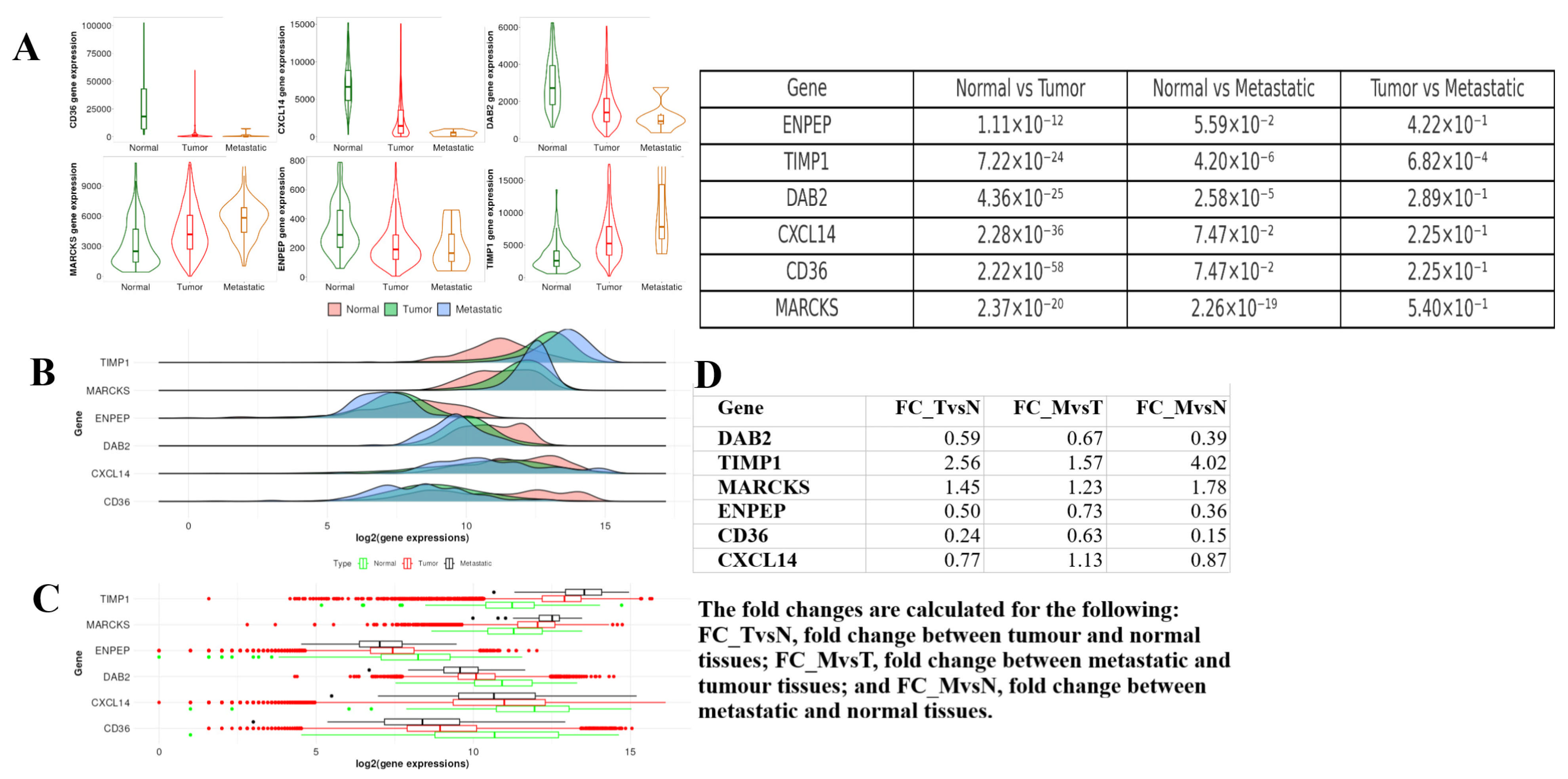
2.7. Tumor, Normal, and Metastatic (TNM) Analysis
2.8. Cancer Hallmark Enrichment Analysis
2.9. Result of miRNAs Associated with Genes
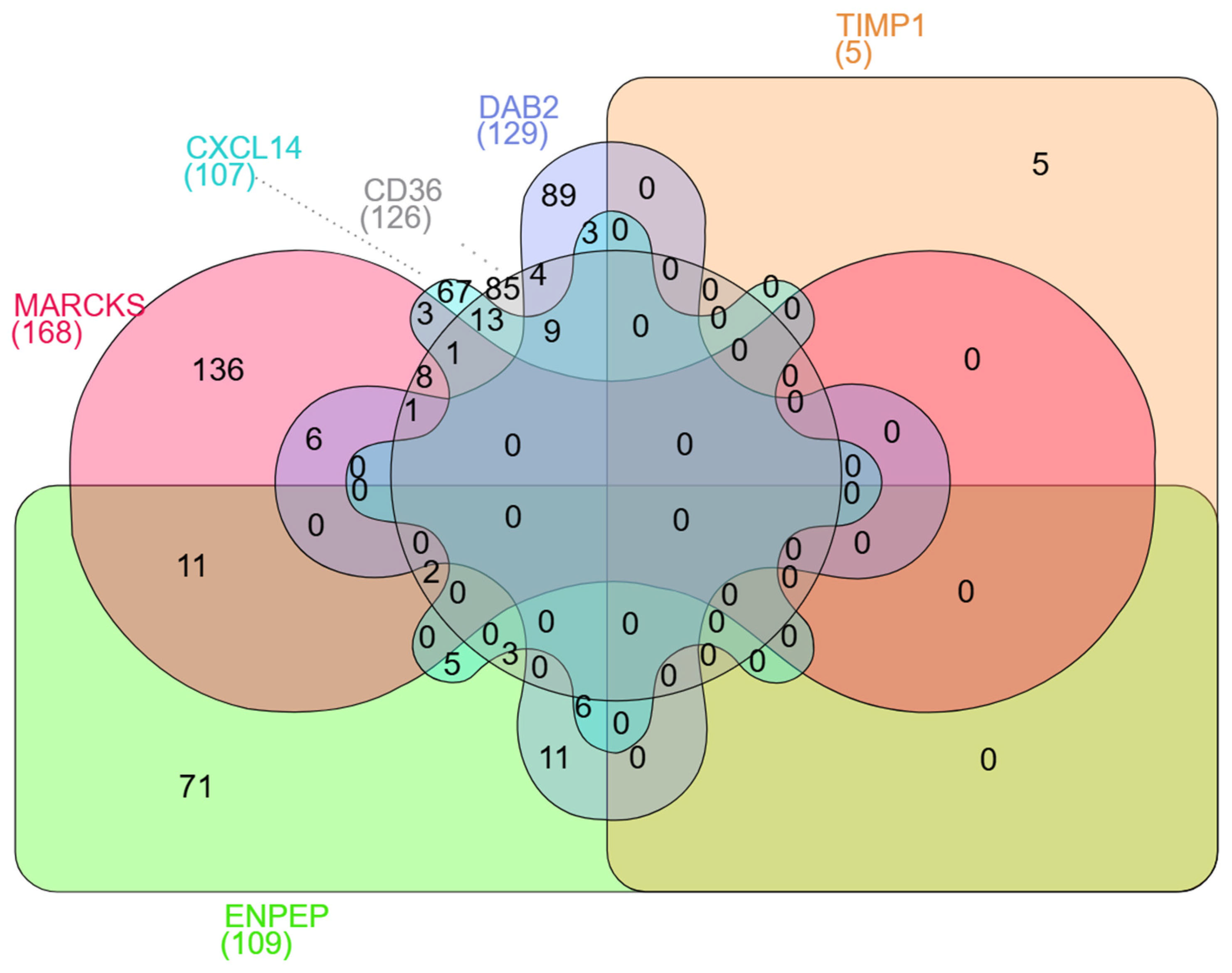
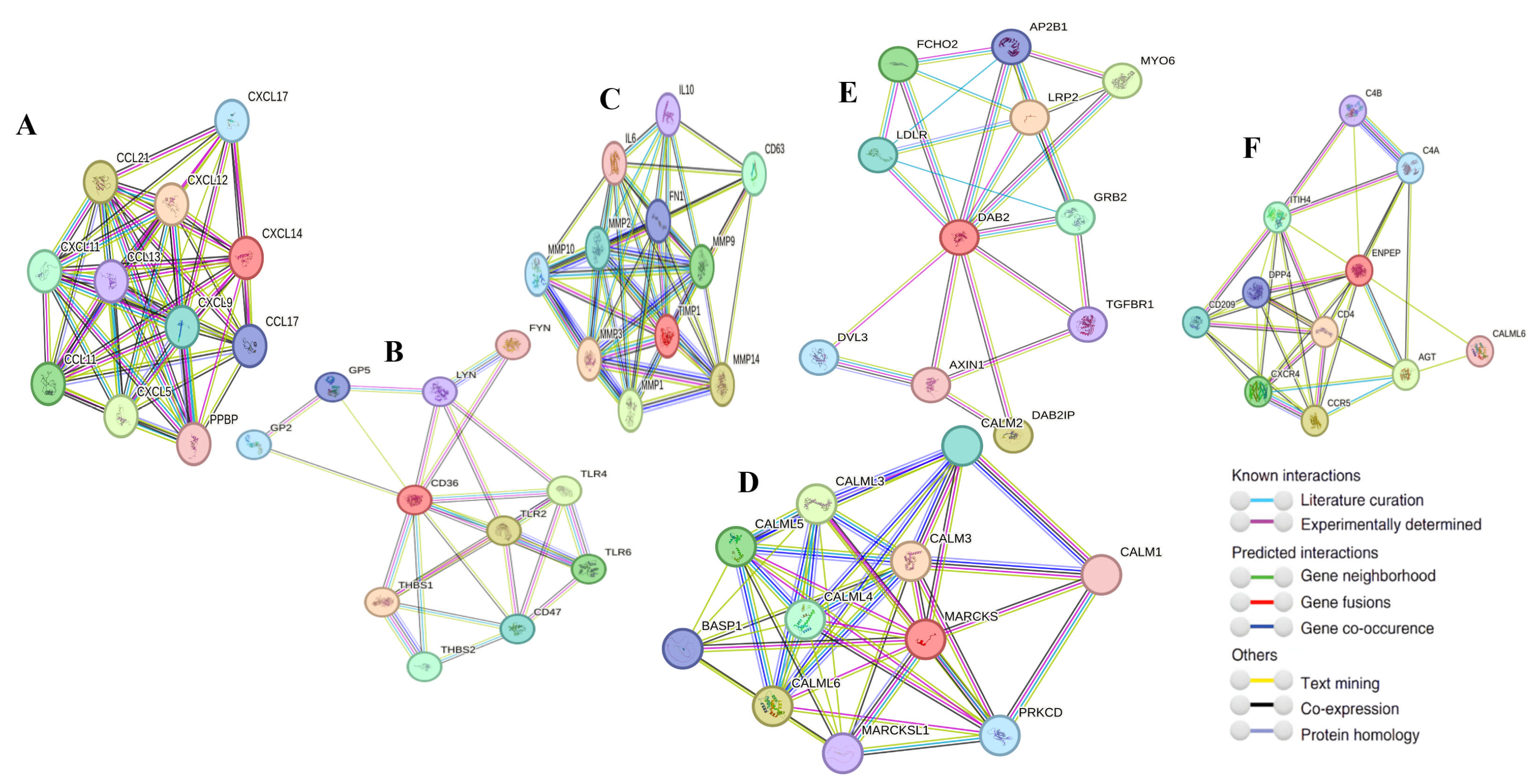
2.10. Gene–Gene Interaction
3. Discussion
Limitation
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Full Name |
| BC | Breast cancer |
| EV | Extracellular vesicle |
| miRNA | MicroRNA |
| lncRNA | Long non-coding RNA |
| IHC | Immunohistochemistry |
| OS | Overall survival |
| GEPIA2 | Gene Expression Profiling Interactive Analysis 2 |
| HPA | Human Protein Atlas |
| UALCAN | University of Alabama at Birmingham CANcer data analysis portal |
| TNM | Tumor, Normal, and Metastatic |
| KM Plotter | Kaplan–Meier Plotter |
| ENCORI | Encyclopedia of RNA Interactomes |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| PAM50 | Prediction Analysis of Microarray 50 |
| TNBC | Triple negative breast cancer |
| ER | Estrogen receptor |
| PR | Progesterone receptor |
| HER2 | Human Epidermal Growth Factor Receptor 2 |
| TIMP1 | Tissue inhibitor of metalloproteinases 1 |
| CD36 | Cluster of Differentiation 36 |
| MARCKS | Myristoylated Alanine-Rich C Kinase Substrate |
| DAB2 | Disabled Homolog 2 |
| CXCL14 | C-X-C Motif Chemokine Ligand 14 |
| ENPEP | Glutamyl Aminopeptidase |
| miR-181b-5p | MicroRNA-181b-5p |
| miR-222-3p | MicroRNA-222-3p |
| miRDB | MicroRNA Target Prediction Database |
| TargetScan | TargetScan microRNA target prediction tool |
| TCGA | The Cancer Genome Atlas |
References
- Lopez-Gonzalez, L.; Sanchez, C.A.; Sanchez, C.C.; Roberts, C.E.; Espinosa, J.; Pekarek, T.; Fraile-Martinez, O.; García-Montero, C.; Rodriguez-Slocker, A.M.; Jiménez-Álvarez, L.; et al. Exploring Biomarkers in Breast Cancer: Hallmarks of Diagnosis, Treatment, and Follow-Up in Clinical Practice. Medicina 2024, 60, 168. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, L.; Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2022, 95, 20211033. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, L.; Samal, P.; Sahoo, S.; Sahoo, B.; Kumar, P.A.; Mahanta, S.; Rath, S.K.; Arakha, M. Nanoparticle-mediated diagnosis, treatment, and prevention of breast cancer. Nanoscale Adv. 2024, 6, 3699–3713. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Yu, J.; Bu, J.; Ai, F.; Wang, Y.; Lin, J.; Zhu, X. Extracellular vesicles in the treatment and diagnosis of breast cancer: A status update. Front. Endocrinol. 2023, 14, 1202493. [Google Scholar] [CrossRef]
- Bandu, R.; Oh, J.; Kim, K. Extracellular vesicle proteins as breast cancer biomarkers: Mass spectrometry-based analysis. Proteomics 2024, 24, 2300062. [Google Scholar] [CrossRef]
- Lopez, K.; Lai, S.; Lopez, G.E.; Dávila, R.; Shuck, S. Extracellular vesicles: A dive into their role in the tumor microenvironment and cancer progression. Front. Cell Dev. Biol. 2023, 11, 1154576. [Google Scholar] [CrossRef]
- Andre, M.; Caobi, A.; Miles, J.; Vashist, A.; Ruiz, M.; Raymond, A. Diagnostic potential of exosomal extracellular vesicles in oncology. BMC Cancer 2024, 24, 322. [Google Scholar] [CrossRef]
- Kumar, M.; Baba, S.; Sadida, H.; Marzooqi, S.; Jerobin, J.; Altemani, H.; Algehainy, N.; Alanazi, M.A.; Abou-Samra, A.B.; Kumar, R.; et al. Extracellular vesicles as tools and targets in therapy for diseases. Signal Transduct. Target. Ther. 2024, 9, 27. [Google Scholar] [CrossRef]
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer-implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, D1241–D1244. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Hutter, C.; Zenklusen, J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018, 173, 283–285. [Google Scholar] [CrossRef]
- Ahmed, S.; Espinoza-Sánchez, N.; El-Damen, A.; Fahim, S.; Badawy, M.; Greve, B.; El-Shinawi, M.; Götte, M.; Ibrahim, S.A. Small extracellular vesicle-encapsulated miR-181b-5p, miR-222-3p and let-7a-5p: Next generation plasma biopsy-based diagnostic biomarkers for inflammatory breast cancer. PLoS ONE 2021, 16, e0250642. [Google Scholar] [CrossRef]
- Warren, L.; Guo, H.; Regan, M.; Nakhlis, F.; Yeh, E.; Jacene, H.; Hirshfield-Bartek, J.; Overmoyer, B.A.; Bellon, J.R. Inflammatory breast cancer and development of brain metastases: Risk factors and outcomes. Breast Cancer Res. Treat. 2015, 151, 225–232. [Google Scholar] [CrossRef]
- Uemura, M.; French, J.; Hess, K.; Liu, D.; Raghav, K.; Hortobagyi, G.; Arun, B.K.; Valero, V.; Ueno, N.T.; Alvarez, R.H.; et al. Development of CNS metastases and survival in patients with inflammatory breast cancer. Cancer 2018, 124, 2299–2305. [Google Scholar] [CrossRef]
- Guerrero-Rodríguez, S.; Mata-Cruz, C.; Pérez-Tapia, S.; Velasco-Velázquez, M. Role of CD36 in cancer progression, stemness, and targeting. Front. Cell Dev. Biol. 2022, 10, 1079076. [Google Scholar] [CrossRef]
- Stirzaker, C.; Song, J.; Davidson, B.; Clark, S. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004, 64, 3871–3877. [Google Scholar] [CrossRef] [PubMed]
- DeFilippis, R.; Chang, H.; Dumont, N.; Rabban, J.; Chen, Y.; Fontenay, G.; Berman, H.K.; Gauthier, M.L.; Zhao, J.; Hu, D.; et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer Discov. 2012, 2, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Chan, C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci. Rep. 2016, 6, 18669. [Google Scholar] [CrossRef] [PubMed]
- Sjöberg, E.; Augsten, M.; Bergh, J.; Jirström, K.; Östman, A. Expression of the chemokine CXCL14 in the tumour stroma is an independent marker of survival in breast cancer. Br. J. Cancer 2016, 114, 1117–1124. [Google Scholar] [CrossRef]
- Gibbs, C.; So, J.; Ahad, A.; Michalowski, A.; Son, D.; Li, Y. CXCL14 Attenuates Triple-Negative Breast Cancer Progression by Regulating Immune Profiles of the Tumor Microenvironment in a T Cell-Dependent Manner. Int. J. Mol. Sci. 2022, 23, 9314. [Google Scholar] [CrossRef]
- Gu, X.; Ou, Z.; Lin, F.; Yang, X.; Luo, J.; Shen, Z.; Shao, Z. Expression of CXCL14 and its anticancer role in breast cancer. Breast Cancer Res. Treat. 2012, 135, 725–735. [Google Scholar] [CrossRef]
- Schrohl, A.; Meijer-van, G.M.; Holten-Andersen, M.; Christensen, I.; Look, M.; Mouridsen, H.; Brünner, N.; Foekens, J.A. Primary tumor levels of tissue inhibitor of metalloproteinases-1 are predictive of resistance to chemotherapy in patients with metastatic breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 7054–7058. [Google Scholar] [CrossRef]
- Agnello, L.; d’Argenio, A.; Caliendo, A.; Nilo, R.; Zannetti, A.; Fedele, M.; Camorani, S.; Cerchia, L. Tissue Inhibitor of Metalloproteinases-1 Overexpression Mediates Chemoresistance in Triple-Negative Breast Cancer Cells. Cells 2023, 12, 1809. [Google Scholar] [CrossRef]
- Nabholz, J.; Abrial, C.; Mouret-Reynier, M.; Dauplat, M.; Weber, B.; Gligorov, J.; Forest, A.M.; Tredan, O.; Vanlemmens, L.; Petit, T.; et al. Multicentric neoadjuvant phase II study of panitumumab combined with an anthracycline/taxane-based chemotherapy in operable triple-negative breast cancer: Identification of biologically defined signatures predicting treatment impact. Ann. Oncol. 2014, 25, 1570–1577. [Google Scholar] [CrossRef]
- Cheng, G.; Fan, X.; Hao, M.; Wang, J.; Zhou, X.; Sun, X. Higher levels of TIMP-1 expression are associated with a poor prognosis in triple-negative breast cancer. Mol. Cancer 2016, 15, 30. [Google Scholar] [CrossRef]
- Keskin, S.E.; Akkoyunlu, D.S.; Yılmaz, M.; Şimşek, T.; Güler, S.; Çine, N.; Cantürk, Z.; Savlı, H. Quantification of MMP-2 and TIMP-1 expressions in breast cancer. İstanb. J. Pharm. 2023, 53, 211–218. [Google Scholar] [CrossRef]
- Xu, S.; Zhu, J.; Wu, Z. Loss of Dab2 Expression in Breast Cancer Cells Impairs Their Ability to Deplete TGF-β and Induce Tregs Development via TGF-β. PLoS ONE 2014, 9, e91709. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Price, Z.; Lokman, N.; Yoshihara, M.; Kajiyama, H.; Oehler, M.; Ricciardelli, C. Disabled-2 (DAB2): A Key Regulator of Anti- and Pro-Tumorigenic Pathways. Int. J. Mol. Sci. 2023, 24, 696. [Google Scholar] [CrossRef]
- Papageorgis, P.; Stylianopoulos, T. Role of TGFβ in regulation of the tumor microenvironment and drug delivery (Review). Int. J. Oncol. 2015, 46, 933–943. [Google Scholar] [CrossRef]
- Wang, D.; Sang, Y.; Sun, T.; Kong, P.; Zhang, L.; Dai, Y.; Cao, Y.; Tao, Z.; Liu, W. Emerging roles and mechanisms of microRNA-222-3p in human cancer (Review). Int. J. Oncol. 2021, 58, 20. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, W.; Lin, Y.; Yin, K.; Zhou, L.; Du, Y.; Yan, T.; Lu, J. Downregulated circulating microRNAs after surgery: Potential noninvasive biomarkers for diagnosis and prognosis of early breast cancer. Cell Death Discov. 2018, 4, 87. [Google Scholar] [CrossRef]
- Costello, J.; Frühwald, M.; Smiraglia, D.; Rush, L.; Robertson, G.; Gao, X.; Wright, F.A.; Feramisco, J.D.; Peltomäki, P.; Lang, J.C.; et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2020, 24, 132–138. [Google Scholar] [CrossRef]
- Esteller, M.; Corn, P.; Baylin, S.; Herman, J. A gene hypermethylation profile of human cancer. Cancer Res. 2001, 61, 3225–3229. [Google Scholar]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Tóth, E.Á.; Turiák, L.; Visnovitz, T.; Cserép, C.; Mázló, A.; Sódar, B.W.; Försönits, A.I.; Petővári, G.; Sebestyén, A.; Komlósi, Z.; et al. Formation of a protein corona on the surface of extracellular vesicles in blood plasma. J. Extracell. Vesicles 2021, 10, e12140. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.; Tűzesi, Á.; Grau, G.E.; Buckland, M.E.; Alexander, K.L. Understanding the extracellular vesicle surface for clinical molecular biology. J. Extracell. Vesicles 2022, 11, e12260. [Google Scholar] [CrossRef]
- Jung, T.; Castellana, D.; Klingbeil, P.; Hernández, I.C.; Vitacolonna, M.; Orlicky, D.J.; Roffler, S.R.; Brodt, P.; Zöller, M. CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia 2009, 11, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Jordan, K.R.; Hall, J.K.; Schedin, T.; Borakove, M.; Xian, J.J.; Dzieciatkowska, M.; Lyons, T.R.; Schedin, P.; Hansen, K.C.; Borges, V.F. Extracellular vesicles from young women’s breast cancer patients drive increased invasion of non-malignant cells via the Focal Adhesion Kinase pathway: A proteomic approach. Breast Cancer Res. 2020, 22, 128. [Google Scholar] [CrossRef]
- Zhou, X.; Su, M.; Lu, J.; Li, D.; Niu, X.; Wang, Y. CD36: The Bridge between Lipids and Tumors. Molecules 2024, 29, 531. [Google Scholar] [CrossRef]
- enito-Martín, A.; Jasiulionis, M.G.; García-Silva, S. Extracellular vesicles and melanoma: New perspectives on tumor microenvironment and metastasis. Front. Cell Dev. Biol. 2023, 10, 1061982. [Google Scholar]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Jézéquel, P.; Frénel, J.; Campion, L.; Guérin-Charbonnel, C.; Gouraud, W.; Ricolleau, G.; Campone, M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database 2013, 2013, bas060. [Google Scholar] [CrossRef]
- Jézéquel, P.; Gouraud, W.; Ben, A.F.; Guérin-Charbonnel, C.; Juin, P.; Lasla, H.; Campone, M. bc-GenExMiner 4.5: New mining module computes breast cancer differential gene expression analyses. Database 2021, 2021, baab007. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.; Bashel, B.; Balasubramanya, S.; Creighton, C.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Lánczky, A.; Menyhárt, O.; Győrffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar]
- Li, J.; Liu, S.; Zhou, H.; Qu, L.; Yang, J. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Evangelista, J.; Xie, Z.; Marino, G.; Nguyen, N.; Clarke, D.; Ma’ayan, A. Enrichr-KG: Bridging enrichment analysis across multiple libraries. Nucleic Acids Res. 2023, 51, W168–W179. [Google Scholar] [CrossRef]
- Menyhart, O.; Kothalawala, W.; Győrffy, B. A gene set enrichment analysis for the cancer hallmarks. J. Pharm. Anal. 2024, 14, 101065. [Google Scholar] [CrossRef]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]





| Cancer Hallmark | Overlap | p-Value | Adjusted p-Value | Odds Ratio | Hallmark vs. Hallmark | Genes |
|---|---|---|---|---|---|---|
| Sustaining Proliferative Signaling | 4/3574 | 0.03172 | 0.06175 | 5.76 | 0.69 | TIMP1; ENPEP; CD36; CXCL14 |
| Genome Instability | 0/1 | nan | 1.0 | nan | 0.0 | nan |
| Evading Growth Suppressors | 5/3288 | 0.00248 | 0.01012 | 13.08 | 0.93 | TIMP1; CXCL14; DAB2; ENPEP; MARCKS |
| Evading Immune Destruction | 1/749 | 0.26464 | 0.26464 | 5.19 | 0.82 | CXCL14 |
| Sustained Angiogenesis | 3/796 | 0.00264 | 0.01012 | 17.9 | 2.31 | TIMP1; CD36; CXCL14 |
| Tissue Invasion and Metastasis | 3/2318 | 0.05115 | 0.07672 | 5.48 | 0.79 | TIMP1; ENPEP; CD36 |
| Tumor-Promoting Inflammation | 2/769 | 0.03431 | 0.06175 | 10.3 | 1.6 | CD36; CXCL14 |
| Resisting Cell Death | 4/1941 | 0.00337 | 0.01012 | 12.13 | 1.26 | TIMP1; DAB2; CD36; CXCL14 |
| Reprogramming Energy Metabolism | 1/740 | 0.26185 | 0.26464 | 5.26 | 0.83 | CD36 |
| Replicative Immortality | 1/547 | 0.19983 | 0.25692 | 7.21 | 1.12 | CXCL14 |
| Gene Groups | miRNAs Related to Genes |
|---|---|
| CD36 and DAB2 | hsa-miR-4729, hsa-miR-651-5p, hsa-miR-5579-3p, hsa-miR-4520-2-3p |
| DAB2 and CXCL14 | hsa-miR-4713-5p, hsa-miR-548t-5p, hsa-miR-548az-5p |
| CD36 and CXCL14 | hsa-miR-4719, hsa-miR-340-5p, hsa-miR-651-3p, hsa-miR-4477b, hsa-miR-1277-5p, hsa-miR-150-5p, hsa-miR-153-5p, hsa-miR-6854-5p, hsa-miR-548ar-3p, hsa-miR-548a-3p, hsa-miR-548f-3p, hsa-miR-548az-3p, hsa-miR-548e-3p |
| MARCKS and CXCL14 | hsa-miR-3121-5p, hsa-miR-5088-3p, hsa-miR-3143 |
| ENPEP and CXCL14 | hsa-miR-3163, hsa-miR-3662, hsa-miR-106a-3p, hsa-miR-142-5p, hsa-miR-5590-3p |
| ENPEP and CD36 | hsa-miR-548e-5p, hsa-miR-4659a-3p, hsa-miR-4659b-3p |
| CD36 and MARCKS | hsa-miR-1537-5p, hsa-miR-4276, hsa-miR-545-3p, hsa-miR-134-3p, hsa-miR-3124-3p, hsa-miR-655-3p, hsa-miR-374c-5p, hsa-miR-576-5p |
| ENPEP and MARCKS | hsa-miR-9-5p, hsa-miR-641, hsa-miR-3617-5p, hsa-miR-520d-5p, hsa-miR-524-5p, hsa-miR-551b-5p, hsa-miR-4668-3p, hsa-miR-3606-3p, hsa-miR-513a-3p, hsa-miR-513c-3p, hsa-miR-4490 |
| MARCKS and DAB2 | hsa-miR-6878-5p, hsa-miR-6825-5p, hsa-miR-664b-3p, hsa-miR-579-3p, hsa-miR-3925-3p, hsa-let-7c-3p |
| ENPEP and DAB2 | hsa-miR-1297, hsa-miR-26a-5p, hsa-miR-26b-5p, hsa-miR-4465, hsa-miR-376a-5p, hsa-miR-3150b-3p, hsa-miR-4784, hsa-miR-3977, hsa-miR-6768-3p, hsa-miR-15b-3p, hsa-miR-4428 |
| ENPEP and DAB2 and CXCL14 | hsa-miR-186-5p, hsa-miR-320d, hsa-miR-320c, hsa-miR-320b, hsa-miR-4429, hsa-miR-9-3p |
| CD36 and MARCKS and DAB2 | hsa-miR-5696 |
| CD36, MARCKS and CXCL14 | hsa-miR-410-3p |
| CD36, DAB2 and CXCL14 | hsa-miR-539-5p, hsa-miR-526b-3p, hsa-miR-93-5p, hsa-miR-20b-5p, hsa-miR-106b-5p, hsa-miR-20a-5p, hsa-miR-17-5p, hsa-miR-519d-3p, hsa-miR-106a-5p |
| ENPEP, CD36 and MARCKS | hsa-miR-4328, hsa-miR-548c-3p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayan, D.; Bozkurt Polat, S.B.; Ozmen, E.; Gul, M.A. Comparison Bioinformatic Analysis of Extracellular Vesicles-Related Genes and MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2025, 26, 5906. https://doi.org/10.3390/ijms26125906
Ayan D, Bozkurt Polat SB, Ozmen E, Gul MA. Comparison Bioinformatic Analysis of Extracellular Vesicles-Related Genes and MicroRNAs in Breast Cancer. International Journal of Molecular Sciences. 2025; 26(12):5906. https://doi.org/10.3390/ijms26125906
Chicago/Turabian StyleAyan, Durmus, Serife Buket Bozkurt Polat, Esma Ozmen, and Mehmet Ali Gul. 2025. "Comparison Bioinformatic Analysis of Extracellular Vesicles-Related Genes and MicroRNAs in Breast Cancer" International Journal of Molecular Sciences 26, no. 12: 5906. https://doi.org/10.3390/ijms26125906
APA StyleAyan, D., Bozkurt Polat, S. B., Ozmen, E., & Gul, M. A. (2025). Comparison Bioinformatic Analysis of Extracellular Vesicles-Related Genes and MicroRNAs in Breast Cancer. International Journal of Molecular Sciences, 26(12), 5906. https://doi.org/10.3390/ijms26125906








