Pro-Apoptotic Activity of 1-(4,5,6,7-Tetrabromo-1H-benzimidazol-1-yl)propan-2-one, an Intracellular Inhibitor of PIM-1 Kinase in Acute Lymphoblastic Leukemia and Breast Cancer Cells
Abstract
1. Introduction
2. Results
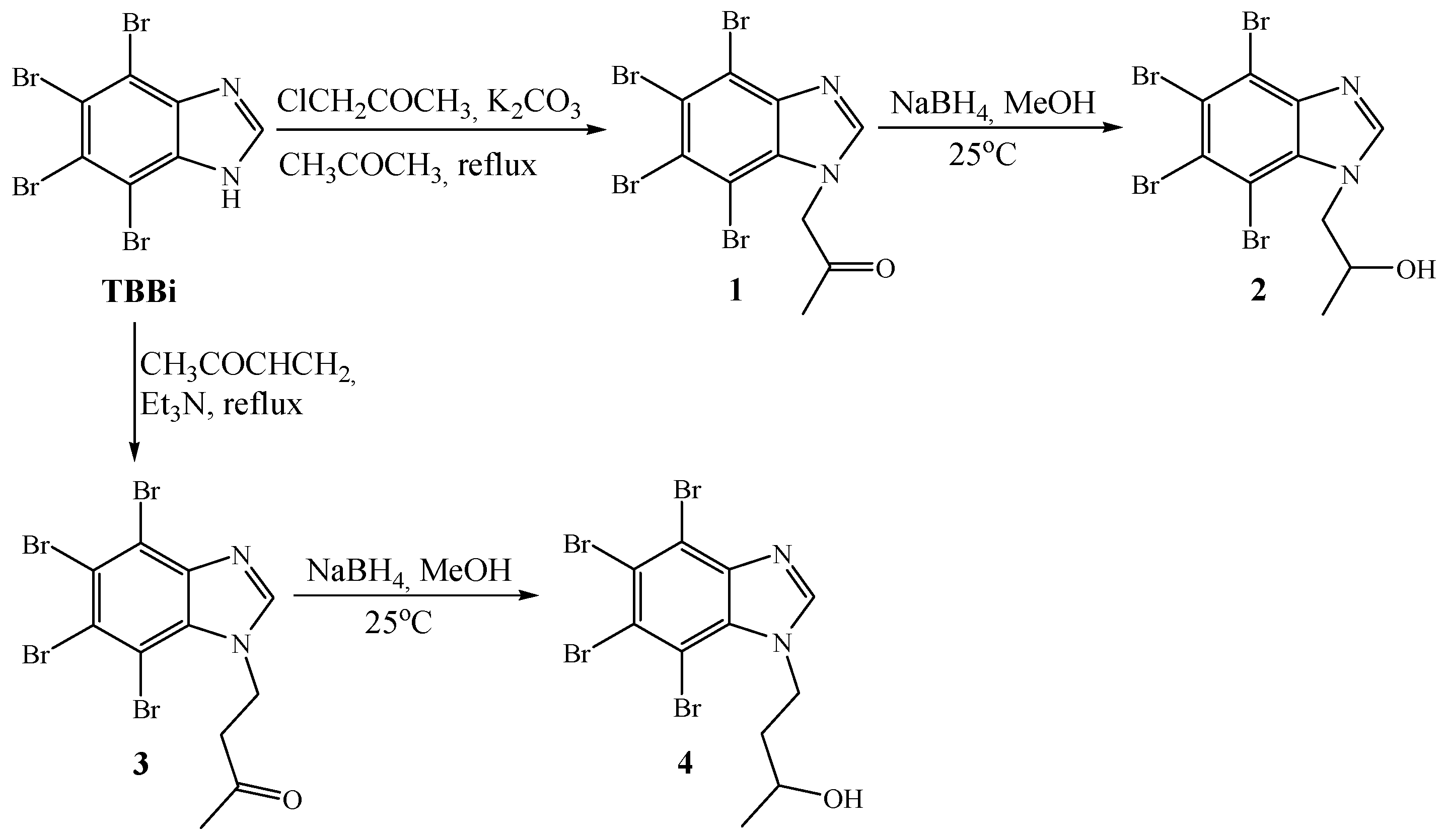
2.1. Synthesis
2.2. Biological Evaluation
2.2.1. Determination of the Inhibitory Activity of Compounds 1–4 Towards CK2 and PIM-1 Kinases
2.2.2. Cytotoxic Effect of TBBi Derivatives Toward Cancer Cell Lines, CCRF-CEM, MCF-7, and MDA-MB-231, and Non-Cancerous Vero Cells
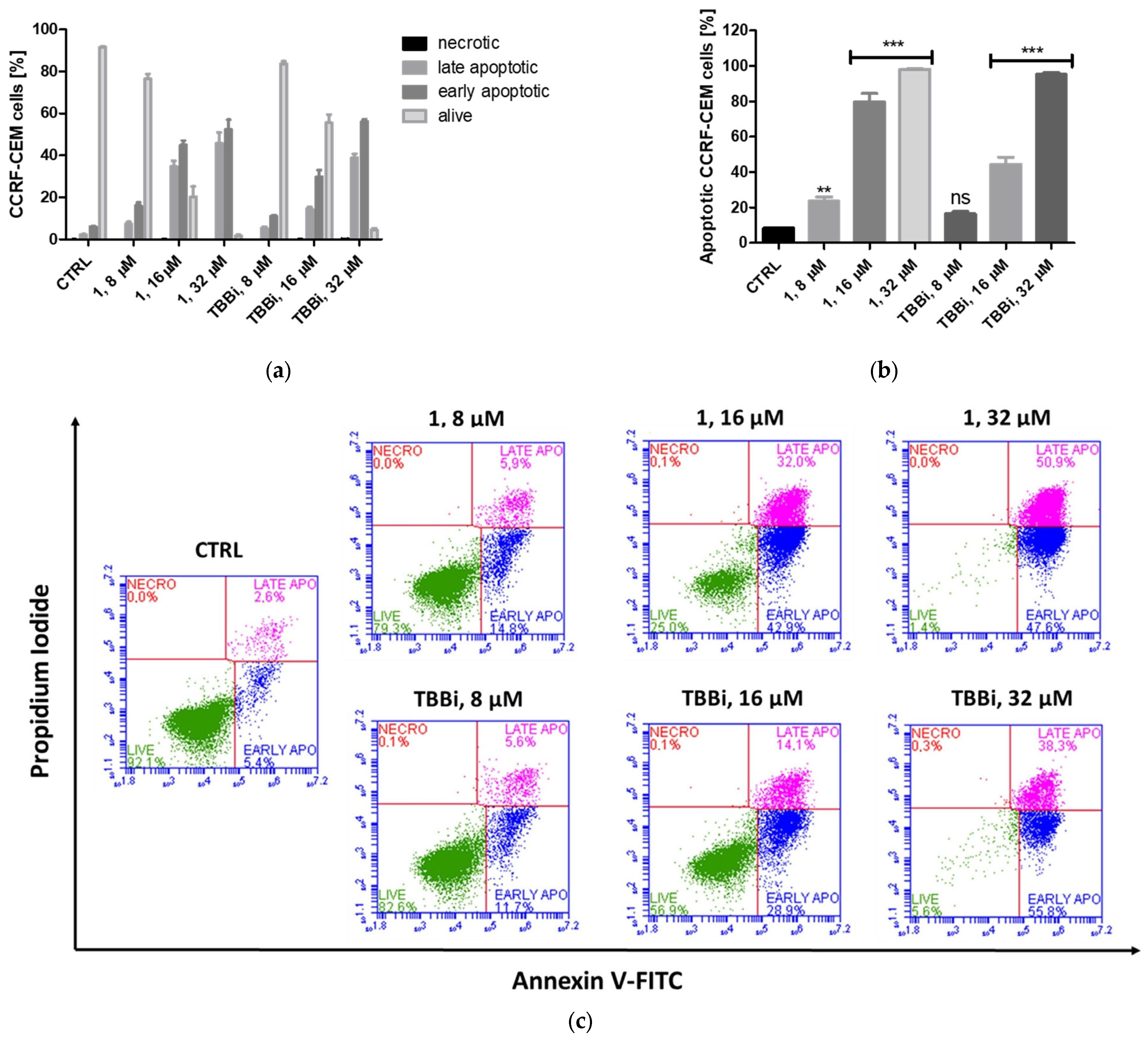
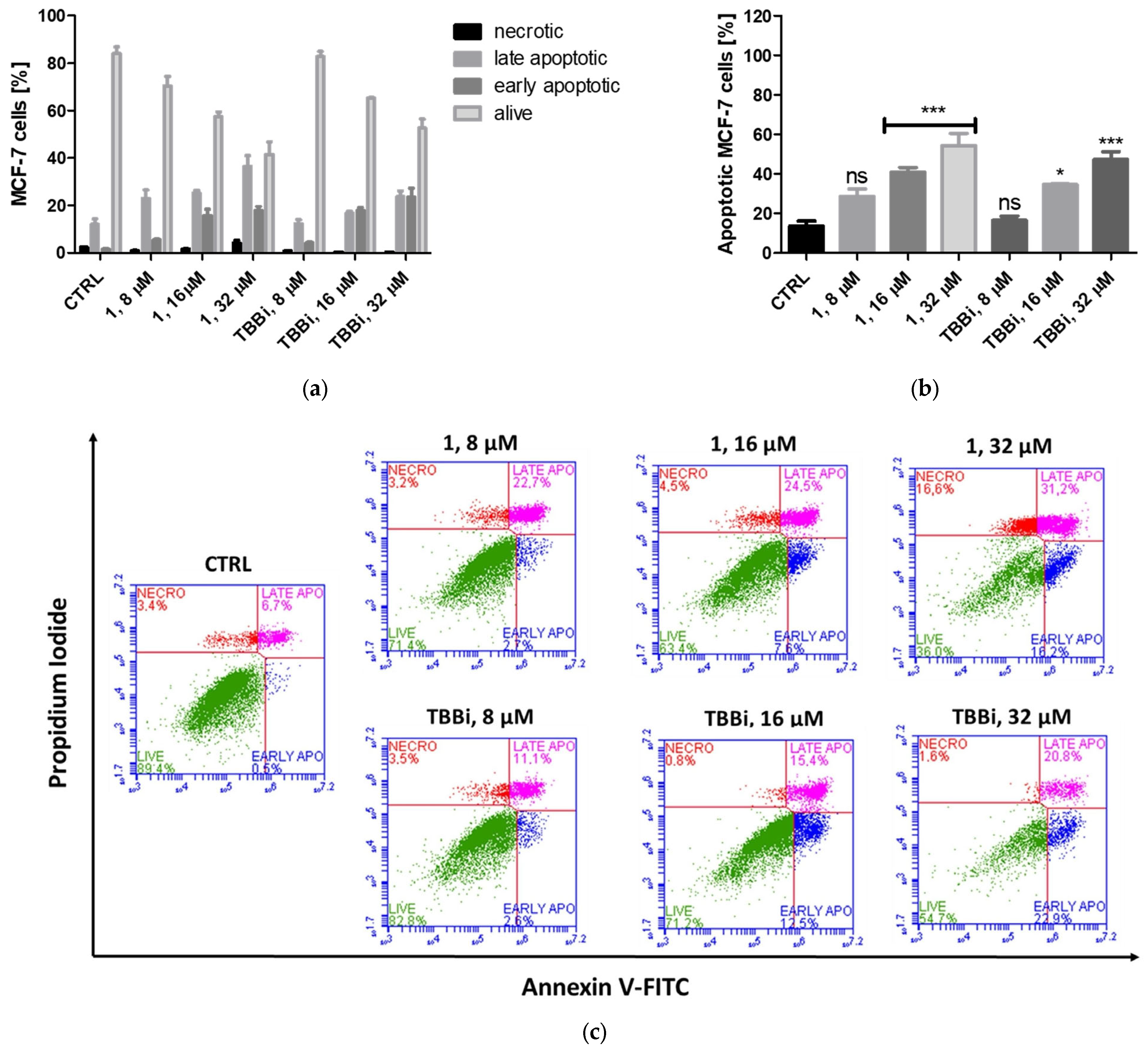
2.2.3. Induction of Apoptosis in CCRF-CEM and MCF-7 Cells
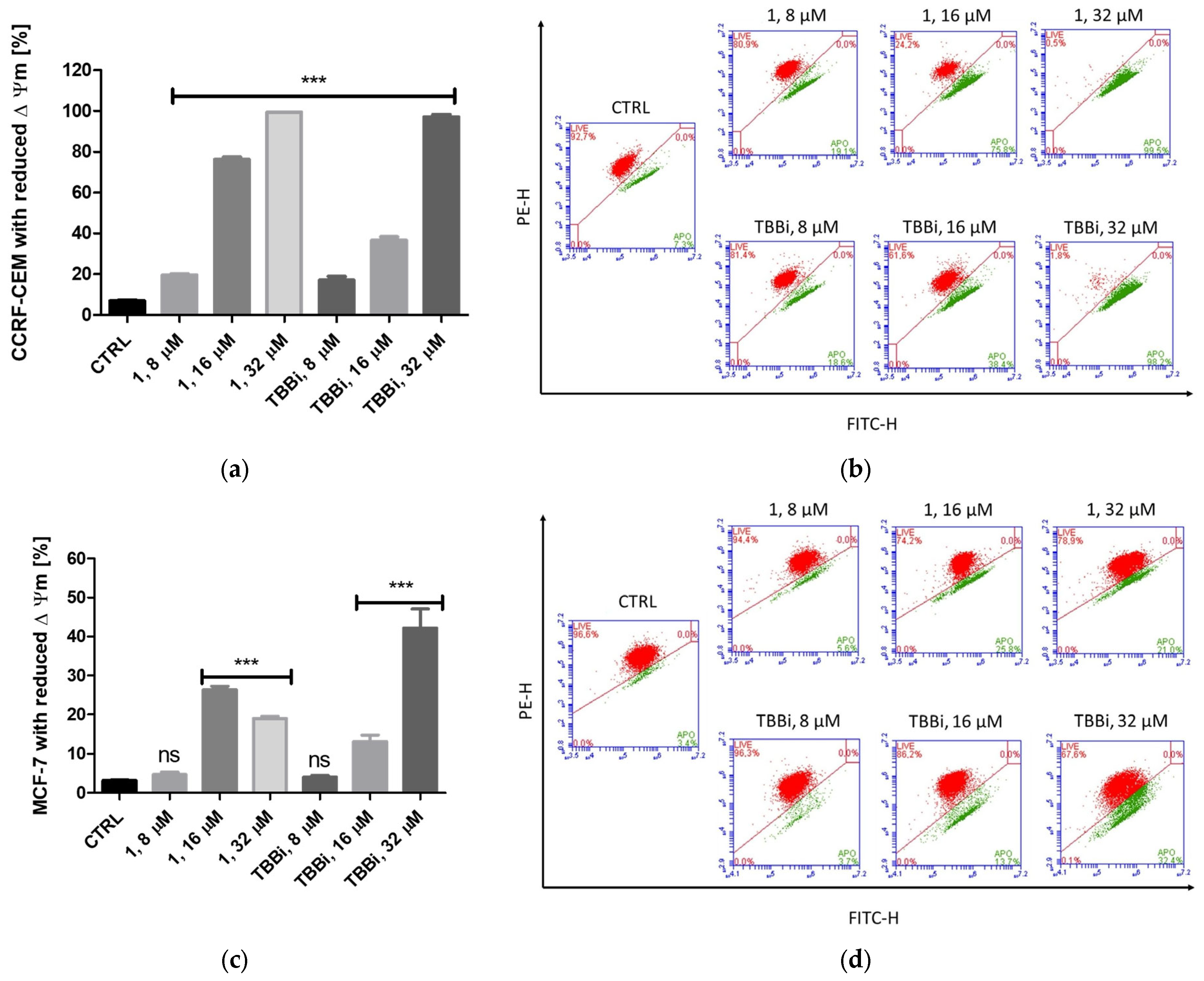
2.2.4. Mitochondrial Membrane Potential (ΔΨm) in CCRF-CEM and MCF-7
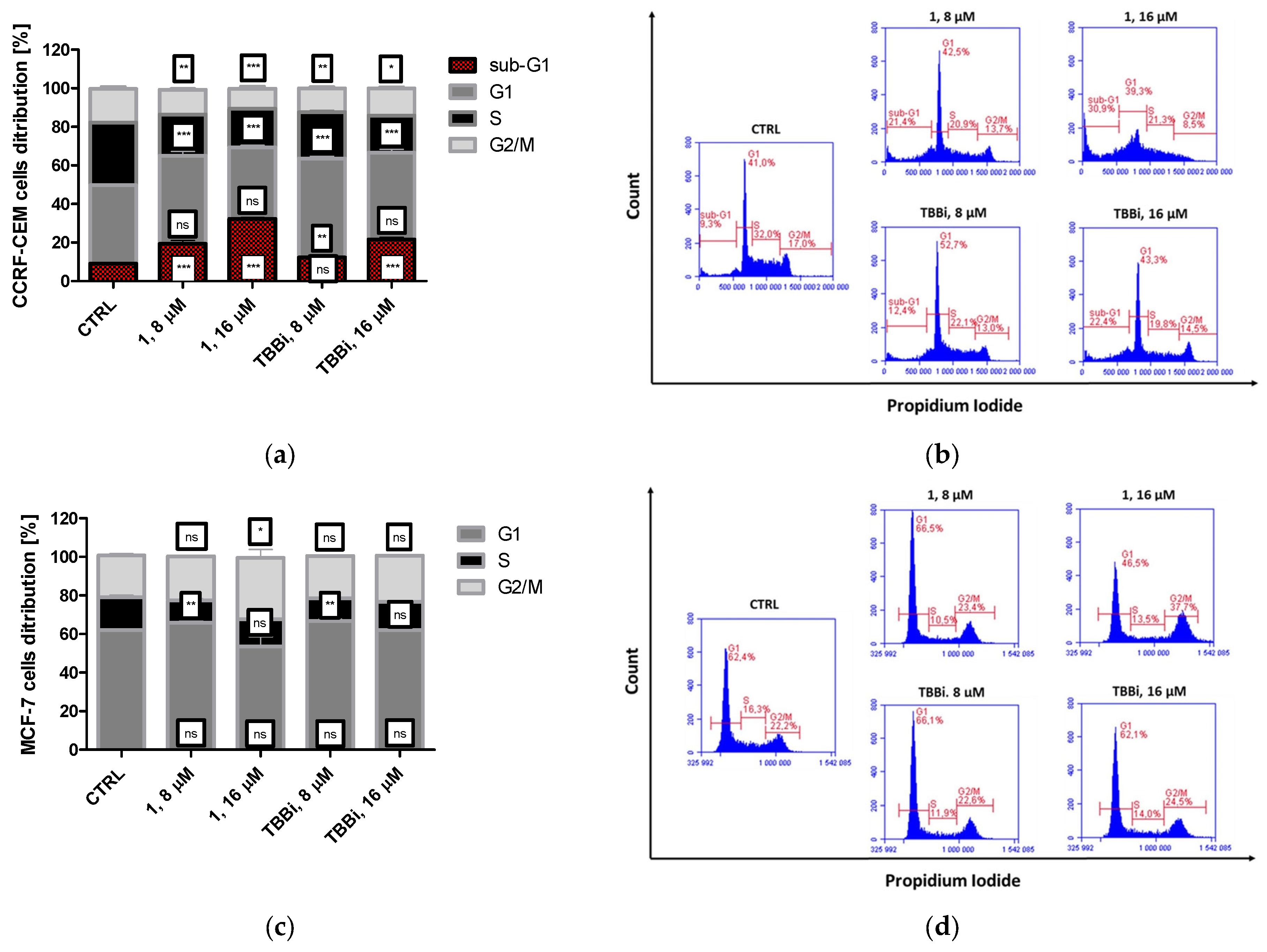
2.2.5. The Effect of Cpd. 1 and TBBi on Cell Cycle Progression in CCRF-CEM and MCF-7 Cells
2.2.6. Activation of Caspase-3 in CCRF-CEM
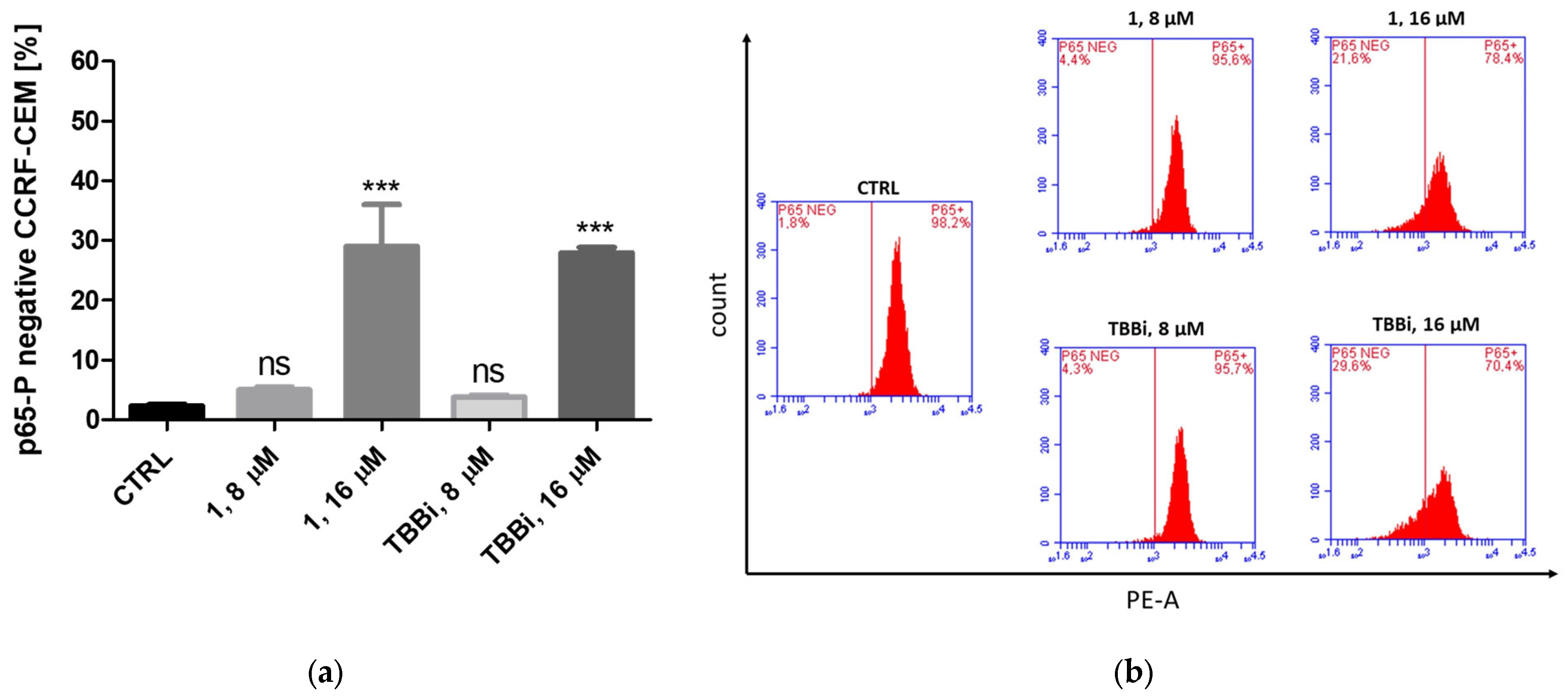
2.2.7. Intracellular Inhibition of CK2 and PIM-1 Kinases in CCRF-CEM and MCF-7 Cells
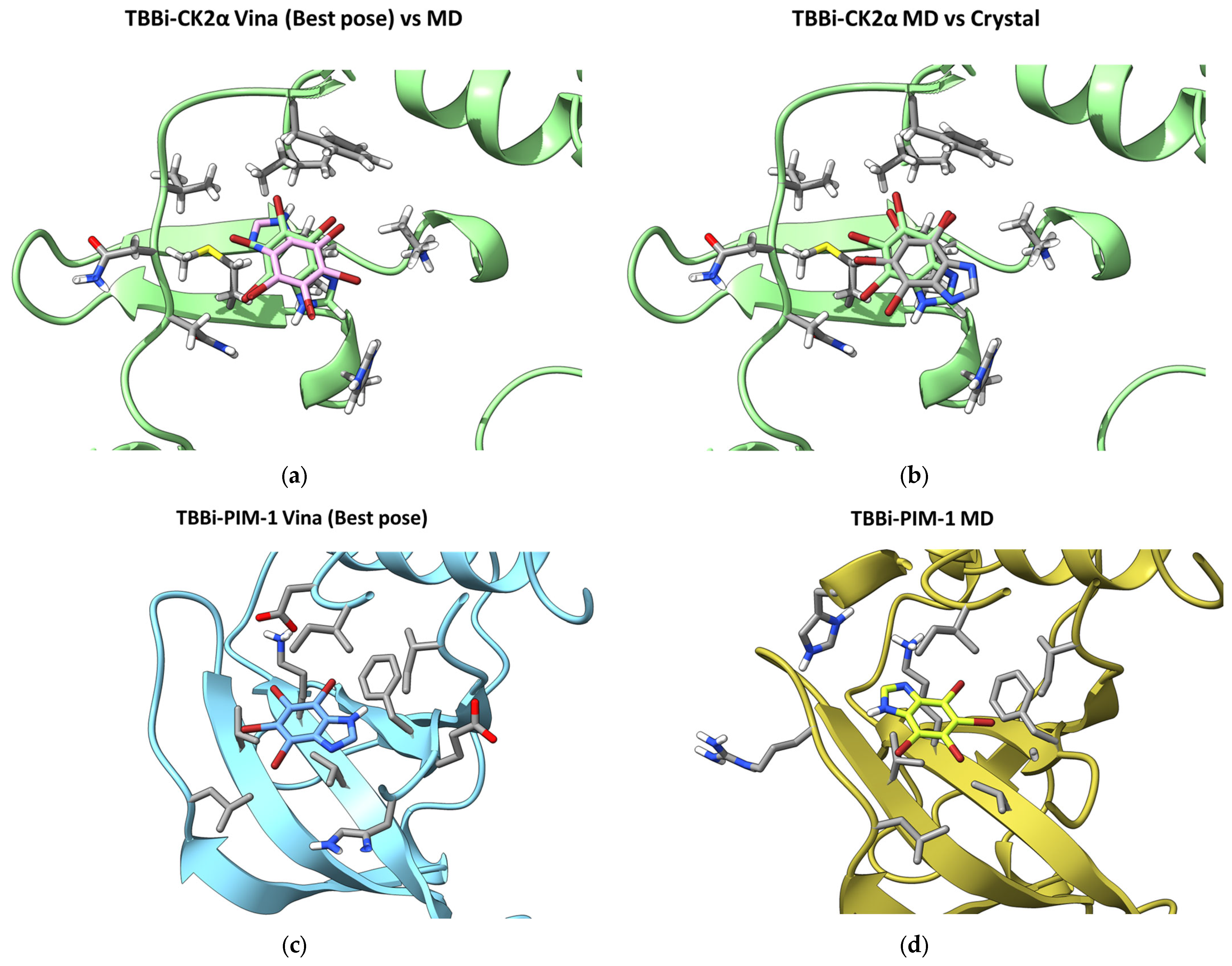
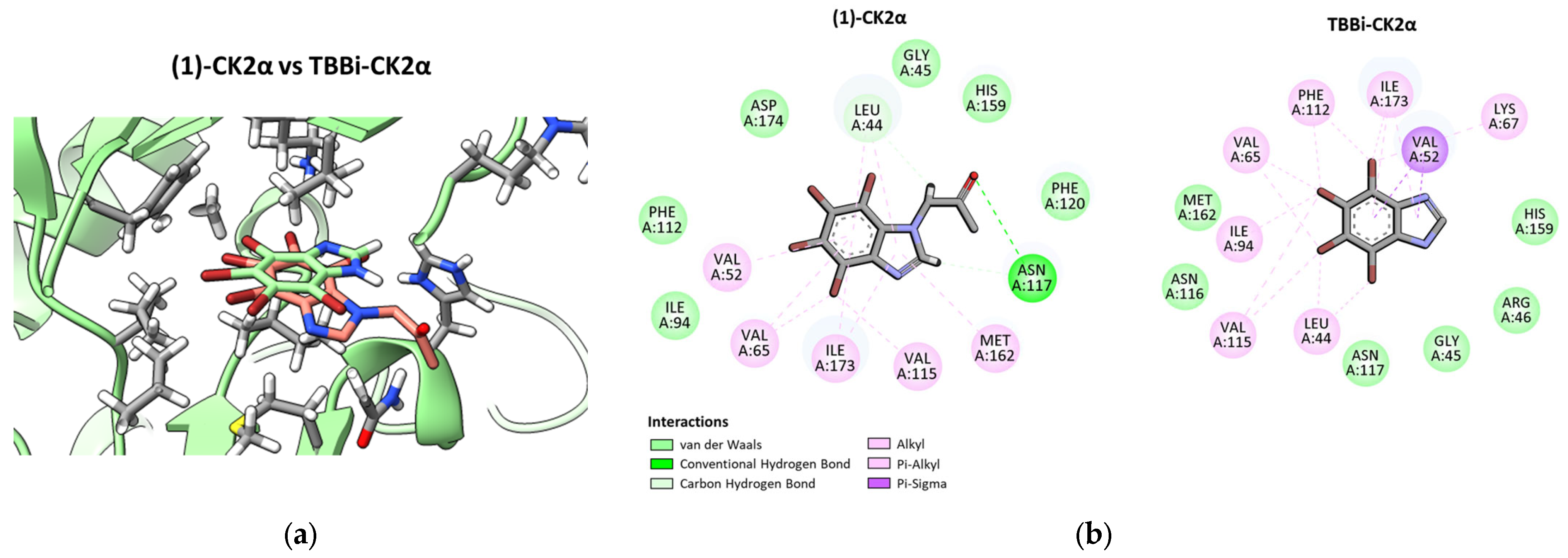
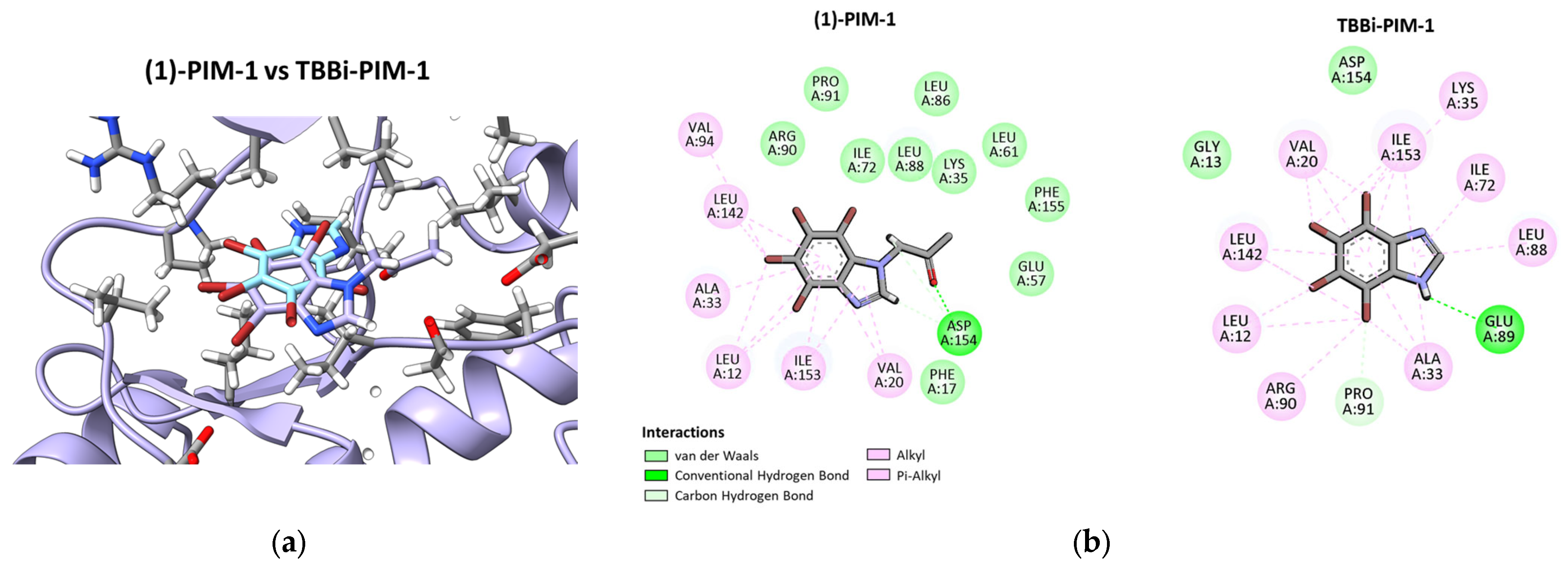
2.3. Molecular Modeling
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Chemical Study
4.2.1. Synthesis of 1-(4,5,6,7-Tetrabromo-1H-benzimidazol-1-yl)propan-2-one (1)
4.2.2. Synthesis of 1-(4,5,6,7-Tetrabromo-1H-benzimidazol-1-yl)propan-2-ol (2)
4.3. Biological Assays
4.3.1. Reagents and Antibodies
4.3.2. Cloning, Expression, and Purification of Human CK2α, holoCK2, and PIM-1
4.3.3. Inhibition of Recombinant CK2 and PIM-1
4.3.4. Cell Culture and Agents’ Treatment
4.3.5. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT)-Based Viability Assay
4.3.6. Detection of Apoptosis by Annexin V/Propidium Iodide (PI) Labelling
4.3.7. Mitochondrial Membrane Potential (ΔΨm) Assay
4.3.8. Caspase-3 Activation
4.3.9. Detection of Cell Cycle Progression by Flow Cytometry
4.3.10. Western Blotting
4.3.11. Densitometry
4.3.12. PhosFlow Analysis
4.3.13. Statistical Evaluation
4.4. In Silico Studies
4.4.1. Molecular Docking
4.4.2. Molecular Dynamics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Attwood, M.M.; Fabbro, D.; Sokolov, A.V.; Knapp, S.; Schiöth, H.B. Trends in kinase drug discovery: Targets, indications and inhibitor design. Nat. Rev. Drug Discov. 2021, 20, 839–861. [Google Scholar] [CrossRef]
- Ghani, M.U.; Shi, J.; Du, Y.; Zhong, L.; Cui, H. A comprehensive review on the dynamics of protein kinase CK2 in cancer development and optimizing therapeutic strategies. Int. J. Biol Macromol. 2024, 280, 135814. [Google Scholar] [CrossRef] [PubMed]
- Narlik-Grassow, M.; Blanco-Aparicio, C.; Carnero, A. The PIM family of serine/threonine kinases in cancer. Med. Res. Rev. 2014, 34, 136–159. [Google Scholar] [CrossRef] [PubMed]
- Trembley, J.H.; Wang, G.; Unger, G.; Slaton, J.; Ahmed, K. Protein kinase CK2 in health and disease: CK2: A key player in cancer biology. Cell Mol. Life Sci. 2009, 66, 1858–1867. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yang, Y.; Yuan, Y.; Liu, B. Targeting PIM kinases in cancer therapy: An update on pharmacological small-molecule inhibitors. Eur. J. Med. Chem. 2024, 264, 116016. [Google Scholar] [CrossRef]
- Trembley, J.H.; Chen, Z.; Unger, G.; Slaton, J.; Kren, B.T.; Van Waes, C.; Ahmed, K. Emergence of protein kinase CK2 as a key target in cancer therapy. Biofactors 2010, 36, 187–195. [Google Scholar] [CrossRef]
- Padi, S.K.R.; Luevano, L.A.; An, N.; Pandey, R.; Singh, N.; Song, J.H.; Aster, J.C.; Yu, X.Z.; Mehrotra, S.; Kraft, A.S. Targeting the PIM protein kinases for the treatment of a T-cell acute lymphoblastic leukemia subset. Oncotarget 2017, 8, 30199–30216. [Google Scholar] [CrossRef]
- La Starza, R.; Messina, M.; Gianfelici, V.; Pierini, V.; Matteucci, C.; Pierini, T.; Limongi, M.Z.; Vitale, A.; Roti, G.; Chiaretti, S.; et al. High PIM1 expression is a biomarker of T-cell acute lymphoblastic leukemia with JAK/STAT activation or t(6;7)(p21;q34)/TRB@-PIM1 rearrangement. Leukemia 2018, 32, 1807–1810. [Google Scholar] [CrossRef]
- Rout, A.K.; Dehury, B.; Parida, S.N.; Rout, S.S.; Jena, R.; Kaushik, N.; Kaushik, N.K.; Pradhan, S.K.; Sahoo, C.R.; Singh, A.K.; et al. A review on structure-function mechanism and signaling pathway of serine/threonine protein PIM kinases as a therapeutic target. Int. J. Biol. Macromol. 2024, 270, 132030. [Google Scholar] [CrossRef]
- Nuñez de Villavicencio-Diaz, T.; Rabalski, A.J.; Litchfield, D.W. Protein Kinase CK2: Intricate Relationships within Regulatory Cellular Networks. Pharmaceuticals 2017, 10, 27. [Google Scholar] [CrossRef]
- Rabalski, A.J.; Gyenis, L.; Litchfield, D.W. Molecular Pathways: Emergence of Protein Kinase CK2 (CSNK2) as a Potential Target to Inhibit Survival and DNA Damage Response and Repair Pathways in Cancer Cells. Clin. Cancer Res. 2016, 22, 2840–2847. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G. The Development of CK2 Inhibitors: From Traditional Pharmacology to in Silico Rational Drug Design. Pharmaceuticals 2017, 10, 26. [Google Scholar] [CrossRef]
- Cai, Z.; Luo, Q.; Zhang, Y.; Meng, R.; Tian, M. CK2 in the spotlight: Decoding its role in hematological malignancies and therapeutic applications. Discov. Oncol. 2025, 16, 965. [Google Scholar] [CrossRef] [PubMed]
- Giusiano, S.; Cochet, C.; Filhol, O.; Duchemin-Pelletier, E.; Secq, V.; Bonnier, P.; Carcopino, X.; Boubli, L.; Birnbaum, D.; Garcia, S.; et al. Protein kinase CK2alpha subunit over-expression correlates with metastatic risk in breast carcinomas: Quantitative immunohistochemistry in tissue microarrays. Eur. J. Cancer 2011, 47, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Guo, Y.; Huo, Y.; Shi, J.; Hou, Y. Regulation of cancer progression by CK2: An emerging therapeutic target. Med Oncol. 2024, 41, 94. [Google Scholar] [CrossRef]
- Atkinson, E.L.; Iegre, J.; Brear, P.D.; Zhabina, E.A.; Hyvönen, M.; Spring, D.R. Downfalls of Chemical Probes Acting at the Kinase ATP-Site: CK2 as a Case Study. Molecules 2021, 26, 1977. [Google Scholar] [CrossRef]
- Borad, M.J.; Bai, L.Y.; Richards, D.; Mody, K.; Hubbard, J.; Rha, S.Y.; Soong, J.; McCormick, D.; Tse, E.; O’Brien, D.; et al. Silmitasertib plus gemcitabine and cisplatin first-line therapy in locally advanced/metastatic cholangiocarcinoma: A Phase 1b/2 study. Hepatology 2023, 77, 760–773. [Google Scholar] [CrossRef]
- Perea, S.E.; Baladrón, I.; Valenzuela, C.; Perera, Y. CIGB-300: A peptide-based drug that impairs the Protein Kinase CK2-mediated phosphorylation. Semin. Oncol. 2018, 45, 58–67. [Google Scholar] [CrossRef]
- Lopez-Ramos, M.; Prudent, R.; Moucadel, V.; Sautel, C.F.; Barette, C.; Lafanechere, L.; Mouawad, L.; Grierson, D.; Schmidt, F.; Florent, J.C.; et al. New potent dual inhibitors of CK2 and Pim kinases: Discovery and structural insights. FASEB J. 2010, 24, 3171–3185. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G.; Girardi, C.; Ranchio, A.; Lolli, G.; Sarno, S.; Orzeszko, A.; Kazimierczuk, Z.; Battistutta, R.; Ruzzene, M.; Pinna, L.A. Cell-permeable dual inhibitors of protein kinases CK2 and PIM-1: Structural features and pharmacological potential. Cell Mol. Life Sci. 2014, 71, 3173–3185. [Google Scholar] [CrossRef]
- Zandomeni, R.; Zandomeni, M.C.; Shugar, D.; Weinmann, R. Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J. Biol. Chem. 1986, 261, 3414–3419. [Google Scholar] [CrossRef] [PubMed]
- Zien, P.; Duncan, J.S.; Skierski, J.; Bretner, M.; Litchfield, D.W.; Shugar, D. Tetrabromobenzotriazole (tbbt) and tetrabromobenzimidazole (tbbz) as selective inhibitors of protein kinase CK2: Evaluation of their effects on cells and different molecular forms of human CK2. Biochim. Et Biophys. Acta Proteins Proteom. 2005, 1754, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Pagano, M.A.; Meggio, F.; Ruzzene, M.; Andrzejewska, M.; Kazimierczuk, Z.; Pinna, L.A. 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: A novel powerful and selective inhibitor of protein kinase CK2. Biochem. Biophys. Res. Commun. 2004, 321, 1040–1044. [Google Scholar] [CrossRef]
- Pagano, M.A.; Andrzejewska, M.; Ruzzene, M.; Sarno, S.; Cesaro, L.; Bain, J.; Elliott, M.; Meggio, F.; Kazimierczuk, Z.; Pinna, L.A. Optimization of protein kinase CK2 inhibitors derived from 4,5,6,7-tetrabromobenzimidazole. J. Med. Chem. 2004, 47, 6239–6247. [Google Scholar] [CrossRef]
- Schneider, C.C.; Hessenauer, A.; Götz, C.; Montenarh, M. DMAT, an inhibitor of protein kinase CK2 induces reactive oxygen species and DNA double strand breaks. Oncol. Rep. 2009, 21, 1593–1597. [Google Scholar] [CrossRef][Green Version]
- Martínez, R.; Geronimo, B.D.; Pastor, M.; Zapico, J.M.; Coderch, C.; Panchuk, R.; Skorokhyd, N.; Maslyk, M.; Ramos, A.; de Pascual-Teresa, B. Multitarget Anti-cancer Agents Based on Histone Deacetylase and Protein Kinase CK2 inhibitors. Molecules 2020, 25, 1497. [Google Scholar] [CrossRef]
- Wińska, P.; Wielechowska, M.; Koronkiewicz, M.; Borowiecki, P. Synthesis and Anti-cancer Activity of Novel Dual Inhibitors of Human Protein Kinases CK2 and PIM-1. Pharmaceutics 2023, 15, 1991. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.C.; Kartarius, S.; Montenarh, M.; Orzeszko, A.; Kazimierczuk, Z. Modified tetrahalogenated benzimidazoles with CK2 inhibitory activity are active against human prostate cancer cells LNCaP in vitro. Bioorg. Med. Chem. 2012, 20, 4390–4396. [Google Scholar] [CrossRef]
- Koronkiewicz, M.; Chilmonczyk, Z.; Kazimerczuk, Z.; Orzeszko, A. Deoxynucleosides with benzimidazoles as aglycone moiety are potent anti-cancer agents. Eur. J. Pharmacol. 2018, 820, 146–155. [Google Scholar] [CrossRef]
- Koronkiewicz, M.; Kazimierczuk, Z.; Orzeszko, A. Antitumor activity of the protein kinase inhibitor 1-(β-D-2′-deoxyribofuranosyl)-4,5,6,7-tetrabromo-1H-benzimidazole in breast cancer cell lines. BMC Cancer 2022, 22, 1069. [Google Scholar] [CrossRef]
- Łukowska-Chojnacka, E.; Wińska, P.; Wielechowska, M.; Poprzeczko, M.; Bretner, M. Synthesis of novel polybrominated benzimidazole derivatives-potential CK2 inhibitors with anti-cancer and pro-apoptotic activity. Bioorg. Med. Chem. 2016, 24, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Wińska, P.; Skierka, K.; Łukowska-Chojnacka, E.; Koronkiewicz, M.; Cieśla, J.; Bretner, M. Effect of Simultaneous Inhibition of Protein Kinase CK2 and Thymidylate Synthase in Leukemia and Breast Cancer Cells. Anti-Cancer Res. 2018, 38, 4617–4627. [Google Scholar] [CrossRef]
- Chojnacki, K.; Wińska, P.; Karatsai, O.; Koronkiewicz, M.; Milner-Krawczyk, M.; Wielechowska, M.; Rędowicz, M.J.; Bretner, M.; Borowiecki, P. Synthesis of Novel Acyl Derivatives of 3-(4,5,6,7-Tetrabromo-1H-benzimidazol-1-yl)propan-1-ols-Intracellular TBBi-Based CK2 Inhibitors with Pro-apoptotic Properties. Int. J. Mol. Sci. 2021, 22, 6261. [Google Scholar] [CrossRef] [PubMed]
- Łukowska-Chojnacka, E.; Fedorov, E.; Kowalkowska, A.; Wielechowska, M.; Sobiepanek, A.; Koronkiewicz, M.; Wińska, P. Synthesis and evaluation of anti-cancer activity of new 4,5,6,7-tetrabromo-1H-benzimidazole derivatives. Bioorg. Chem. 2024, 153, 107880. [Google Scholar] [CrossRef]
- Łukowska-Chojnacka, E.; Staniszewska, M.; Bondaryk, M.; Maurin, J.K.; Bretner, M. Lipase-Catalyzed Kinetic Resolution of Novel Antifungal N-Substituted Benzimidazole Derivatives. Chirality 2016, 28, 347–354. [Google Scholar] [CrossRef]
- Battistutta, R.; Mazzorana, M.; Cendron, L.; Bortolato, A.; Sarno, S.; Kazimierczuk, Z.; Zanotti, G.; Moro, S.; Pinna, L.A. The ATP-Binding Site of Protein Kinase CK2 Holds a Positive Electrostatic Area and Conserved Water Molecules. ChemBioChem 2007, 8, 1749–1890. [Google Scholar] [CrossRef]
- Khan, Y.; Rizvi, S.; Raza, A.; Khan, A.; Hussain, S.; Khan, N.U.; Alshammari, S.O.; Alshammari, Q.A.; Alshammari, A.; Ellakwa, D.E. Tailored therapies for triple-negative breast cancer: Current landscape and future perceptions. Naunyn Schmiedebergs Arch. Pharmacol. 2025, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, E.; Armour, S.; Harris, M.; Thompson, C.B. Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ. 2003, 10, 709–717. [Google Scholar] [CrossRef]
- Porter, A.G.; Jänicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef]
- Plesca, D.; Mazumder, S.; Almasan, A. DNA damage response and apoptosis. Methods Enzymol. 2008, 446, 107–122. [Google Scholar] [CrossRef]
- Wang, S.; He, M.; Li, L.; Liang, Z.; Zou, Z.; Tao, A. Cell-in-cell death is not restricted by caspase-3 deficiency in mcf-7 cells. J. Breast Cancer 2016, 19, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Izeradjene, K.; Douglas, L.; Delaney, A.; Houghton, J.A. Casein kinase ii (CK2) enhances death-inducing signaling complex (disc) activity in trail-induced apoptosis in human colon carcinoma cell lines. Oncogene 2005, 24, 2050–2058. [Google Scholar] [CrossRef]
- Hori, M.; Nogami, T.; Itabashi, M.; Yoshim, F.; Ono, H.; Koizumi, S. Expression of Bcl-2 in human breast cancer: Correlation between hormone receptor status, p53 protein accumulation and DNA strand breaks associated with apoptosis. Pathol. Int. 1997, 47, 757–762. [Google Scholar] [CrossRef]
- Zha, J.; Harada, H.; Yang, E.; Jockel, J.; Korsmeyer, S.J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 Not BCL-X(L). Cell 1996, 87, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Lilly, M.; Sandholm, J.; Cooper, J.J.; Koskinen, P.J.; Kraft, A. The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene 1999, 18, 4022–4031. [Google Scholar] [CrossRef]
- Macdonald, A.; Campbell, D.G.; Toth, R.; McLauchlan, H.; Hastie, C.J.; Arthur, J.S. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 2006, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Pierre, F.; Chua, P.C.; O’Brien, S.E.; Siddiqui-Jain, A.; Bourbon, P.; Haddach, M.; Michaux, J.; Nagasawa, J.; Schwaebe, M.K.; Stefan, E.; et al. Pre-clinical characterization of CX-4945, a potent and selective small molecule inhibitor of CK2 for the treatment of cancer. Mol. Cell. Biochem. 2011, 356, 37–43. [Google Scholar] [CrossRef]
- Li, Y.; Dowbenko, D.; Lasky, L.A. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J. Biol. Chem. 2002, 277, 11352–11361. [Google Scholar] [CrossRef]
- Fan, R.F.; Lu, Y.; Fang, Z.G.; Guo, X.Y.; Chen, Y.X.; Xu, Y.C.; Lei, Y.M.; Liu, K.F.; Lin, D.J.; Liu, L.L.; et al. PIM-1 kinase inhibitor SMI-4a exerts antitumor effects in chronic myeloid leukemia cells by enhancing the activity of glycogen synthase kinase 3β. Mol. Med. Rep. 2017, 16, 4603–4612. [Google Scholar] [CrossRef]
- Malinen, M.; Jääskeläinen, T.; Pelkonen, M.; Heikkinen, S.; Väisänen, S.; Kosma, V.M.; Nieminen, K.; Mannermaa, A.; Palvimo, J.J. Proto-oncogene PIM-1 is a novel estrogen receptor target associating with high grade breast tumors. Mol. Cell Endocrinol. 2013, 365, 270–276. [Google Scholar] [CrossRef]
- Horiuchi, D.; Camarda, R.; Zhou, A.Y.; Yau, C.; Momcilovic, O.; Balakrishnan, S.; Corella, A.N.; Eyob, H.; Kessenbrock, K.; Lawson, D.A.; et al. PIM1 kinase inhibition as a targeted therapy against triple-negative breast tumors with elevated MYC expression. Nat. Med. 2016, 22, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- El-Hachem, N.; Haibe-Kains, B.; Khalil, A.; Kobeissy, F.H.; Nemer, G. AutoDock and AutoDockTools for Protein-Ligand Docking: Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1(BACE1) as a Case Study. Methods Mol. Biol. 2017, 1598, 391–403. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- openmm/pdbfixer. (10 April 2025). Python. OpenMM. Available online: https://github.com/openmm/pdbfixer (accessed on 11 April 2025).
- Talagayev, V.; Chen, Y.; Doering, N.P.; Obendorf, L.; Denzinger, K.; Puls, K.; Lam, K.; Liu, S.; Wolf, C.A.; Noonan, T.; et al. OpenMMDL-Simplifying the Complex: Building, Simulating, and Analyzing Protein-Ligand Systems in OpenMM. J. Chem. Inf. Model. 2025, 65, 1967–1978. [Google Scholar] [CrossRef]
- Eastman, P.; Galvelis, R.; Peláez, R.P.; Abreu, C.R.A.; Farr, S.E.; Gallicchio, E.; Gorenko, A.; Henry, M.M.; Hu, F.; Huang, J.; et al. OpenMM 8: Molecular Dynamics Simulation with Machine Learning Potentials. J. Phys. Chem. B. 2024, 128, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Price, D.J.; Brooks, C.L., 3rd. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef] [PubMed]
- Gowers, R.J.; Linke, M.; Barnoud, J.; Reddy, T.; Melo, M.N.; Seyler, S.L.; Domanski, J.J.; Dotson, D.L.; Buchoux, S.; Kenney, I.M.; et al. MDAnalysis: A Python Package for the Rapid Analysis of Molecular Dynamics Simulations. In Proceedings of the 15th Python in Science Conference, Austin, TX, USA, 11–17 July 2016. [Google Scholar] [CrossRef]
- Systèmes, D. “Free Download: BIOVIA Discovery Studio Visualizer,” Dassault Systèmes. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 11 April 2025).




| CK2α | PIM-1 | ||||
|---|---|---|---|---|---|
| Cpd. | Cpd. Concentration [μM] | Residual Activity [%] | IC50 [μM] | Residual Activity [%] | IC50 [μM] |
| 1 | 2 | 58.5 ± 2.2 | 6.65 ± 0.74 | 22.4 ± 0.9 | 0.52 ± 0.02 |
| 5 | 32.3 ± 0.3 | 10.3 ± 0.7 | |||
| 20 | 10.2 ± 0.6 | 2.9 ± 0.2 | |||
| 2 | 2 | 70.9 ± 0.9 | 11.05 ± 0.15 | 58.0 ± 2.4 | 2.41 ± 0.29 |
| 5 | 41.8 ± 2.3 | 26.4 ± 1.7 | |||
| 20 | 20.1 ± 1.2 | 9.3 ± 0.3 | |||
| 3 | 2 | 101.7 ± 8.9 | - | 82.5 ± 4.2 | - |
| 5 | 89.2 ± 8.6 | 67.4 ± 0.7 | |||
| 20 | 82.8 ± 15.4 | 62.6 ± 0.8 | |||
| 4 | 2 | 72.1 ± 3.0 | - | 66.9 ± 1.1 | 3.08 ± 0.07 |
| 5 | 21.3 ± 1.9 | 29.1 ± 0.1 | |||
| 20 | 15.6 ± 0.3 | 21.4 ± 0.2 | |||
| TBBi | 1.16 ± 0.09 | 0.53 ± 0.04 | |||
| Cpd. | IC50 [µM] | SI * | |||||
|---|---|---|---|---|---|---|---|
| CCRF-CEM | MCF-7 | MDA-MB-231 | Vero | CCRF-CEM | MCF-7 | MDA-MB-231 | |
| 1 | 12.43 ± 0.82 | 17.09 ± 2.07 | 21.20 ± 0.76 | 20.20 ± 3.59 | 1.62 | 1.18 | 0.95 |
| 2 | 58.24 ± 5.86 | 18.42 ± 0.41 | 28.17 ± 3.27 | 96.06 ± 4.76 | 1.65 | 5.21 | 3.4 |
| 4 | 75.26 ± 10.30 | 41.86 ± 8.37 | >200 | 132.00 ± 19.36 | 1.75 | 3.15 | - |
| TBBi | 17.56 ± 0.67 | 12.61 ± 2.72 | 29.71 ± 2.23 | 24.80 ± 0.90 | 1.41 | 1.97 | 0.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wińska, P.; Wielechowska, M.; Milewski, Ł.; Siedlecki, P.; Łukowska-Chojnacka, E. Pro-Apoptotic Activity of 1-(4,5,6,7-Tetrabromo-1H-benzimidazol-1-yl)propan-2-one, an Intracellular Inhibitor of PIM-1 Kinase in Acute Lymphoblastic Leukemia and Breast Cancer Cells. Int. J. Mol. Sci. 2025, 26, 5897. https://doi.org/10.3390/ijms26125897
Wińska P, Wielechowska M, Milewski Ł, Siedlecki P, Łukowska-Chojnacka E. Pro-Apoptotic Activity of 1-(4,5,6,7-Tetrabromo-1H-benzimidazol-1-yl)propan-2-one, an Intracellular Inhibitor of PIM-1 Kinase in Acute Lymphoblastic Leukemia and Breast Cancer Cells. International Journal of Molecular Sciences. 2025; 26(12):5897. https://doi.org/10.3390/ijms26125897
Chicago/Turabian StyleWińska, Patrycja, Monika Wielechowska, Łukasz Milewski, Paweł Siedlecki, and Edyta Łukowska-Chojnacka. 2025. "Pro-Apoptotic Activity of 1-(4,5,6,7-Tetrabromo-1H-benzimidazol-1-yl)propan-2-one, an Intracellular Inhibitor of PIM-1 Kinase in Acute Lymphoblastic Leukemia and Breast Cancer Cells" International Journal of Molecular Sciences 26, no. 12: 5897. https://doi.org/10.3390/ijms26125897
APA StyleWińska, P., Wielechowska, M., Milewski, Ł., Siedlecki, P., & Łukowska-Chojnacka, E. (2025). Pro-Apoptotic Activity of 1-(4,5,6,7-Tetrabromo-1H-benzimidazol-1-yl)propan-2-one, an Intracellular Inhibitor of PIM-1 Kinase in Acute Lymphoblastic Leukemia and Breast Cancer Cells. International Journal of Molecular Sciences, 26(12), 5897. https://doi.org/10.3390/ijms26125897







