Association of Gene Expression Profiles in HPV-Positive Head and Neck Squamous Cell Carcinoma with Patient Outcome: In Search of Prognostic Biomarkers
Abstract
1. Introduction
2. Results
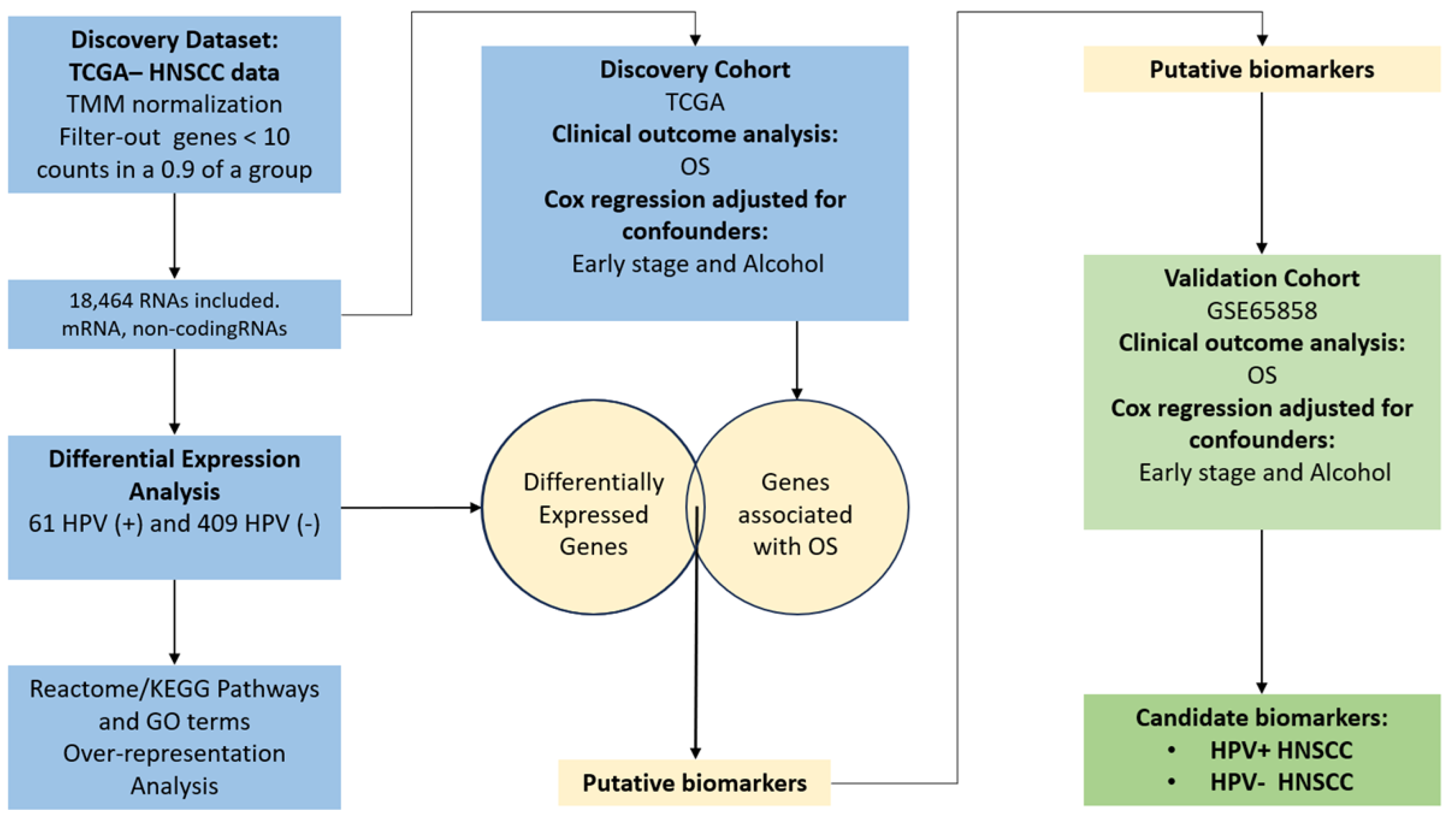
2.1. Selection of TCGA HNSCC Samples According to HPV-Status for the Discovery Cohort
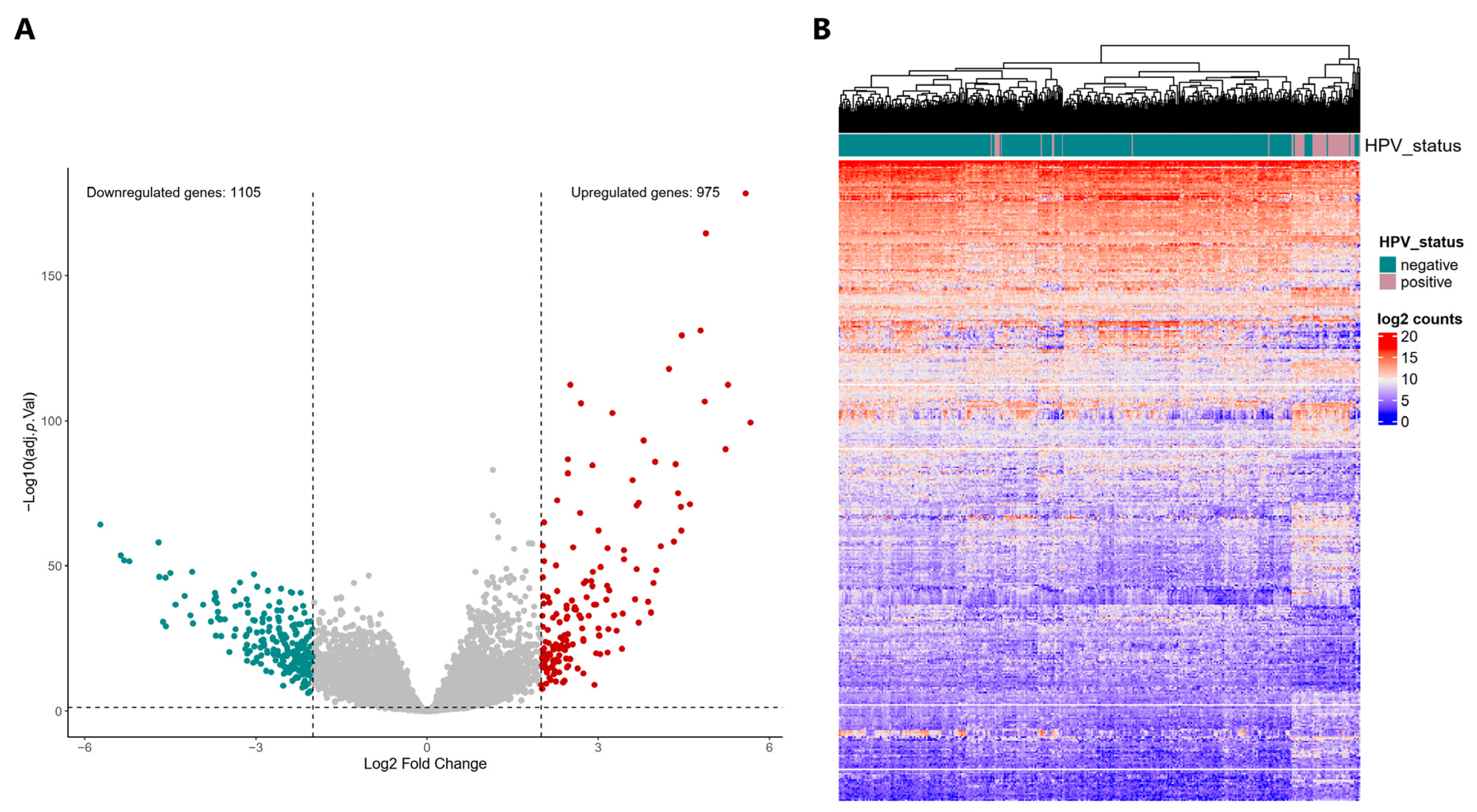
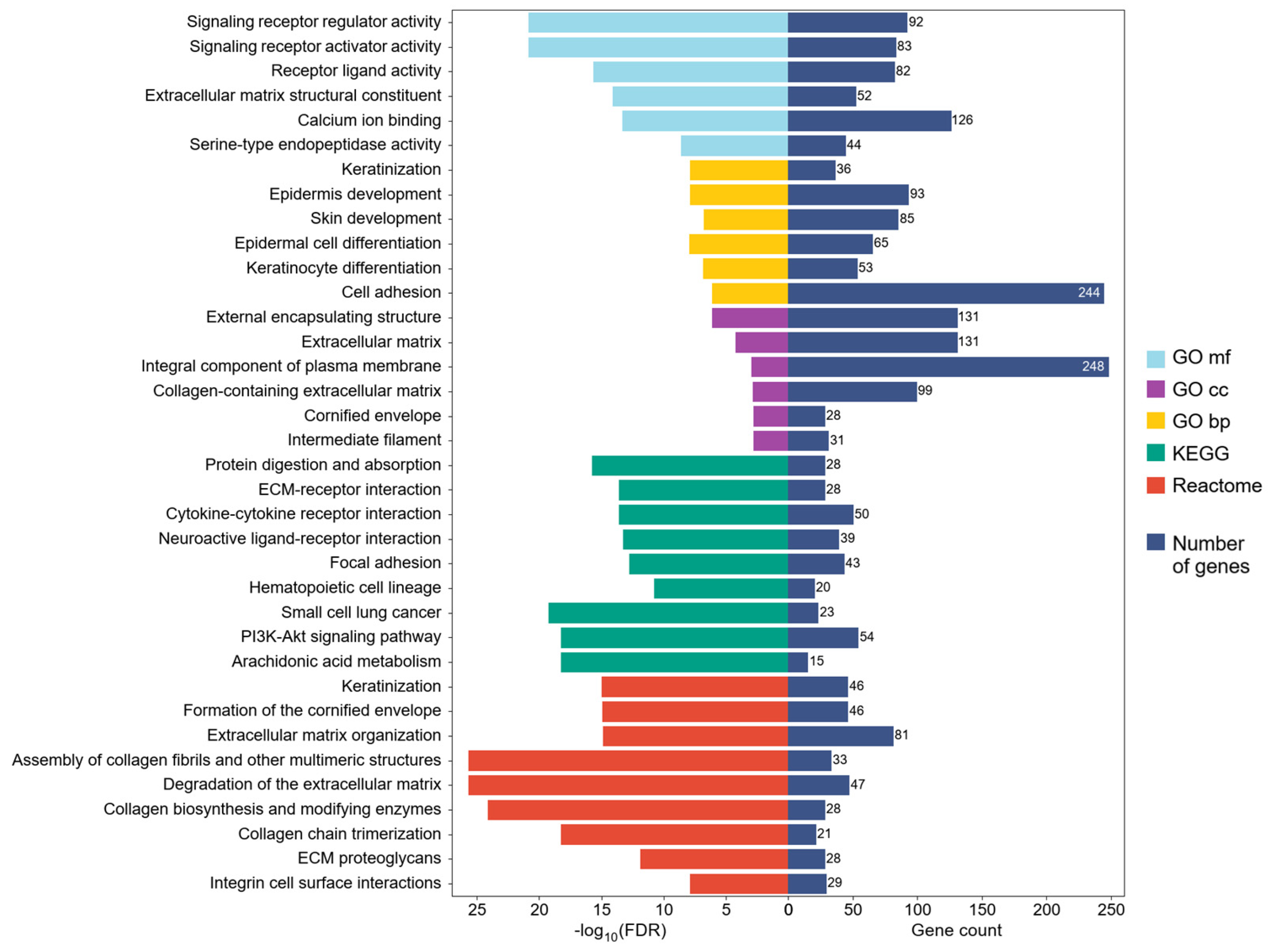
2.2. Differential Expression Profiles in HPV-Positive HNSCC and Their Potential Implication in Biological Processes, Molecular Functions, and Signaling Pathways
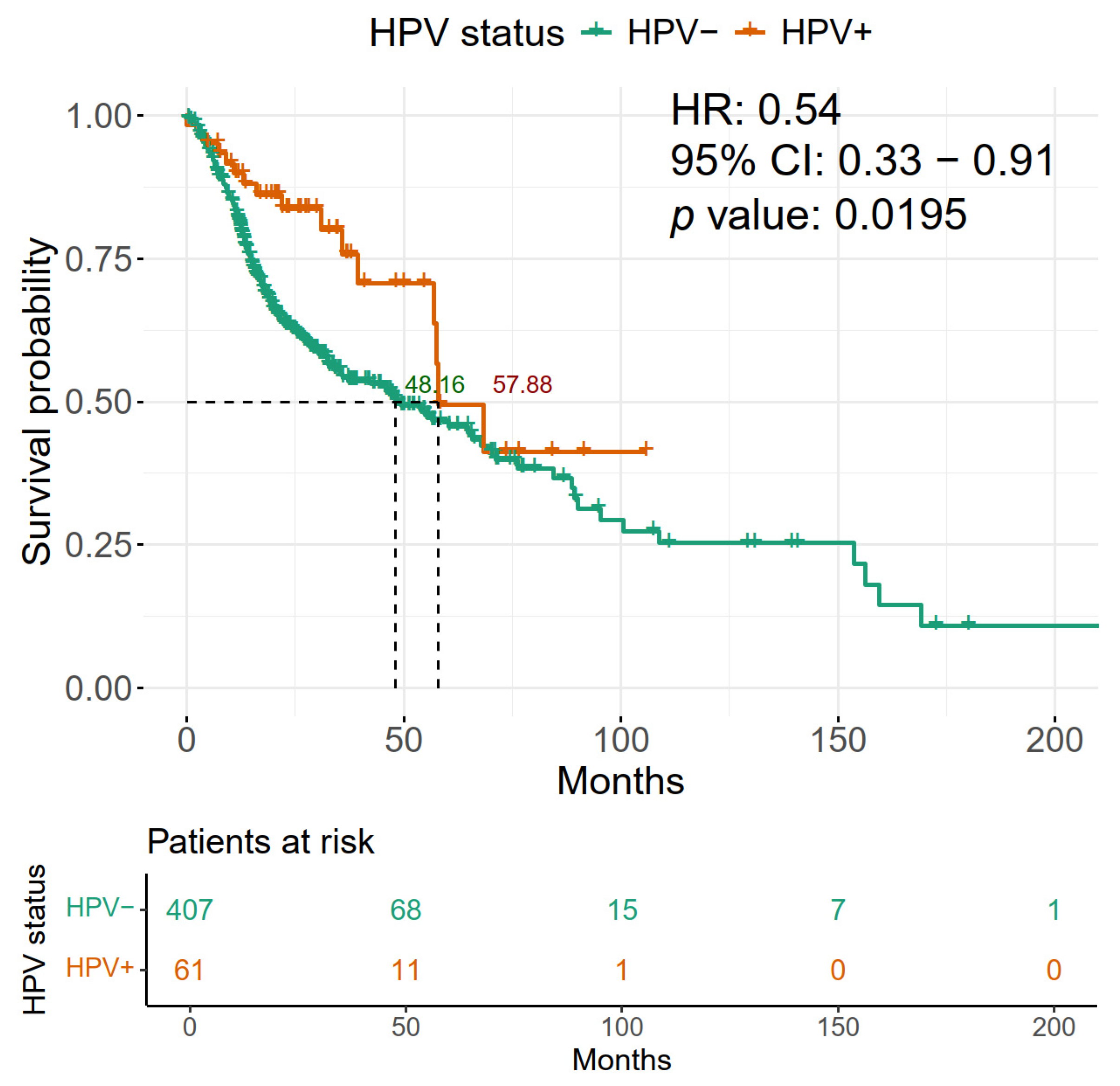
2.3. HPV Positivity of HNSCC Is Associated with Improved Overall Survival
2.4. Differentially Regulated Genes in HPV-Positive HNSCC Show an Association with Patient Overall Survival
2.5. Confounding Factors Analysis Using Multivariate Cox Models Reveals Independent Biomarkers for HPV-Positive and HPV-Negative HNSCC Patients
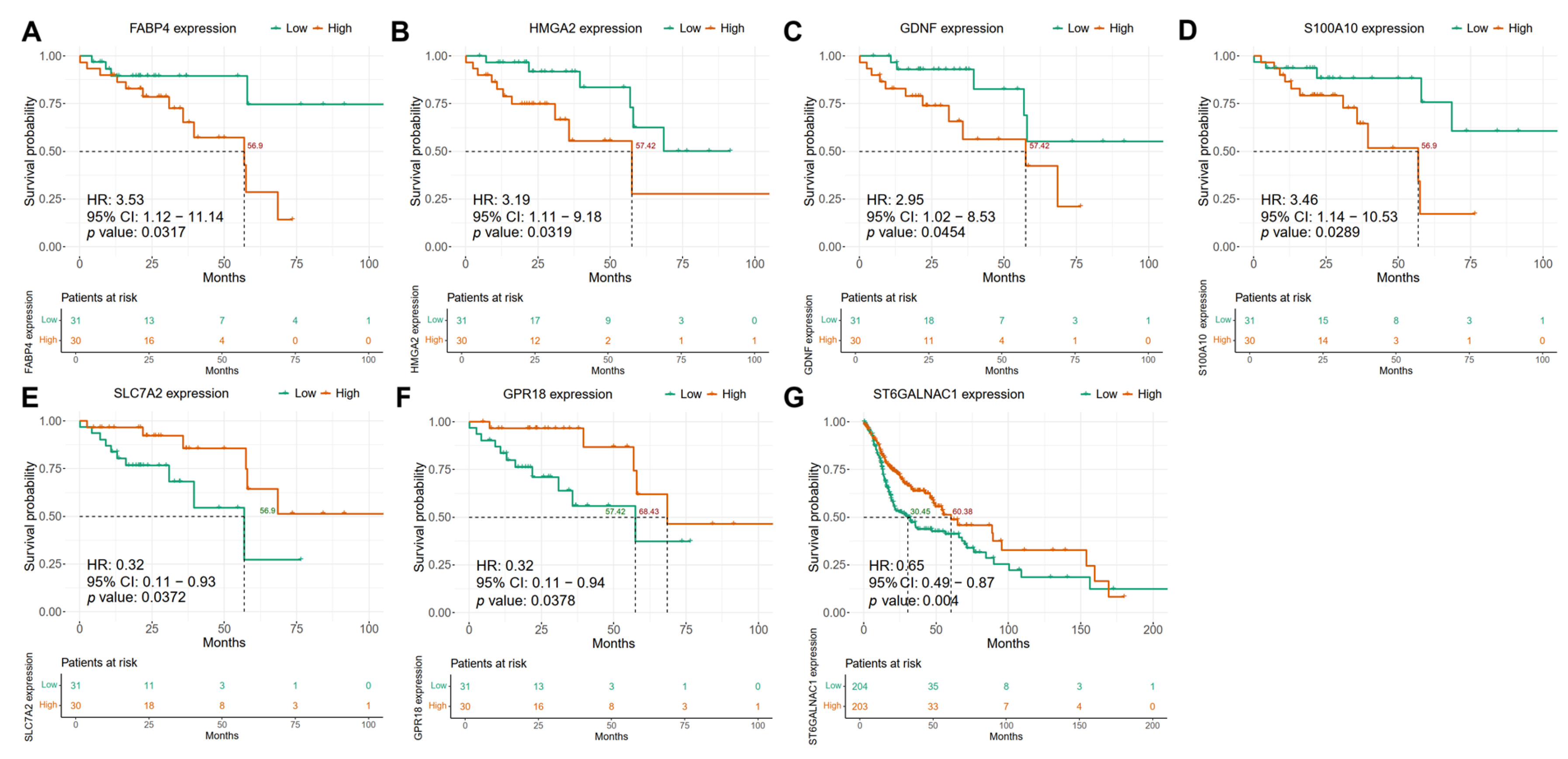
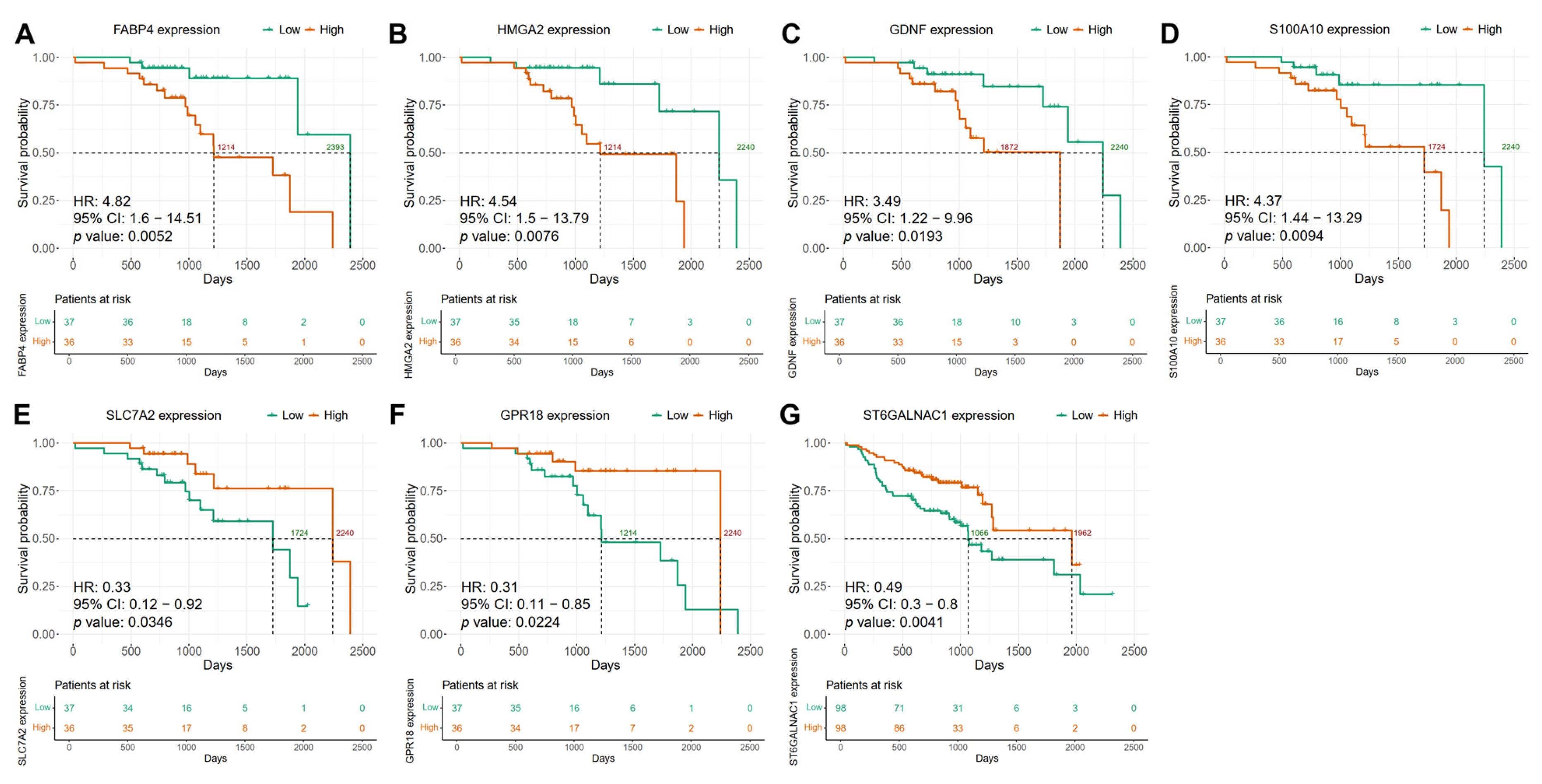
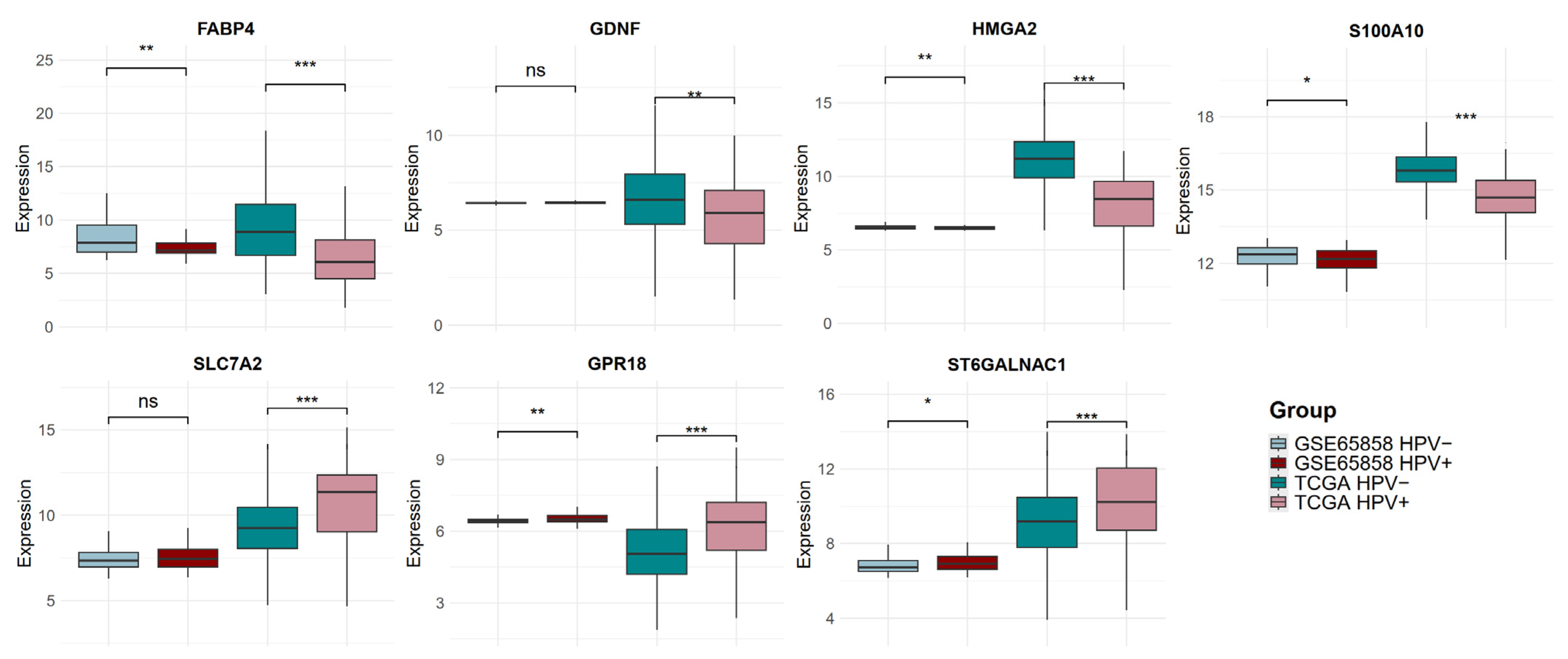
2.6. The Expression of Candidate Biomarker Genes Is Associated with the Clinical Outcome of Patients with HNSCC in the Discovery and Validation Cohorts
3. Discussion
4. Methods
4.1. Data Collection and Pre-Processing of HNSCC Transcriptomics
4.2. Data Normalization and Differential Gene Expression Analysis of HNSCC HPV-Positive vs. HPV-Negative HNSCC
4.3. Function Annotation of Differentially Expressed Genes in HPV-Positive HNSCC
4.4. Clinical Outcome Analysis in HPV-Positive HNSCC from TCGA and Validation Cohorts
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer Cancer Today. Data Visualization Tools for Exploring the Global Cancer Burden in 2022. Available online: https://gco.iarc.fr/today/en (accessed on 27 April 2025).
- Berthiller, J.; Straif, K.; Agudo, A.; Ahrens, W.; Dos Santos, A.B.; Boccia, S.; Cadoni, G.; Canova, C.; Castellsague, X.; Chen, C.; et al. Low Frequency of Cigarette Smoking and the Risk of Head and Neck Cancer in the INHANCE Consortium Pooled Analysis. Int. J. Epidemiol. 2016, 45, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Di Credico, G.; Polesel, J.; Dal Maso, L.; Pauli, F.; Torelli, N.; Luce, D.; Radoï, L.; Matsuo, K.; Serraino, D.; Brennan, P.; et al. Alcohol Drinking and Head and Neck Cancer Risk: The Joint Effect of Intensity and Duration. Br. J. Cancer 2020, 123, 1456–1463. [Google Scholar] [CrossRef] [PubMed]
- Guha, N.; Warnakulasuriya, S.; Vlaanderen, J.; Straif, K. Betel Quid Chewing and the Risk of Oral and Oropharyngeal Cancers: A Meta-Analysis with Implications for Cancer Control. Int. J. Cancer 2014, 135, 1433–1443. [Google Scholar] [CrossRef]
- Mork, J.; Lie, A.K.; Glattre, E.; Clark, S.; Hallmans, G.; Jellum, E.; Koskela, P.; Møller, B.; Pukkala, E.; Schiller, J.T.; et al. Human Papillomavirus Infection as a Risk Factor for Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2001, 344, 1125–1131. [Google Scholar] [CrossRef]
- Kreimer, A.R.; Clifford, G.M.; Boyle, P.; Franceschi, S. Human Papillomavirus Types in Head and Neck Squamous Cell Carcinomas Worldwide: A Systematic Review. Cancer Epidemiol. Biomark. Prev. 2005, 14, 467–475. [Google Scholar] [CrossRef]
- Ndiaye, C.; Mena, M.; Alemany, L.; Arbyn, M.; Castellsagué, X.; Laporte, L.; Bosch, F.X.; de Sanjosé, S.; Trottier, H. HPV DNA, E6/E7 MRNA, and P16INK4a Detection in Head and Neck Cancers: A Systematic Review and Meta-Analysis. Lancet Oncol. 2014, 15, 1319–1331. [Google Scholar] [CrossRef]
- Vani, N.V.; Rama, R.; Madhanagopal, R.; Vijayalakshmi, R.; Swaminathan, R. Human Papillomavirus-Attributable Head and Neck Cancers in India—A Systematic Review and Meta-Analysis. JCO Glob. Oncol. 2024, 10, e2300464. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human Papillomavirus and Rising Oropharyngeal Cancer Incidence in the United States. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef]
- Rautava, J.; Syrjänen, S. Biology of Human Papillomavirus Infections in Head and Neck Carcinogenesis. Head Neck Pathol. 2012, 6 (Suppl. S1), 3–15. [Google Scholar] [CrossRef]
- Qin, T.; Li, S.; Henry, L.E.; Liu, S.; Sartor, M.A. Molecular Tumor Subtypes of HPV-Positive Head and Neck Cancers: Biological Characteristics and Implications for Clinical Outcomes. Cancers 2021, 13, 2721. [Google Scholar] [CrossRef] [PubMed]
- Fakhry, C.; Westra, W.H.; Li, S.; Cmelak, A.; Ridge, J.A.; Pinto, H.; Forastiere, A.; Gillison, M.L. Improved Survival of Patients with Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma in a Prospective Clinical Trial. J. Natl. Cancer Inst. 2008, 100, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Wuerdemann, N.; Wittekindt, C.; Sharma, S.J.; Prigge, E.S.; Reuschenbach, M.; Gattenlöhner, S.; Klussmann, J.P.; Wagner, S. Risk Factors for Overall Survival Outcome in Surgically Treated Human Papillomavirus-Negative and Positive Patients with Oropharyngeal Cancer. Oncol. Res. Treat. 2017, 40, 320–327. [Google Scholar] [CrossRef]
- Yang, M.-Q.; Liu, Y.-C.; Sui, J.-D.; Jin, F.; Li, D.; Zhang, L.; Wang, N.-H.; Xie, Y.; Wang, Y.; Wu, Y.-Z. Reduced-Dose Radiation in Human Papillomavirus-Associated Oropharyngeal Carcinoma Can Improve Outcome: A Systematic Review and Meta-Analysis. Ann. Transl. Med. 2022, 10, 1391. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; McBride, S.M.; Riaz, N.; Kang, J.J.; Spielsinger, D.J.; Waldenberg, T.; Gelblum, D.; Yu, Y.; Chen, L.C.; Zakeri, K.; et al. Evaluation of Substantial Reduction in Elective Radiotherapy Dose and Field in Patients with Human Papillomavirus-Associated Oropharyngeal Carcinoma Treated with Definitive Chemoradiotherapy. JAMA Oncol. 2022, 8, 364–372. [Google Scholar] [CrossRef]
- Muñoz-Bello, J.O.; Romero-Córdoba, S.L.; García-Chávez, J.N.; González-Espinosa, C.; Langley, E.; Lizano, M. Potential Transcript-Based Biomarkers Predicting Clinical Outcomes of HPV-Positive Head and Neck Squamous Cell Carcinoma Patients. Cells 2024, 13, 1107. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, L.; Qiu, J.; Xu, J.; Yang, X.; Zhang, Y.; Tian, X.; Guan, X.; Cen, X.; Zhao, Y. Tumor Biomarkers for Diagnosis, Prognosis and Targeted Therapy. Signal Transduct. Target. Ther. 2024, 9, 132. [Google Scholar] [CrossRef]
- Das, S.; Dey, M.K.; Devireddy, R.; Gartia, M.R. Biomarkers in Cancer Detection, Diagnosis, and Prognosis. Sensors 2023, 24, 37. [Google Scholar] [CrossRef]
- Nska, M.K.; Kolenda, T.; Guglas, K.; Nska, J.S.; Teresiak, A.; Zniak, R.B.; Mackiewicz, A.; Mackiewicz, J.; Lamperska, K. PRINS LncRNA Is a New Biomarker Candidate for HPV Infection and Prognosis of Head and Neck Squamous Cell Carcinomas. Diagnostics 2020, 10, 762. [Google Scholar] [CrossRef]
- Méndez-Matías, G.; Velázquez-Velázquez, C.; Castro-Oropeza, R.; Mantilla-Morales, A.; Ocampo-Sandoval, D.; Burgos-González, A.; Heredia-Gutiérrez, C.; Alvarado-Cabrero, I.; Sánchez-Sandoval, R.; Barco-Bazán, A.; et al. Prevalence of HPV in Mexican Patients with Head and Neck Squamous Carcinoma and Identification of Potential Prognostic Biomarkers. Cancers 2021, 13, 5602. [Google Scholar] [CrossRef]
- Luo, X.J.; Zheng, M.; Cao, M.X.; Zhang, W.L.; Huang, M.C.; Dai, L.; Tang, Y.L.; Liang, X.H. Distinguishable Prognostic MiRNA Signatures of Head and Neck Squamous Cell Cancer with or Without HPV Infection. Front. Oncol. 2021, 10, 614487. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA Sequencing: New Technologies and Applications in Cancer Research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Nulton, T.J.; Olex, A.L.; Dozmorov, M.; Morgan, I.M.; Windle, B. Analysis of The Cancer Genome Atlas Sequencing Data Reveals Novel Properties of the Human Papillomavirus 16 Genome in Head and Neck Squamous Cell Carcinoma. Oncotarget 2017, 8, 17684–17699. [Google Scholar] [CrossRef]
- Campbell, J.D.; Yau, C.; Bowlby, R.; Liu, Y.; Brennan, K.; Fan, H.; Taylor, A.M.; Wang, C.; Walter, V.; Akbani, R.; et al. Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas. Cell Rep. 2018, 23, 194–212.e6. [Google Scholar] [CrossRef]
- Bubie, A.; Zoulim, F.; Testoni, B.; Miles, B.; Posner, M.; Villanueva, A.; Losic, B. Landscape of Oncoviral Genotype and Co-Infection via Human Papilloma and Hepatitis B Viral Tumor in Situ Profiling. iScience 2021, 24, 102368. [Google Scholar] [CrossRef]
- Wichmann, G.; Rosolowski, M.; Krohn, K.; Kreuz, M.; Boehm, A.; Reiche, A.; Scharrer, U.; Halama, D.; Bertolini, J.; Bauer, U.; et al. The Role of HPV RNA Transcription, Immune Response-Related Gene Expression and Disruptive TP53 Mutations in Diagnostic and Prognostic Profiling of Head and Neck Cancer. Int. J. Cancer 2015, 137, 2846–2857. [Google Scholar] [CrossRef]
- Albers, A.E.; Qian, X.; Kaufmann, A.M.; Coordes, A. Meta Analysis: HPV and P16 Pattern Determines Survival in Patients with HNSCC and Identifies Potential New Biologic Subtype. Sci. Rep. 2017, 7, 16715. [Google Scholar] [CrossRef]
- Sharkey Ochoa, I.; O’Regan, E.; Toner, M.; Kay, E.; Faul, P.; O’Keane, C.; O’Connor, R.; Mullen, D.; Nur, M.; O’Murchu, E.; et al. The Role of HPV in Determining Treatment, Survival, and Prognosis of Head and Neck Squamous Cell Carcinoma. Cancers 2022, 14, 4321. [Google Scholar] [CrossRef]
- Sabatini, M.E.; Chiocca, S. Human Papillomavirus as a Driver of Head and Neck Cancers. Br. J. Cancer 2020, 122, 306–314. [Google Scholar] [CrossRef]
- Reyes-Hernández, D.O.; Morán-Torres, A.; Jimenez-Lima, R.; Cano-Valdez, A.M.; Cortés-González, C.C.; Castro-Muñoz, L.J.; Olmedo-Nieva, L.; Maldonado-Frías, S.; Pazos-Salazar, N.G.; de Jesús Marín-Aquíno, J.; et al. HPV Prevalence and Predictive Biomarkers for Oropharyngeal Squamous Cell Carcinoma in Mexican Patients. Pathogens 2022, 11, 1527. [Google Scholar] [CrossRef]
- Rischin, D.; Young, R.J.; Fisher, R.; Fox, S.B.; Le, Q.T.; Peters, L.J.; Solomon, B.; Choi, J.; O’Sullivan, B.; Kenny, L.M.; et al. Prognostic Significance of P16INK4A and Human Papillomavirus in Patients with Oropharyngeal Cancer Treated on TROG 02.02 Phase III Trial. J. Clin. Oncol. 2010, 28, 4142–4148. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human Papillomavirus and Survival of Patients with Oropharyngeal Cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef]
- Christopherson, K.M.; Moreno, A.C.; Elgohari, B.; Gross, N.; Ferrarotto, R.; Mohamed, A.S.R.; Brandon Gunn, G.; Goepfert, R.P.; Mott, F.E.; Shah, S.J.; et al. Outcomes after Salvage for HPV-Positive Recurrent Oropharyngeal Cancer Treated with Primary Radiation. Oral Oncol. 2021, 113, 105125. [Google Scholar] [CrossRef]
- Ferris, R.L.; Flamand, Y.; Weinstein, G.S.; Li, S.; Quon, H.; Mehra, R.; Garcia, J.J.; Chung, C.H.; Gillison, M.L.; Duvvuri, U.; et al. Phase II Randomized Trial of Transoral Surgery and Low-Dose Intensity Modulated Radiation Therapy in Resectable P161 Locally Advanced Oropharynx Cancer: An ECOG-ACRIN Cancer Research Group Trial (E3311). J. Clin. Oncol. 2022, 40, 138–149. [Google Scholar] [CrossRef]
- Mesía, R.; Taberna, M. HPV-Related Oropharyngeal Carcinoma de-Escalation Protocols. Lancet Oncol. 2017, 18, 704–705. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Z.; Qiu, S.; Wang, R. Therapeutic Strategies of Different HPV Status in Head and Neck Squamous Cell Carcinoma. Int. J. Biol. Sci. 2021, 17, 1104–1118. [Google Scholar] [CrossRef]
- Marur, S.; Li, S.; Cmelak, A.J.; Gillison, M.L.; Zhao, W.J.; Ferris, R.L.; Westra, W.H.; Gilbert, J.; Bauman, J.E.; Wagner, L.I.; et al. E1308: Phase II Trial of Induction Chemotherapy Followed by Reduced-Dose Radiation and Weekly Cetuximab in Patients With HPV-Associated Resectable Squamous Cell Carcinoma of the Oropharynx- ECOG-ACRIN Cancer Research Group. J. Clin. Oncol. 2017, 35, 490–497. [Google Scholar] [CrossRef]
- Chen, A.M.; Felix, C.; Wang, P.C.; Hsu, S.; Basehart, V.; Garst, J.; Beron, P.; Wong, D.; Rosove, M.H.; Rao, S.; et al. Reduced-Dose Radiotherapy for Human Papillomavirus-Associated Squamous-Cell Carcinoma of the Oropharynx: A Single-Arm, Phase 2 Study. Lancet Oncol. 2017, 18, 803–811. [Google Scholar] [CrossRef]
- O’Sullivan, B.; Huang, S.H.; Siu, L.L.; Waldron, J.; Zhao, H.; Perez-Ordonez, B.; Weinreb, I.; Kim, J.; Ringash, J.; Bayley, A.; et al. Deintensification Candidate Subgroups in Human Papillomavirus-Related Oropharyngeal Cancer According to Minimal Risk of Distant Metastasis. J. Clin. Oncol. 2013, 31, 543–550. [Google Scholar] [CrossRef]
- Augustin, J.G.; Lepine, C.; Morini, A.; Brunet, A.; Veyer, D.; Brochard, C.; Mirghani, H.; Péré, H.; Badoual, C. HPV Detection in Head and Neck Squamous Cell Carcinomas: What Is the Issue? Front. Oncol. 2020, 10, 1751. [Google Scholar] [CrossRef]
- Hinić, S.; Rich, A.; Anayannis, N.V.; Cabarcas-Petroski, S.; Schramm, L.; Meneses, P.I. Gene Expression and DNA Methylation in Human Papillomavirus Positive and Negative Head and Neck Squamous Cell Carcinomas. Int. J. Mol. Sci. 2022, 23, 967. [Google Scholar] [CrossRef]
- Furuhashi, M.; Hotamisligil, G.S. Fatty Acid-Binding Proteins: Role in Metabolic Diseases and Potential as Drug Targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef]
- Hao, J.; Yan, F.; Zhang, Y.; Triplett, A.; Zhang, Y.; Schultz, D.A.; Sun, Y.; Zeng, J.; Silverstein, K.A.T.; Zheng, Q.; et al. Expression of Adipocyte/Macrophage Fatty Acid-Binding Protein in Tumor-Associated Macrophages Promotes Breast Cancer Progression. Cancer Res. 2018, 78, 2343–2355. [Google Scholar] [CrossRef]
- Harjes, U.; Bridges, E.; Gharpure, K.M.; Roxanis, I.; Sheldon, H.; Miranda, F.; Mangala, L.S.; Pradeep, S.; Lopez-Berestein, G.; Ahmed, A.; et al. Antiangiogenic and Tumour Inhibitory Effects of Downregulating Tumour Endothelial FABP4. Oncogene 2017, 36, 912–921. [Google Scholar] [CrossRef]
- Tang, Z.; Shen, Q.; Xie, H.; Zhou, X.; Li, J.; Feng, J.; Liu, H.; Wang, W.; Zhang, S.; Ni, S. Elevated Expression of FABP3 and FABP4 Cooperatively Correlates with Poor Prognosis in Non-Small Cell Lung Cancer (NSCLC). Oncotarget 2016, 7, 46253–46262. [Google Scholar] [CrossRef]
- Yang, J.; Liu, X.; Shao, Y.; Zhou, H.; Pang, L.; Zhu, W. Diagnostic, Prognostic, and Immunological Roles of FABP4 in Pancancer: A Bioinformatics Analysis. Comput. Math. Methods Med. 2022, 2022, 3764914. [Google Scholar] [CrossRef]
- Mathis, C.; Lascombe, I.; Monnien, F.; Bittard, H.; Kleinclauss, F.; Bedgedjian, I.; Fauconnet, S.; Valmary-Degano, S. Down-Regulation of A-FABP Predicts Non-Muscle Invasive Bladder Cancer Progression: Investigation with a Long Term Clinical Follow-Up. BMC Cancer 2018, 18, 1239. [Google Scholar] [CrossRef]
- Zhong, C.Q.; Zhang, X.P.; Ma, N.; Zhang, E.B.; Li, J.J.; Jiang, Y.B.; Gao, Y.Z.; Yuan, Y.M.; Lan, S.Q.; Xie, D.; et al. FABP4 Suppresses Proliferation and Invasion of Hepatocellular Carcinoma Cells and Predicts a Poor Prognosis for Hepatocellular Carcinoma. Cancer Med. 2018, 7, 2629–2640. [Google Scholar] [CrossRef]
- Saizonou, I.; Lascombe, I.; Monnien, F.; Bedgedjian, I.; Kleinclauss, F.; Algros, M.P.; Fauconnet, S. Concomitant Decrease of E- and A-FABP Expression Predicts Worse Survival in Urothelial Bladder Cancer Patients. Sci. Rep. 2024, 14, 15390. [Google Scholar] [CrossRef]
- Li, C.; Ma, Y.; Fei, F.; Zheng, M.; Li, Z.; Zhao, Q.; Du, J.; Liu, K.; Lu, R.; Zhang, S. Critical Role and Its Underlying Molecular Events of the Plasminogen Receptor, S100A10 in Malignant Tumor and Non-Tumor Diseases. J. Cancer 2020, 11, 826–836. [Google Scholar] [CrossRef]
- Zhou, X.; Shi, M.; Cao, J.; Yuan, T.; Yu, G.; Chen, Y.; Fang, W.; Li, H. S100 Calcium Binding Protein A10, A Novel Oncogene, Promotes the Proliferation, Invasion, and Migration of Hepatocellular Carcinoma. Front. Genet. 2021, 12, 695036. [Google Scholar] [CrossRef]
- Bydoun, M.; Sterea, A.; Liptay, H.; Uzans, A.; Huang, W.Y.; Rodrigues, G.J.; Weaver, I.C.G.; Gu, H.; Waisman, D.M. S100A10, a Novel Biomarker in Pancreatic Ductal Adenocarcinoma. Mol. Oncol. 2018, 12, 1895–1916. [Google Scholar] [CrossRef]
- Ma, K.; Chen, S.; Chen, X.; Yang, C.; Yang, J. S100A10 Is a New Prognostic Biomarker Related to the Malignant Molecular Features and Immunosuppression Process of Adult Gliomas. World Neurosurg. 2022, 165, e650–e663. [Google Scholar] [CrossRef]
- Vignali, R.; Marracci, S. HMGA Genes and Proteins in Development and Evolution. Int. J. Mol. Sci. 2020, 21, 654. [Google Scholar] [CrossRef]
- Binabaj, M.M.; Soleimani, A.; Rahmani, F.; Avan, A.; Khazaei, M.; Fiuji, H.; Soleimanpour, S.; Ryzhikov, M.; Ferns, G.A.; Bahrami, A.; et al. Prognostic Value of High Mobility Group Protein A2 (HMGA2) over-Expression in Cancer Progression. Gene 2019, 706, 131–139. [Google Scholar] [CrossRef]
- Yamazaki, H.; Mori, T.; Yazawa, M.; Maeshima, A.M.; Matsumoto, F.; Yoshimoto, S.; Ota, Y.; Kaneko, A.; Tsuda, H.; Kanai, Y. Stem Cell Self-Renewal Factors Bmi1 and HMGA2 in Head and Neck Squamous Cell Carcinoma: Clues for Diagnosis. Lab. Investig. 2013, 93, 1331–1338. [Google Scholar] [CrossRef]
- Li, Q.; Chen, W.; Song, M.; Chen, W.; Yang, Z.; Yang, A. Weighted Gene Co-Expression Network Analysis and Prognostic Analysis Identifies Hub Genes and the Molecular Mechanism Related to Head and Neck Squamous Cell Carcinoma. Cancer Biol. Ther. 2019, 20, 750–759. [Google Scholar] [CrossRef]
- Chang, K.P.; Lin, S.J.; Liu, S.C.; Yi, J.S.; Chien, K.Y.; Chi, L.M.; Kao, H.K.; Liang, Y.; Lin, Y.T.; Chang, Y.S.; et al. Low-Molecular-Mass Secretome Profiling Identifies HMGA2 and MIF as Prognostic Biomarkers for Oral Cavity Squamous Cell Carcinoma. Sci. Rep. 2015, 5, 11689. [Google Scholar] [CrossRef]
- Günther, K.; Foraita, R.; Friemel, J.; Günther, F.; Bullerdiek, J.; Nimzyk, R.; Markowski, D.N.; Behrens, T.; Ahrens, W. The Stem Cell Factor HMGA2 Is Expressed in Non-HPV-Associated Head and Neck Squamous Cell Carcinoma and Predicts Patient Survival of Distinct Subsites. Cancer Epidemiol. Biomark. Prev. 2017, 26, 197–205. [Google Scholar] [CrossRef]
- Qiao, L.; Liang, Z.; Ma, P.; Zhang, S.; Sun, C.; Luo, W.; Yu, L. HMGA2 Interacts with KAT6A to Regulate MMPs Chromatin Architecture and Promote Triple-Negative Breast Cancer Metastasis. Front. Immunol. 2025, 16, 1590368. [Google Scholar] [CrossRef]
- Li, Z.; Wu, X.; Li, J.; Yu, S.; Ke, X.; Yan, T.; Zhu, Y.; Cheng, J.; Yang, J. HMGA2-Snai2 Axis Regulates Tumorigenicity and Stemness of Head and Neck Squamous Cell Carcinoma. Exp. Cell Res. 2022, 418, 113271. [Google Scholar] [CrossRef]
- Airaksinen, M.S.; Saarma, M. The GDNF Family: Signalling, Biological Functions and Therapeutic Value. Nat. Rev. Neurosci. 2002, 3, 383–394. [Google Scholar] [CrossRef]
- Mahato, A.K.; Sidorova, Y.A. Glial Cell Line-Derived Neurotrophic Factors (GFLs) and Small Molecules Targeting RET Receptor for the Treatment of Pain and Parkinson’s Disease. Cell Tissue Res. 2020, 382, 147–160. [Google Scholar] [CrossRef]
- Lin, C.; Cao, W.; Ren, Z.; Tang, Y.; Zhang, C.; Yang, R.; Chen, Y.; Liu, Z.; Peng, C.; Wang, L.; et al. GDNF Secreted by Nerves Enhances PD-L1 Expression via JAK2-STAT1 Signaling Activation in HNSCC. Oncoimmunology 2017, 6, e1353860. [Google Scholar] [CrossRef]
- Cao, H.; He, Q.; von Eyben, R.; Bloomstein, J.; Nambiar, D.K.; Viswanathan, V.; Aggarwal, S.; Kwok, S.; Liang, R.; Koong, A.J.; et al. The Role of Glial Cell Derived Neurotrophic Factor in Head and Neck Cancer. PLoS ONE 2020, 15, 229311. [Google Scholar] [CrossRef]
- Sheng, L.; Luo, Q.; Chen, L. Special Section on New Era of Transporter Science: Unraveling the Functional Role of Orphan Transporters-Minireview Amino Acid Solute Carrier Transporters in Inflammation and Autoimmunity. Drug Metab. Dispos. 2022, 50, 1228–1237. [Google Scholar] [CrossRef]
- Rashid, K.; Ahmad, A.; Liang, L.; Liu, M.; Cui, Y.; Liu, T. Solute Carriers as Potential Oncodrivers or Suppressors: Their Key Functions in Malignant Tumor Formation. Drug Discov. Today 2021, 26, 1689–1701. [Google Scholar] [CrossRef]
- Song, K.; Li, Y.; Yang, K.; Lu, T.; Wang, M.; Wang, Z.; Liu, C.; Yu, M.; Wang, M.; Cheng, Z.; et al. Regulatory Effects of SLC7A2-CPB2 on Lymphangiogenesis: A New Approach to Suppress Lymphatic Metastasis in HNSCC. Cancer Med. 2024, 13, e70273. [Google Scholar] [CrossRef]
- Console-Bram, L.; Brailoiu, E.; Brailoiu, G.C.; Sharir, H.; Abood, M.E. Activation of GPR18 by Cannabinoid Compounds: A Tale of Biased Agonism. Br. J. Pharmacol. 2014, 171, 3908–3917. [Google Scholar] [CrossRef]
- Kohno, M.; Hasegawa, H.; Inoue, A.; Muraoka, M.; Miyazaki, T.; Oka, K.; Yasukawa, M. Identification of N-Arachidonylglycine as the Endogenous Ligand for Orphan G-Protein-Coupled Receptor GPR18. Biochem. Biophys. Res. Commun. 2006, 347, 827–832. [Google Scholar] [CrossRef]
- Honkisz-Orzechowska, E.; Łażewska, D.; Baran, G.; Kieć-Kononowicz, K. Uncovering the Power of GPR18 Signalling: How RvD2 and Other Ligands Could Have the Potential to Modulate and Resolve Inflammation in Various Health Disorders. Molecules 2024, 29, 1258. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, Q.; Yao, L.; Wang, S.; Zhang, Z. Comprehensive Analysis Reveals Novel Gene Signature in Head and Neck Squamous Cell Carcinoma: Predicting Is Associated with Poor Prognosis in Patients. Transl. Cancer Res. 2020, 9, 5882–5892. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Verdegaal, E.M.E.; Siderius, M.; Bebelman, J.P.; Smit, M.J.; Leurs, R.; Willemze, R.; Tensen, C.P.; Osanto, S. Quantitative Expression Profiling of G-Protein-Coupled Receptors (GPCRs) in Metastatic Melanoma: The Constitutively Active Orphan GPCR GPR18 as Novel Drug Target. Pigment Cell Melanoma Res. 2011, 24, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Sewell, R.; Bäckström, M.; Dalziel, M.; Gschmeissner, S.; Karlsson, H.; Noll, T.; Gätgens, J.; Clausen, H.; Hansson, G.C.; Burchell, J.; et al. The ST6GalNAc-I Sialyltransferase Localizes throughout the Golgi and Is Responsible for the Synthesis of the Tumor-Associated Sialyl-Tn O-Glycan in Human Breast Cancer. J. Biol. Chem. 2006, 281, 3586–3594. [Google Scholar] [CrossRef]
- Wang, W.Y.; Cao, Y.X.; Zhou, X.; Wei, B.; Zhan, L.; Sun, S.Y. Stimulative Role of ST6GALNAC1 in Proliferation, Migration and Invasion of Ovarian Cancer Stem Cells via the Akt Signaling Pathway. Cancer Cell Int. 2019, 19, 86. [Google Scholar] [CrossRef]
- Ogawa, T.; Hirohashi, Y.; Murai, A.; Nishidate, T.; Okita, K.; Wang, L.; Ikehara, Y.; Satoyoshi, T.; Usui, A.; Kubo, T.; et al. ST6GALNAC1 Plays Important Roles in Enhancing Cancer Stem Phenotypes of Colorectal Cancer via the Akt Pathway. Oncotarget 2017, 8, 112550–112564. [Google Scholar] [CrossRef]
- Ahmad, M.S.; Braoudaki, M.; Siddiqui, S.S. Differential Expression of ST6GALNAC1 and ST6GALNAC2 and Their Clinical Relevance to Colorectal Cancer Progression. PLoS ONE 2024, 19, 311212. [Google Scholar] [CrossRef]
- Luo, Y.; Cao, H.; Lei, C.; Liu, J. ST6GALNAC1 Promotes the Invasion and Migration of Breast Cancer Cells via the EMT Pathway. Genes Genom. 2023, 45, 1367–1376. [Google Scholar] [CrossRef]
- Broad Institute of MIT & Harvard Firebrowse. Available online: http://firebrowse.org/ (accessed on 27 April 2025).
- NIH National Cancer Institute GDC Data Portal. Available online: https://portal.gdc.cancer.gov/ (accessed on 27 April 2025).
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Therneau, T. A Package for Survival Analysis in R; Springer International Publishing: Cham, Switzerland, 2024. [Google Scholar]
- Alboukadel, K.; Marcin, K.; Przemyslaw, B.; Scheipl, F. Drawing Survival Curves Using “ggplot2”, Version 0.4.9. R Package Survminer. Datanovia: Montpellier, France, 2021. [CrossRef]
- Wickham, H. ggplot2; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
| Discovery Cohort (TCGA) | ||||||
|---|---|---|---|---|---|---|
| HNSCC | Gene | HR | 95% IC | High Expression p Value | Early-Stage p Value | Alcohol History p Value |
| HPV+ | FABP4 | 3.34 | 1.04–10.73 | 0.0426 | 0.3678 | 0.5836 |
| GDNF | 3.41 | 1.07–10.83 | 0.0379 | 0.9975 | 0.8007 | |
| HMGA2 | 3.35 | 1.08–10.37 | 0.0357 | 0.5047 | 0.9147 | |
| S100A10 † | 3.05 | 0.98–9.51 | 0.0538 | 0.6274 | 0.8405 | |
| SLC7A2 | 0.32 | 0.1–0.99 | 0.0481 | 0.2619 | 0.9952 | |
| GPR18 | 0.25 | 0.07–0.84 | 0.0253 | 0.4677 | 0.6414 | |
| HPV− | ST6GALNAC1 | 0.63 | 0.47–0.85 | 0.0027 | 0.073 | 0.8883 |
| Validation Cohort (GSE65858) | ||||||
| HPV+ | FABP4 | 5.18 | 1.59–16.93 | 0.0065 | 0.9976 | 0.3013 |
| GDNF | 3.19 | 1.12–9.05 | 0.0294 | 0.9975 | 0.8007 | |
| HMGA2 | 3.96 | 1.3–12.1 | 0.0157 | 0.9976 | 0.7485 | |
| S100A10 | 2.97 | 1.05–8.41 | 0.041 | 0.9975 | 0.7149 | |
| SLC7A2 | 0.3 | 0.11–0.83 | 0.0209 | 0.9975 | 0.7638 | |
| GPR18 | 0.34 | 0.12–0.95 | 0.0389 | 0.9975 | 0.8128 | |
| HPV− | ST6GALNAC1 | 0.55 | 0.33–0.9 | 0.018 | 0.0744 | 0.8922 |
| Biomarkers | Function | Alterations in Cancer | Clinical Outcome | References |
|---|---|---|---|---|
| FABP4 (Fatty Acid Binding Protein 4) | Fatty acid binding proteins (FABPs) are intracellular lipid chaperones that reversibly bind hydrophobic ligands and coordinate lipid responses in cells. FABP4, also known as A-FABP, is primarily expressed in adipocytes, macrophages, and dendritic cells. | FABP4 expression in tumor-associated macrophages promotes breast cancer progression. Angiogenesis, growth, and metastasis in ovarian tumor xenografts were significantly inhibited by suppressing endothelial FABP4 expression. | High FABP4 expression is associated with poor OS and relapse-free survival (RFS) in patients with lung, gastric, and colorectal cancers. Loss of FABP4 expression was correlated with poor prognosis in bladder cancer. | [43,44,45,46,47,48,49,50] |
| S100A10 (S100 Calcium Binding Protein A10) | S100A10 belongs to the S100 family of calcium-binding proteins and constitutes the largest subgroup of EF-hand proteins. S100A10, also known as p11, regulates plasminogen activation and plasmin generation. | S100A10 was shown to be positively associated with metastasis, blood vessel development, EMT, hypoxia, and invasion in HNSCC. | High expression is linked to a worse prognosis in patients with hepatocellular carcinoma, pancreatic adenocarcinoma and glioma. Low S100A10 gene expression was associated with good OS in patients with HNSCC. | [51,52,53,54] |
| HMGA2 (High Mobility Group AT-Hook 2) | HMGA2 is a nonhistone chromatin factor that binds to AT-rich DNA sequences affecting chromatin structure, thereby enhancing or suppressing the activity of many transcription factors. HMGA proteins regulate embryonic development, stem cell maintenance, cellular senescence, and tumorigenesis. | HMGA2 facilitates triple-negative breast cancer metastasis by transcriptionally activating matrix metalloproteinases. Specifically, HMGA2 induced chromatin conformation changes to enhance MMP transcriptional activity. HMGA2 is overexpressed in HNSCC and induces the expression of genes associated with the cancer stem cell phenotype. HMGA2 promotes malignant progression of HNSCC and the acquisition of CSC properties by directly regulating Snai2 expression. | Higher HMGA2 expression was associated with poor OS in different types of cancer, including HNSCC. High gene and protein expression were associated with decreased OS and disease-free survival (DFS) in HNSCC patients. HMGA2 expression was an independent predictor of OS and DFS in oral cancer and HNSCC | [55,56,57,58,59,60,61,62] |
| GDNF (Glial Cell Derived Neurotrophic Factor) | GDNF is a member of the GDNF ligand (GFL) family. It binds to GDNF receptor-α1 (GFRα1) and its coreceptor, RET. This growth factor has been implicated in neuronal differentiation and survival of neurons | In HNSCC, GDNF expression was associated with increased perineural invasion and lymphatic metastasis by increasing PD-L1 expression through the JAK2-STAT1 signaling pathway. | High expression was related to decreased overall survival in HNSCC patients. GDNF positivity correlated with poorer OS and PFS, regardless of HPV status, in HNSCC patients. The presence of the GDNF protein in stroma was associated with a significantly higher risk of death only in the HPV-negative HNSCC patients. Higher GDNF gene expression was associated with better clinical outcomes in HPV-negative HNSCC patients. | [63,64,65,66] |
| SLC7A2 (Solute Carrier Family 7 Member 2) | SLC7A2 is a membrane protein that facilitates the cellular transport of cationic amino acids, including lysine and L-arginine. | SLC7A2 is highly expressed in normal tissues compared to HNSCC. The overexpression of SLC7A2 promotes apoptosis in HNSCC cells while effectively inhibiting their growth, proliferation, and metastasis. | High levels of SLC7A2 correlated with prolonged OS in HNSCC patients, and this survival difference was even more prominent in HPV-positive cases. | [67,68,69] |
| GPR18 (G Protein-Coupled Receptor 18) | GPR18 is an atypical cannabinoid receptor capable of binding N-arachidonoyl glycine (NAGly) and resolvin D2 (RvD2), anti-inflammatory compounds that could contribute to limiting tumor spread. | GPR18 is overexpressed in melanoma metastatic sites, is constitutively active, and inhibits apoptosis. | Increased expression of GPR18 correlated with an increase in OS of patients with HNSCC. | [70,71,72,73,74] |
| ST6GALNAC1 (ST6 N-Acetylgalactosaminide Alpha-2,6-Sialyltransferase 1) | The ST6GalNAc-I enzyme is a glycosyltransferase capable of adding sialic acid at the 2,6 linkage to GalNAc linked to serine or threonine, thus creating the STn epitope. | High levels of ST6GALNAC1 in ovarian cancer cells promote cell proliferation, migration, invasion, self-renewal capacity, and tumorigenicity. In breast cancer cells, ST6GALNAC1 expression significantly enhanced cell migration and invasion, and it was found to directly induce the EMT signaling pathway. | ST6GALNAC1 positivity in stage III and IV colorectal cancer samples is significantly associated with short OS. Low ST6GALNAC1 gene expression is associated with poor OS, RFS, and post-progression survival (PPS) in colorectal cancer patients. | [75,76,77,78,79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Chávez, J.N.; Contreras-Paredes, A.; González-Espinosa, C.; Martínez-Ramírez, I.; Langley, E.; Lizano, M.; Muñoz-Bello, J.O. Association of Gene Expression Profiles in HPV-Positive Head and Neck Squamous Cell Carcinoma with Patient Outcome: In Search of Prognostic Biomarkers. Int. J. Mol. Sci. 2025, 26, 5894. https://doi.org/10.3390/ijms26125894
García-Chávez JN, Contreras-Paredes A, González-Espinosa C, Martínez-Ramírez I, Langley E, Lizano M, Muñoz-Bello JO. Association of Gene Expression Profiles in HPV-Positive Head and Neck Squamous Cell Carcinoma with Patient Outcome: In Search of Prognostic Biomarkers. International Journal of Molecular Sciences. 2025; 26(12):5894. https://doi.org/10.3390/ijms26125894
Chicago/Turabian StyleGarcía-Chávez, J. Noé, Adriana Contreras-Paredes, Claudia González-Espinosa, Imelda Martínez-Ramírez, Elizabeth Langley, Marcela Lizano, and J. Omar Muñoz-Bello. 2025. "Association of Gene Expression Profiles in HPV-Positive Head and Neck Squamous Cell Carcinoma with Patient Outcome: In Search of Prognostic Biomarkers" International Journal of Molecular Sciences 26, no. 12: 5894. https://doi.org/10.3390/ijms26125894
APA StyleGarcía-Chávez, J. N., Contreras-Paredes, A., González-Espinosa, C., Martínez-Ramírez, I., Langley, E., Lizano, M., & Muñoz-Bello, J. O. (2025). Association of Gene Expression Profiles in HPV-Positive Head and Neck Squamous Cell Carcinoma with Patient Outcome: In Search of Prognostic Biomarkers. International Journal of Molecular Sciences, 26(12), 5894. https://doi.org/10.3390/ijms26125894






