Full-Length Transcriptome of Testis and Ovary Provides Insights into Alternative Splicing During Gonadal Development in Litopenaeus vannamei
Abstract
1. Introduction
2. Results
2.1. Overview of PacBio Iso-Seq Sequencing
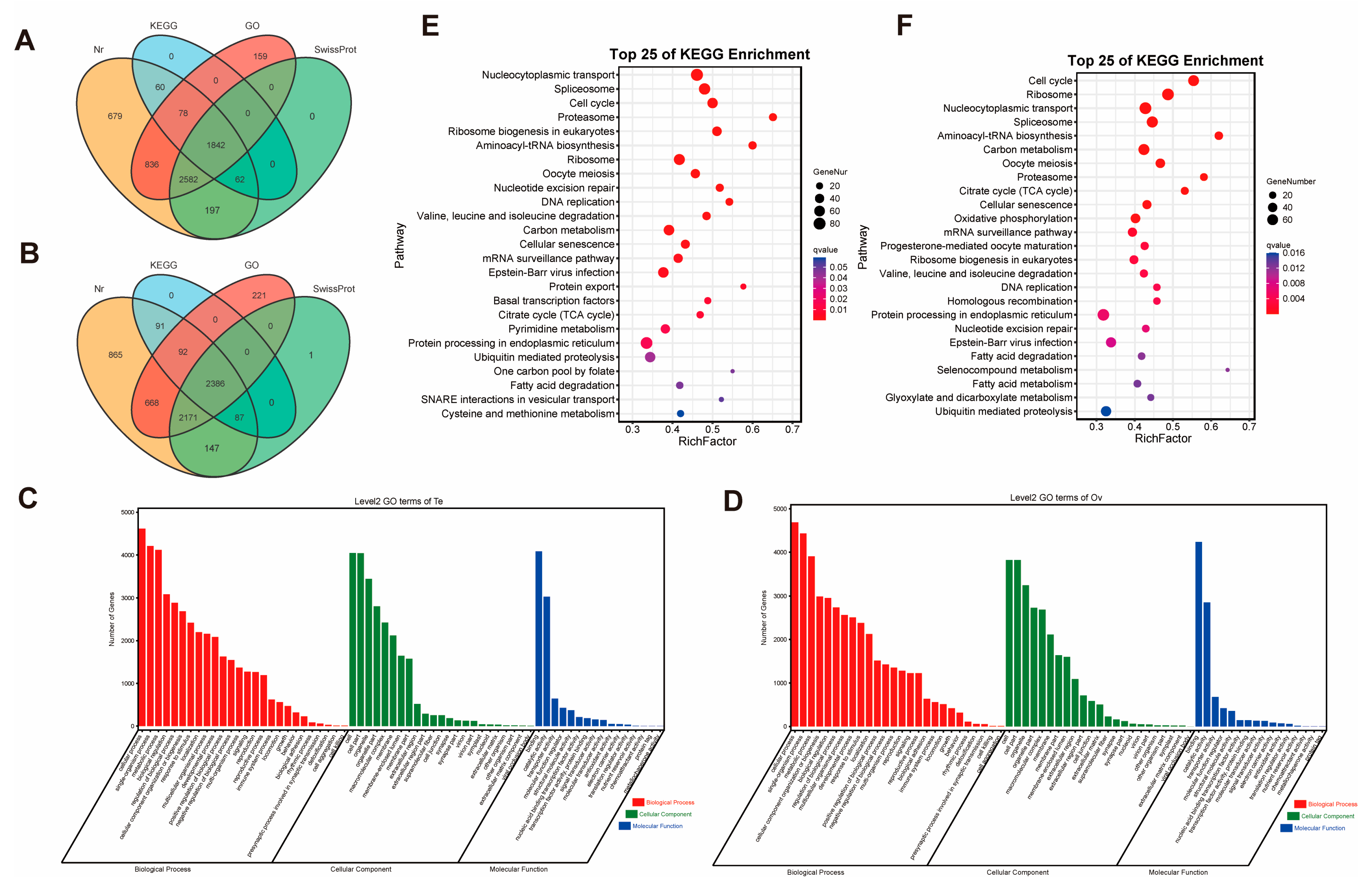
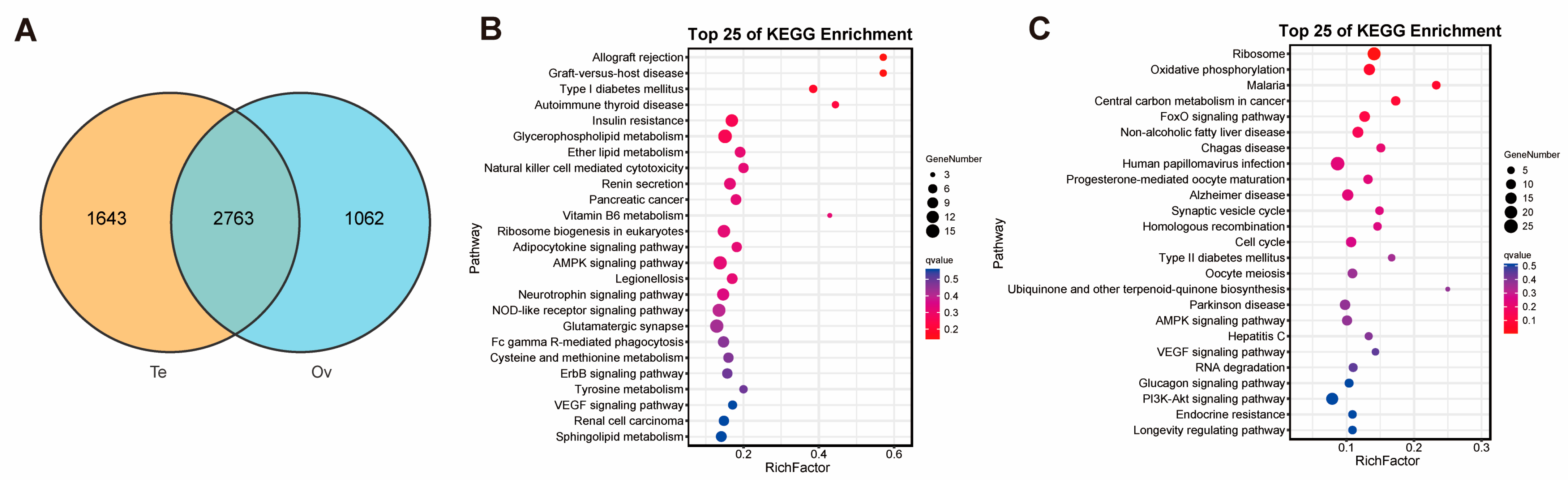
2.2. Comparison of Genes Between Ovary and Testis
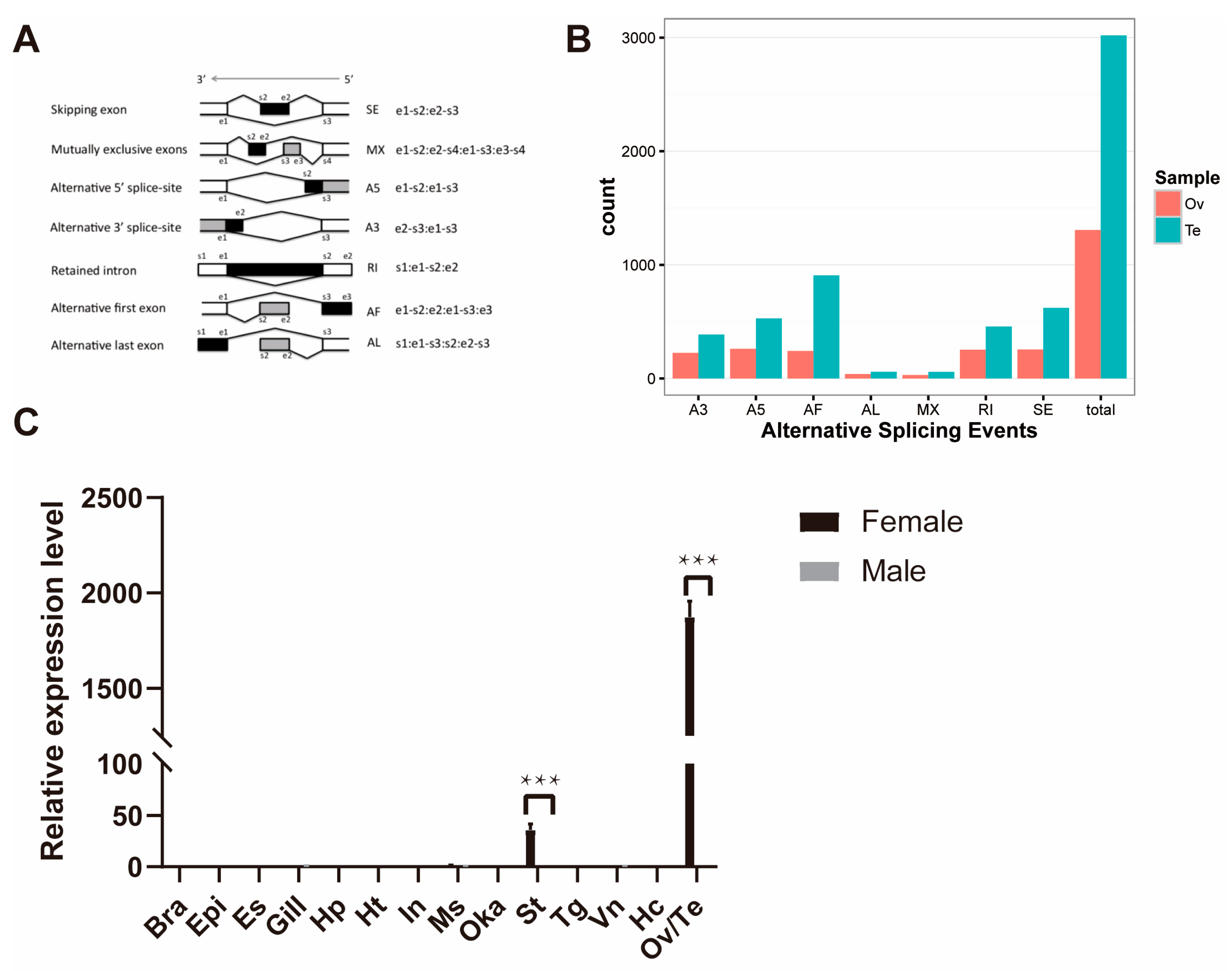
2.3. Alternative Splicing Analysis
2.4. Annotation and Comparison of AS in Ovary and Testis
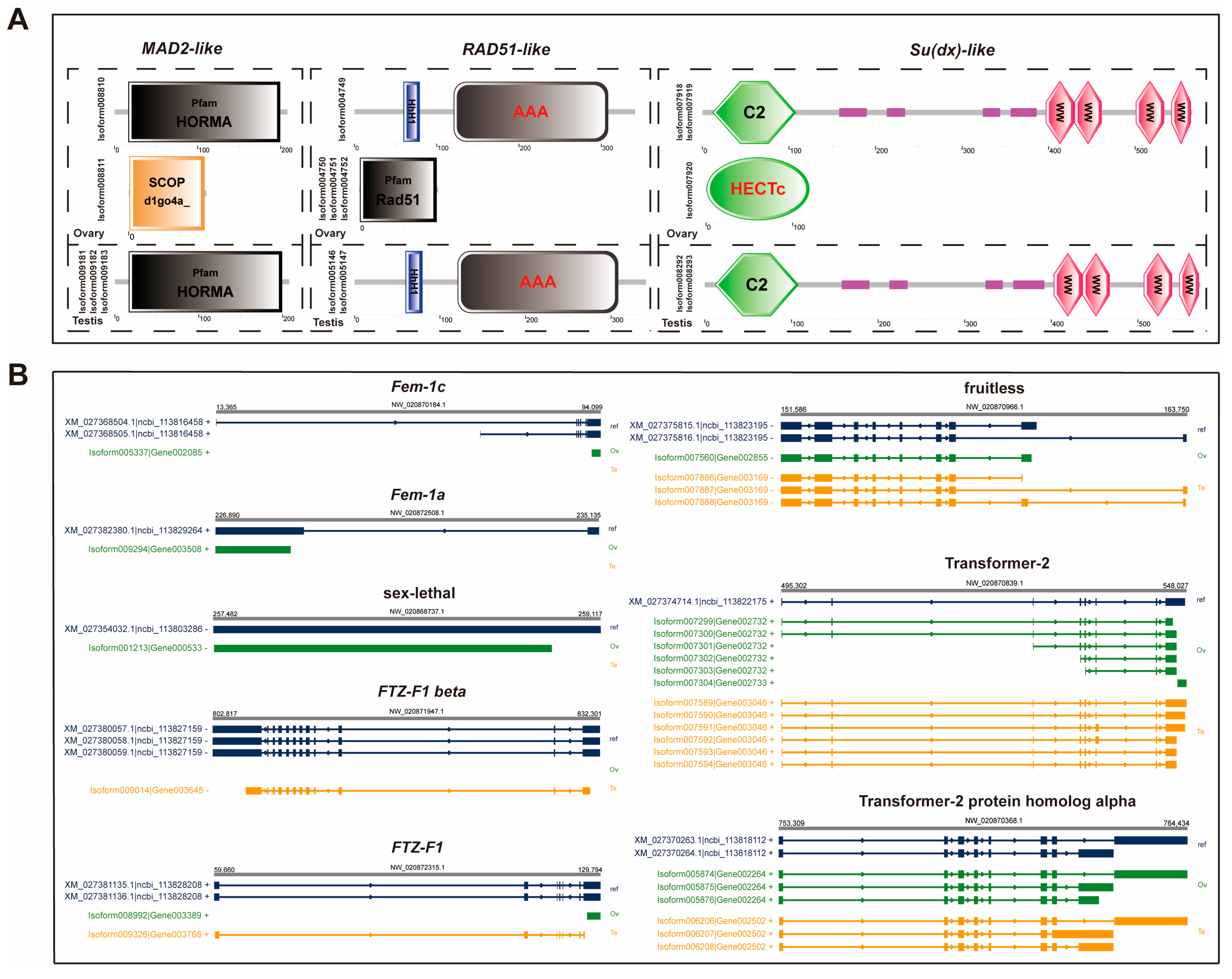
2.5. AS Analysis of the Known Sex-Determination and Differentiation Genes
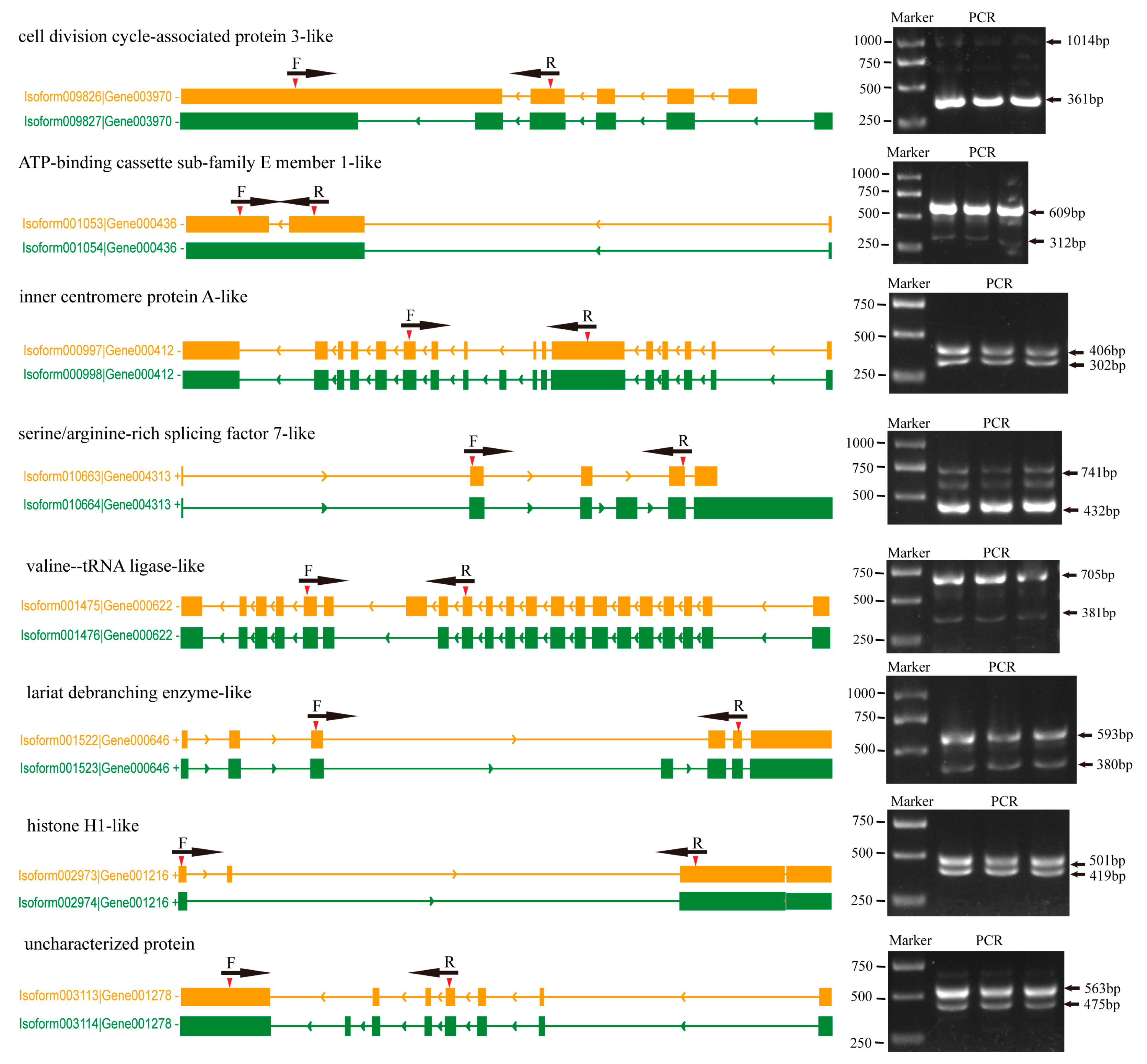
2.6. Verification of the ISO-Seq Results
3. Discussion
4. Materials and Methods
4.1. Animal Material and RNA Sample Preparation
4.2. PacBio Iso-Seq Sequencing
4.3. PacBio Data Processing
4.4. Functional Annotation of Transcripts
4.5. AS Analysis
4.6. Validation Experiments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, Y.; Zhang, X.J.; Yuan, J.B.; Wang, Q.C.; Li, S.H.; Huang, H.; Li, F.H.; Xiang, J.H. Identification of Sex-determining Loci in Pacific White Shrimp Litopeneaus vannamei Using Linkage and Association Analysis. Mar. Biotechnol. 2017, 19, 277–286. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2024—Blue Transformation in Action; FAO: Rome, Italy, 2024. [Google Scholar]
- Peng, J.X.; Wei, P.Y.; Zhang, B.; Zhao, Y.Z.; Zeng, D.G.; Chen, X.L.; Li, M.; Chen, X.H. Gonadal transcriptomic analysis and differentially expressed genes in the testis and ovary of the Pacific white shrimp (Litopenaeus vannamei). BMC Genom. 2015, 16, 1006. [Google Scholar] [CrossRef]
- Gitterle, T.; Rye, M.; Salte, R.; Cock, J.; Johansen, H.; Lozano, C.; Arturo Suárez, J.; Gjerde, B. Genetic (co)variation in harvest body weight and survival in Penaeus (Litopenaeus) vannamei under standard commercial conditions. Aquaculture 2005, 243, 83–92. [Google Scholar] [CrossRef]
- Pérez-Rostro, C.I.; Ibarra, A.M. Heritabilities and genetic correlations of size traits at harvest size in sexually dimorphic Pacific white shrimp (Litopenaeus vannamei) grown in two environments. Aquac. Res. 2003, 34, 1079–1085. [Google Scholar] [CrossRef]
- Levy, T.; Aflalo, E.D.; Sagi, A. Sex Control in Cultured Decapod Crustaceans. In Sex Control in Aquaculture; Wiley: Hoboken, NJ, USA, 2018; pp. 689–704. [Google Scholar]
- Miao, M.; Li, S.H.; Yu, Y.; Li, F.H. LysM-containing proteins function in the resistance of Litopenaeus vannamei against Vibrio parahaemolyticus infection. Dev. Comp. Immunol. 2023, 148, 104900. [Google Scholar] [CrossRef]
- Boonyoung, G.; Panrat, T.; Phongdara, A.; Wanna, W. Evaluation of the relationship between the 14-3-3ε protein and LvRab11 in the shrimp Litopenaeus vannamei during WSSV infection. Sci. Rep. 2021, 11, 19188. [Google Scholar] [CrossRef]
- Wang, Q.C.; Yu, Y.; Zhang, Q.; Luo, Z.; Zhang, X.J.; Xiang, J.H.; Li, F.H. The Polymorphism of LvMMD2 and Its Association with Growth Traits in Litopenaeus vannamei. Mar. Biotechnol. 2020, 22, 564–571. [Google Scholar] [CrossRef]
- Si, S.; Zhang, X.; Yu, Y.; Zhang, X.; Zhong, X.; Yuan, J.; Yang, S.; Li, F. Structure and function analyses of the Mmd2 gene in pacific white shrimp Litopenaeus vannamei. Front. Genet. 2023, 14, 1151193. [Google Scholar] [CrossRef]
- Li, W.; He, P.P.; Zhang, X.Z.; Guan, J.L.; Chen, Y.X.; Zhang, L.; Zhang, B.; Zheng, Y.S.; Li, X.; He, Q.S.; et al. Identification and Characterization of microRNAs in the Gonads of Litopenaeus vannamei Using High-Throughput Sequencing. Fishes 2022, 7, 308. [Google Scholar] [CrossRef]
- Siegfried, K.R. In search of determinants: Gene expression during gonadal sex differentiation. J. Fish Biol. 2010, 76, 1879–1902. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lin, X.; Zhu, Y.; Zhou, D.; Zhang, Y.; Huang, Y.; Chen, H.; Li, G.; Tian, C. A time-course transcriptome analysis of gonads from HongKong catfish (Clarias fuscus) reveals genes and pathways associated with gonadal development. Aquacult Rep. 2024, 37, 102247. [Google Scholar] [CrossRef]
- Wang, T.; Yu, Y.; Li, S.H.; Li, F.H. Molecular mechanisms of sex determination and differentiation in decapod crustaceans for potential aquaculture applications: An overview. Rev. Aquacult. 2024, 16, 1819–1839. [Google Scholar] [CrossRef]
- Huang, X.S.; Feng, B.Y.; Huang, H.Y.; Ye, H.H. In vitro stimulation of vitellogenin expression by insulin in the mud crab, Scylla paramamosain, mediated through PI3K/Akt/TOR pathway. Gen. Comp. Endocr. 2017, 250, 175–180. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Jiang, X.H.; Qi, H.Y.; Xiong, L.W.; Qiu, G.F. A novel SoxB2 gene is required for maturation of sperm nucleus during spermiogenesis in the Chinese mitten crab, Eriocheir sinensis. Sci. Rep. 2016, 6, 32139. [Google Scholar] [CrossRef]
- Levy, T.; Sagi, A. The “IAG-Switch”-A Key Controlling Element in Decapod Crustacean Sex Differentiation. Front. Endocrinol. 2020, 11, 651. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell 2006, 126, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef]
- Kornblihtt, A.R.; Schor, I.E.; Alló, M.; Dujardin, G.; Petrillo, E.; Muñoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165. [Google Scholar] [CrossRef]
- Rogers, T.F.; Palmer, D.H.; Wright, A.E. Sex-Specific Selection Drives the Evolution of Alternative Splicing in Birds. Mol. Biol. Evol. 2021, 38, 519–530. [Google Scholar] [CrossRef]
- Cline, T.W. The Drosophila sex determination signal: How do flies count to two? Trends Genet. 1993, 9, 385–390. [Google Scholar] [CrossRef]
- Bell, L.R.; Horabin, J.I.; Schedl, P.; Cline, T.W. Positive Autoregulation of Sex-Lethal by Alternative Splicing Maintains the Female Determined State in Drosophila. Cell 1991, 65, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, R.N.; Mckeown, M.; Burtis, K.C.; Belote, J.M.; Baker, B.S. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell 1988, 53, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef] [PubMed]
- Penalva, L.O.F.; Sánchez, L. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol. Mol. Biol. Rev. 2003, 67, 343–359. [Google Scholar] [CrossRef]
- Baker, B.S. Sex in Flies: The Splice of Life. Nature 1989, 340, 521–524. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Zhang, X.; Liu, C.; Xiang, J.; Li, F. Genome-Wide Analysis of Alternative Splicing Provides Insights Into Stress Response of the Pacific White Shrimp Litopenaeus vanname. Front. Genet. 2019, 10, 845. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.; Yuan, J.; Li, F. The Responses of Alternative Splicing during Heat Stress in the Pacific White Shrimp Litopenaeus vannamei. Genes 2023, 14, 1473. [Google Scholar] [CrossRef]
- Soñanez-Organis, J.G.; Rodriguez-Armenta, M.; Leal-Rubio, B.; Peregrino-Uriarte, A.B.; Gómez-Jiménez, S.; Yepiz-Plascencia, G. Alternative splicing generates two lactate dehydrogenase subunits differentially expressed during hypoxia via HIF-1 in the shrimp Litopenaeus vannamei. Biochimie 2012, 94, 1250–1260. [Google Scholar] [CrossRef]
- Shi, X.; Zhang, R.; Liu, Z.; Zhao, G.; Guo, J.; Mao, X.; Fan, B. Alternative Splicing Reveals Acute Stress Response of Litopenaeus vannamei at High Alkalinity. Mar. Biotechnol. 2024, 26, 103–115. [Google Scholar] [CrossRef]
- Zhao, S.; Lu, X.; Zhang, Y.; Zhao, X.; Zhong, M.; Li, S.; Lun, J. Identification of a novel alternative splicing variant of hemocyanin from shrimp Litopenaeus vannamei. Immunol. Lett. 2013, 154, 1–6. [Google Scholar] [CrossRef]
- Gao, Y.B.; Xi, F.H.; Zhang, H.X.; Liu, X.Q.; Wang, H.Y.; Zhao, L.Z.; Reddy, A.S.N.; Gu, L.F. Single-Molecule Real-Time (SMRT) Isoform Sequencing (Iso-Seq) in Plants: The Status of the Bioinformatics Tools to Unravel the Transcriptome Complexity. Curr. Bioinform. 2019, 14, 566–573. [Google Scholar] [CrossRef]
- Chow, K.S.; Khoo, J.S.; Mohd-Zainuddin, Z.; Ng, S.M.; Hoh, C.C. Utility of PacBio Iso-Seq for transcript and gene discovery in Hevea latex. J. Rubber Res. 2019, 22, 169–186. [Google Scholar] [CrossRef]
- Huang, J.C.; Chen, W.J.; Wang, Q.Y.; Zhang, Y.X.; Liu, Q.; Yang, D.H. Iso-Seq assembly and functional annotation of full-length transcriptome of turbot (Scophthalmus maximus) during bacterial infection. Mar. Genom. 2022, 63, 100954. [Google Scholar] [CrossRef]
- Pootakham, W.; Uengwetwanit, T.; Sonthirod, C.; Sittikankaew, K.; Karoonuthaisiri, N. A Novel Full-Length Transcriptome Resource for Black Tiger Shrimp (Penaeus monodon) Developed Using Isoform Sequencing (Iso-Seq). Front. Mar. Sci. 2020, 7, 172. [Google Scholar] [CrossRef]
- Jo, E.; Lee, S.J.; Choi, E.; Kim, J.; Lee, J.H.; Park, H. Sex-Biased Gene Expression and Isoform Profile of Brine Shrimp Artemia franciscana by Transcriptome Analysis. Animals 2021, 11, 2630. [Google Scholar] [CrossRef]
- Wang, J.J.; Li, J.T.; Ge, Q.Q.; Li, W.Y.; Li, J. Full-Length Transcriptome Sequencing and Comparative Transcriptomic Analysis Provide Insights Into the Ovarian Maturation of Exopalaemon carinicauda. Front. Mar. Sci. 2022, 9, 906730. [Google Scholar] [CrossRef]
- Alamancos, G.P.; Pagès, A.; Trincado, J.L.; Bellora, N.; Eyras, E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 2015, 21, 1521–1531. [Google Scholar] [CrossRef]
- Adhikari, D.; Liu, K.; Shen, Y. Cdk1 drives meiosis and mitosis through two different mechanisms. Cell Cycle 2012, 11, 2763–2764. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, K.; Zhuan, Q.; Liu, Z.; Meng, L.; Fu, X.; Jia, G.; Hou, Y. Mitochondrial Calcium Disorder Affects Early Embryonic Development in Mice through Regulating the ERK/MAPK Pathway. Oxidative Med. Cell. Longev. 2022, 2022, 8221361. [Google Scholar] [CrossRef]
- Zhang, X.J.; Li, G.Y.; Jiang, H.Y.; Li, L.M.; Ma, J.G.; Li, H.M.; Chen, J.P. Full-length transcriptome analysis of Litopenaeus vannamei reveals transcript variants involved in the innate immune system. Fish Shellfish Immun. 2019, 87, 346–359. [Google Scholar] [CrossRef]
- Guo, X.F.; Zhou, Y.L.; Liu, M.; Wang, Z.W.; Gui, J.F. Integrated application of Iso-seq and RNA-seq provides insights into unsynchronized growth in red swamp crayfish (Procambarus clarkii). Aquacult. Rep. 2022, 22, 101008. [Google Scholar] [CrossRef]
- Tan, K.A.; Xu, P.; Lim, L.S.; Nie, C.H.; Tan, K.; Peng, Y.; Cai, X.H.; Yan, X.Y.; Xu, Y.H.; Kwan, K.Y. Iso-seq and RNA-seq profiling of Fenneropenaeus penicillatus: Unravelling the signaling pathway and genes involved in the ovarian development. Aquacult. Rep. 2023, 32, 101731. [Google Scholar] [CrossRef]
- Cui, W.; Yang, Q.; Zhang, Y.; Farhadi, A.; Fang, H.; Zheng, H.; Li, S.; Zhang, Y.; Ikhwanuddin, M.; Ma, H. Integrative Transcriptome Sequencing Reveals the Molecular Difference of Maturation Process of Ovary and Testis in Mud Crab Scylla paramamosain. Front. Mar. Sci. 2021, 8, 658091. [Google Scholar] [CrossRef]
- Galdones, E.; Bernabe, B.P.; Skory, R.M.; Mackovic, D.; Broadbelt, L.J.; Shea, L.D.; Woodruff, T. Ovarian Expression of Cartilage Oligomeric Matrix Protein as a Potential Biomarker of Antral Follicle Development in the Mouse. Biol. Reprod. 2011, 85, 647. [Google Scholar] [CrossRef]
- Jiang, T.; Li, W.D.; Li, X.Q.; Huang, Y.; Wang, Q.G.; Wang, H.W.; Yang, C.W.; Liu, L.B. Screening Candidate Genes for Ovarian Development and Constructing Regulatory Network in Nesting Chickens by Transcriptome and Proteome. Acta Vet. Et Zootech. Sin. 2024, 55, 4950–4967. [Google Scholar] [CrossRef]
- Huang, K.Y.; Chen, Y.M.; Huo, Z.M.; Qin, Y.J. Advances in the Control of Aquatic Invertebrates’ Gonadal Development. Open J. Fish. Res. 2024, 11, 167–176. [Google Scholar] [CrossRef]
- Tu, D.Y.; Wang, S.D. The Function of Dmrt1 in Sexual Development in Different Species. Open J. Fish. Res. 2024, 11, 56–61. [Google Scholar] [CrossRef]
- Grmai, L.; Jimenez, E.; Baxter, E.; Doren, M.V. Steroid signaling controls sex-specific development in an invertebrate. bioRxiv Prepr. Serv. Biol. 2024, 573099. [Google Scholar] [CrossRef]
- Anand, A.; Villella, A.; Ryner, L.C.; Carlo, T.; Goodwin, S.F.; Song, H.J.; Gailey, D.A.; Morales, A.; Hall, J.C.; Baker, B.S.; et al. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics 2001, 158, 1569–1595. [Google Scholar] [CrossRef]
- Lin, J.L.; Yuan, Y.Y.; Shi, X.; Fang, S.B.; Zhang, Y.; Guan, M.Y.; Xie, Z.F.; Ma, H.Y.; Lin, F. Molecular cloning, characterization and expression profiles of a SoxB2 gene related to gonadal development in mud crab (Scylla paramamosain). Invertebr. Reprod. Dev. 2020, 64, 126–136. [Google Scholar] [CrossRef]
- Toyota, K.; Miyakawa, H.; Hiruta, C.; Sato, T.; Katayama, H.; Ohira, T.; Iguchi, T. Sex Determination and Differentiation in Decapod and Cladoceran Crustaceans: An Overview of Endocrine Regulation. Genes 2021, 12, 305. [Google Scholar] [CrossRef] [PubMed]
- Gempe, T.; Beye, M. Function and evolution of sex determination mechanisms, genes and pathways in insects. Bioessays 2011, 33, 52–60. [Google Scholar] [CrossRef]
- Ma, K.Y.; Liu, Z.Q.; Lin, J.Y.; Li, J.L.; Qiu, G.F. Molecular characterization of a novel ovary-specific gene fem-1 homolog from the oriental river prawn, Macrobrachium nipponense. Gene 2016, 575, 244–252. [Google Scholar] [CrossRef]
- Hashiyama, K.; Hayashi, Y.; Kobayashi, S. Drosophila sex lethal gene initiates female development in germline progenitors. Science 2011, 333, 885–888. [Google Scholar] [CrossRef]
- Chen, L.R.; Zheng, J.B.; Jia, Y.Y.; Li, F.; Gu, Z.M.; Chi, M.L.; Cheng, S.; Liu, S.L.; Jiang, W.P.; Liu, Y.N. Molecular characterization of the Ftz-f1 gene in redclaw crayfish Cherax quadricarinatus and its potential role in ovarian development. Aquac. Res. 2022, 53, 5261–5269. [Google Scholar] [CrossRef]
- Bertho, S.; Pasquier, J.; Pan, Q.W.; Le Trionnaire, G.; Bobe, J.; Postlethwait, J.H.; Pailhoux, E.; Schartl, M.; Herpin, A.; Guiguen, Y. Foxl2 and its relatives are evolutionary conserved players in gonadal sex differentiation. Sex. Dev. 2016, 10, 111–129. [Google Scholar] [CrossRef]
- Bownes, M. The Regulation of the Yolk Protein Genes, a Family of Sex Differentiation Genes in Drosophila Melanogaster. Bioessays 1994, 16, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Chen, L.; Jia, Y.; Chi, M.; Li, F.; Cheng, S.; Liu, S.; Liu, Y.; Gu, Z. Genomic structure, expression, and functional characterization of the Fem-1 gene family in the redclaw crayfish, Cherax quadricarinatus. Gen. Comp. Endocrinol. 2022, 316, 113961. [Google Scholar] [CrossRef]
- Chakraborty, T.; Mohapatra, S.; Matsuyama, M.; Nagahama, Y.; Ohta, K. Sex Lethal Gene Manipulates Gonadal Development of Medaka, Oryzias latipes, through Estrogenic Interventions. Int. J. Mol. Sci. 2022, 23, 5496. [Google Scholar] [CrossRef]
- Dobles, M.; Liberal, V.; Scott, M.L.; Benezra, R.; Sorger, P.K. Chromosome Missegregation and Apoptosis in Mice Lacking the Mitotic Checkpoint Protein Mad2. Cell 2000, 101, 635–645. [Google Scholar] [CrossRef]
- Niault, T.; Hached, K.; Sotillo, R.; Sorger, P.K.; Maro, B.; Benezra, R.; Wassmann, K. Changing Mad2 Levels Affects Chromosome Segregation and Spindle Assembly Checkpoint Control in Female Mouse Meiosis I. PLoS ONE 2007, 2, e1165. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Hong, K.H.; Zhou, Q.Y.; Wang, Y.K.; Li, W.; Liu, X.L.; Hong, X.Y.; Chen, C.; Yu, L.Y.; Zhu, X.P. Transcriptome analysis of gonads and brain of giant freshwater prawn (Macrobrachium rosenbergii): Screening and validation of genes related to germ cell development. Front. Mar. Sci. 2022, 9, 1060594. [Google Scholar] [CrossRef]
- Wilkin, M.B.; Whiteford, R.; Akbar, T.; Hosseini-Alghaderi, S.; Revici, R.; Carbery, A.M.; Baron, M. The First Defined Null Allele of the Notch Regulator, a Suppressor of Deltex: Uncovering Its Novel Roles in Drosophila melanogaster Oogenesis. Biomolecules 2024, 14, 522. [Google Scholar] [CrossRef]
- Gordon, S.P.; Tseng, E.; Salamov, A.; Zhang, J.W.; Meng, X.D.; Zhao, Z.Y.; Kang, D.W.; Underwood, J.; Grigoriev, I.V.; Figueroa, M.; et al. Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLoS ONE 2015, 10, e0132628. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Yuan, J.B.; Sun, Y.M.; Li, S.H.; Gao, Y.; Yu, Y.; Liu, C.Z.; Wang, Q.C.; Lv, X.J.; Zhang, X.X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef]
- Shimizu, K.; Adachi, J.; Muraoka, Y. ANGLE: A sequencing errors resistant program for predicting protein coding regions in unfinished cDNA. J. Bioinform. Comput. Biol. 2006, 4, 649–664. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.J.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
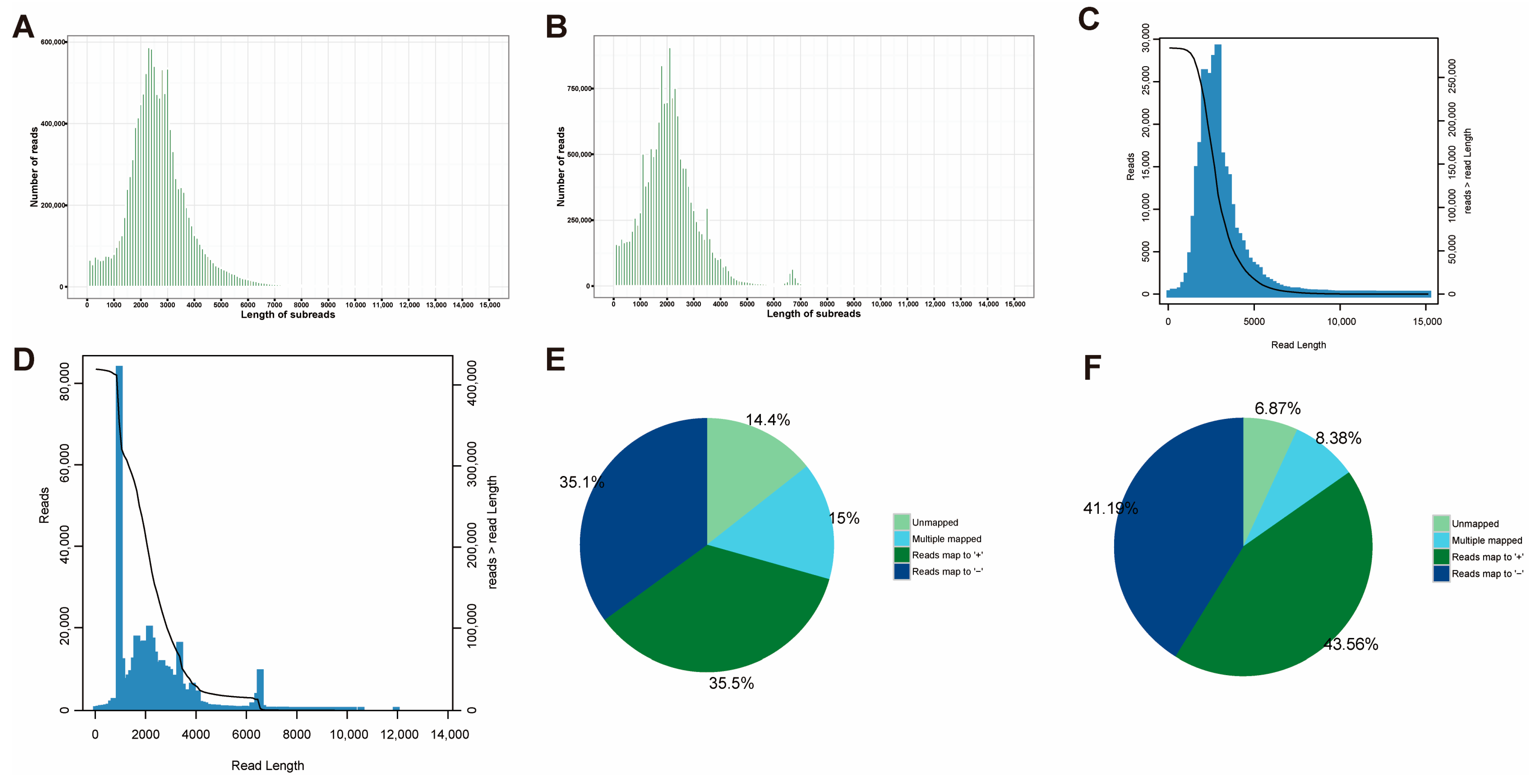
| Subjects | Data | Number (%)/Length (bp) | |
|---|---|---|---|
| Testis | Ovary | ||
| Subreads | Total base (bp) | 32,844,540,629 | 34,014,931,007 |
| Subreads number | 12,098,870 | 15,597,803 | |
| Average length (bp) | 2714 | 2180 | |
| N50 (bp) | 2953 | 2443 | |
| Number of CCSs | Number of CCS reads | 483,391 | 730,978 |
| Full-length non-chimeric, FLNC | 284,013 | 419,324 | |
| Mean full-length non-concatemer read length (bp) | 2946 | 2225 | |
| HQ isoform mapped to genome | Uniquely mapped | 13,349 (70.61%) | 15,734 (84.76%) |
| Multiple mapped | 2838 (15.01%) | 1555 (8.38%) | |
| Unmapped | 2717 (14.37%) | 1275 (6.87%) | |
| Reference transcripts | All mapped genes | 4392 | 3942 |
| All mapped isoforms | 10,830 | 10,648 | |
| Known isoforms | 3983 | 2834 | |
| Novel isoforms | 830 | 690 | |
| New isoforms | 6017 | 7124 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Yu, Y.; Wang, Y.; Li, F. Full-Length Transcriptome of Testis and Ovary Provides Insights into Alternative Splicing During Gonadal Development in Litopenaeus vannamei. Int. J. Mol. Sci. 2025, 26, 5863. https://doi.org/10.3390/ijms26125863
Wang Y, Yu Y, Wang Y, Li F. Full-Length Transcriptome of Testis and Ovary Provides Insights into Alternative Splicing During Gonadal Development in Litopenaeus vannamei. International Journal of Molecular Sciences. 2025; 26(12):5863. https://doi.org/10.3390/ijms26125863
Chicago/Turabian StyleWang, Youyan, Yang Yu, Yue Wang, and Fuhua Li. 2025. "Full-Length Transcriptome of Testis and Ovary Provides Insights into Alternative Splicing During Gonadal Development in Litopenaeus vannamei" International Journal of Molecular Sciences 26, no. 12: 5863. https://doi.org/10.3390/ijms26125863
APA StyleWang, Y., Yu, Y., Wang, Y., & Li, F. (2025). Full-Length Transcriptome of Testis and Ovary Provides Insights into Alternative Splicing During Gonadal Development in Litopenaeus vannamei. International Journal of Molecular Sciences, 26(12), 5863. https://doi.org/10.3390/ijms26125863






