Liver Metabolism at the Crossroads: The Reciprocal Control of Nutrient-Sensing Nuclear Receptors and Autophagy
Abstract
1. Introduction
2. Nutrient-Sensing Nuclear Receptors PPARα and FXR
2.1. Fasting-Activated Nuclear Receptor PPARα
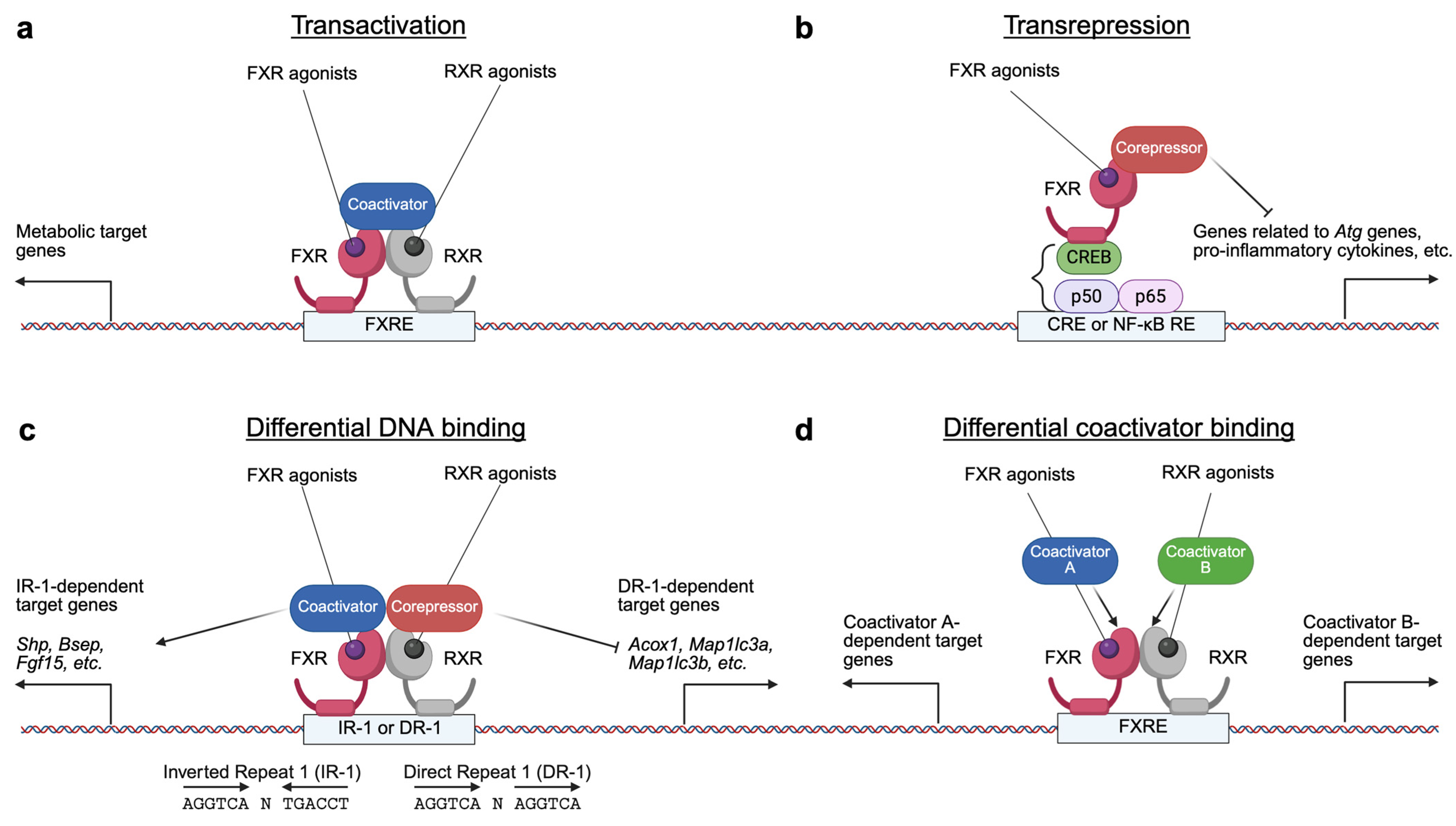
2.2. BAs-Activated Nuclear Receptor FXR
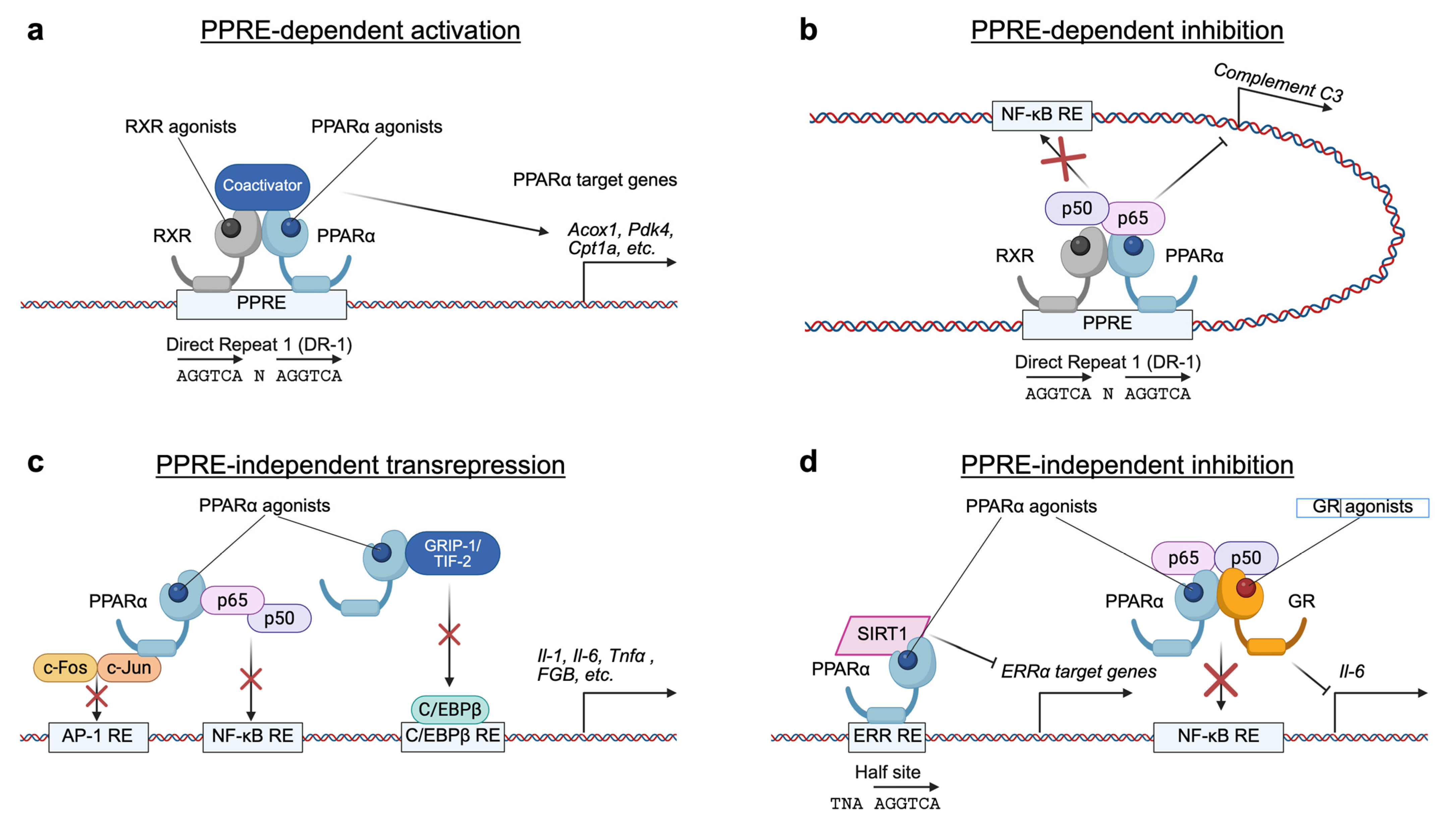
3. Antagonistic Functions of PPARα and FXR in Hepatic Metabolism
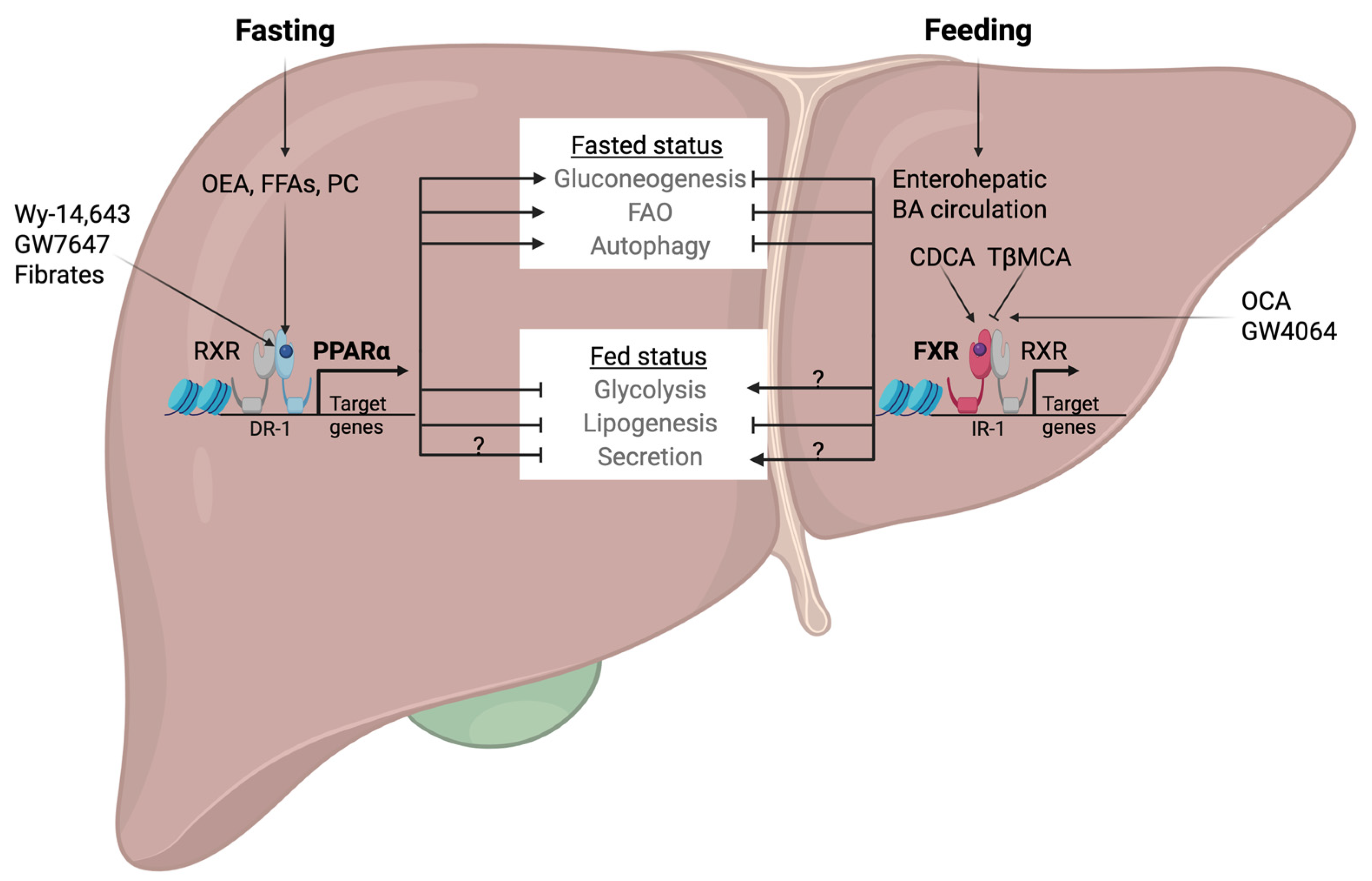
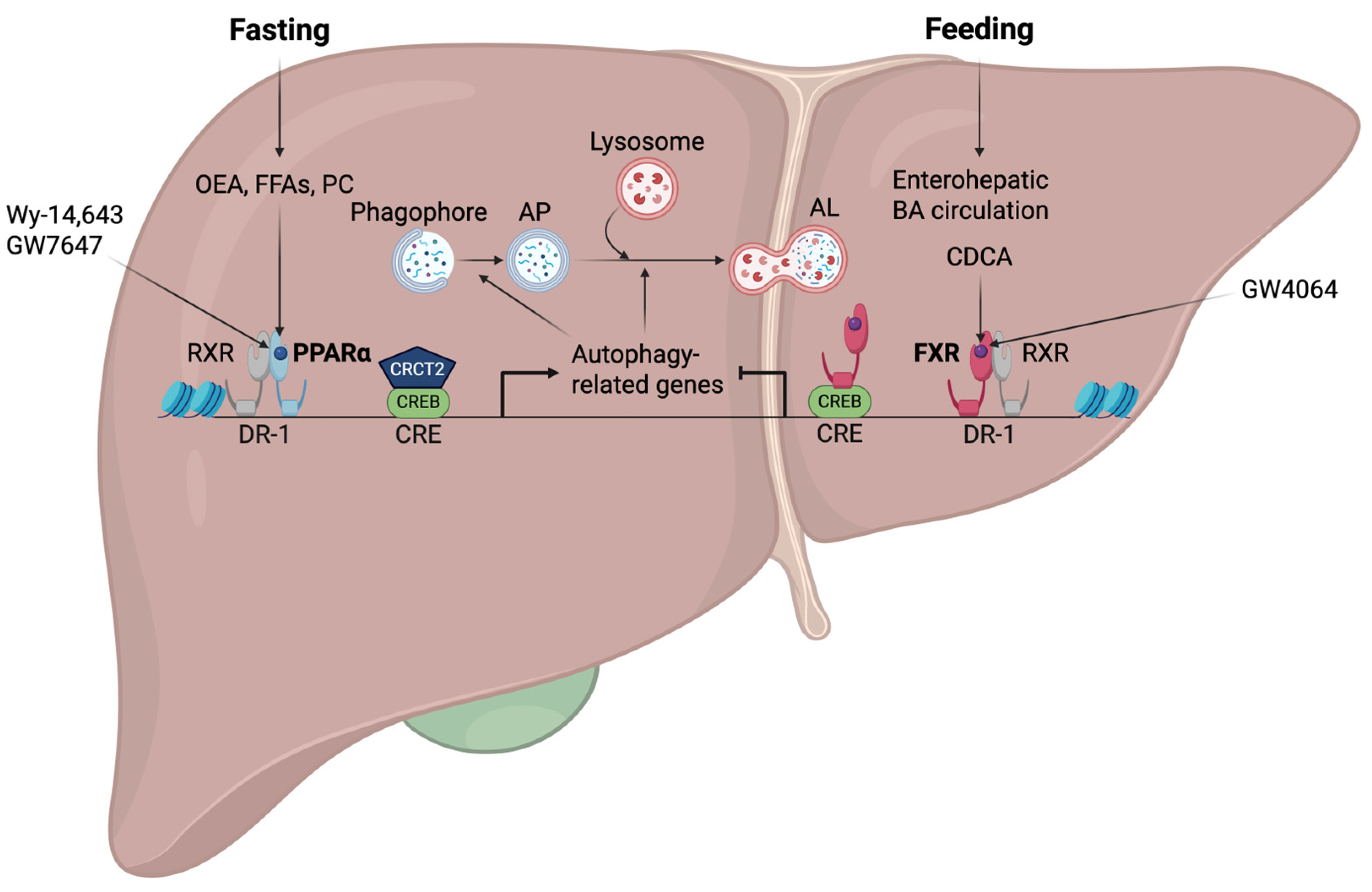
4. Opposing Transcriptional Coordination of Liver Autophagy by PPARα and FXR
5. Phenotypic Abnormalities in Liver-Specific Knockout Mice of Core Autophagy-Related Genes
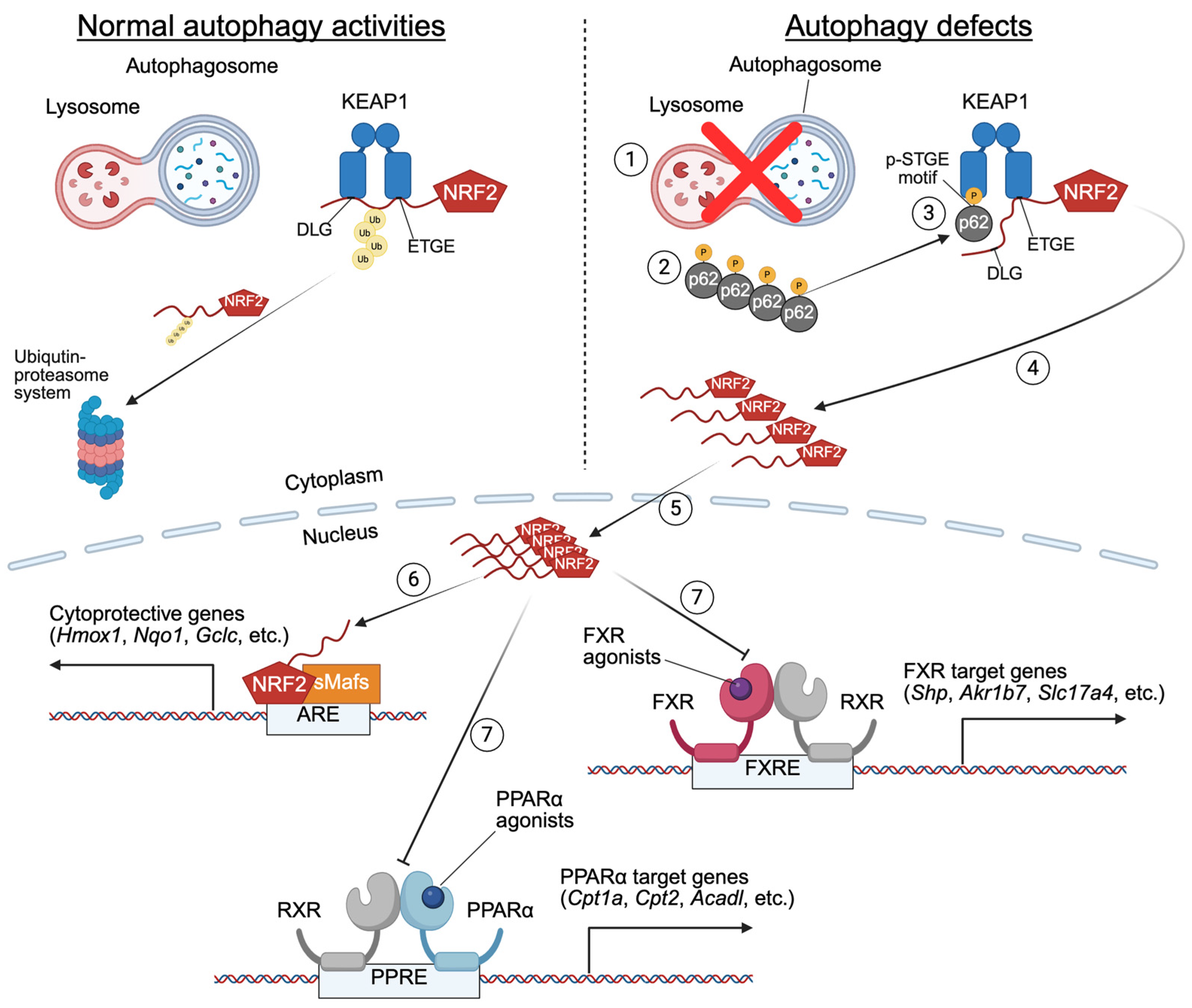
6. Altered Nutrient-Sensing Nuclear Receptor Signaling in Liver-Specific Knockout Mice of Core-Autophagy Genes
7. Bidirectional Regulation Between Autophagy and Nutrient-Sensing Nuclear Receptors PPARα and FXR
8. Clinical Implications and Therapeutic Potential of Targeting the Interplay Between Autophagy and Nutrient-Sensing Nuclear Receptors PPARα and FXR
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2014, 4, 177–197. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Integrated physiology and systems biology of PPARalpha. Mol. Metab. 2014, 3, 354–371. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Li, F.; Gonzalez, F.J. FXR signaling in the enterohepatic system. Mol. Cell Endocrinol. 2013, 368, 17–29. [Google Scholar] [CrossRef]
- Pineda Torra, I.; Claudel, T.; Duval, C.; Kosykh, V.; Fruchart, J.C.; Staels, B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol. Endocrinol. 2003, 17, 259–272. [Google Scholar] [CrossRef]
- Kok, T.; Wolters, H.; Bloks, V.W.; Havinga, R.; Jansen, P.L.; Staels, B.; Kuipers, F. Induction of hepatic ABC transporter expression is part of the PPARalpha-mediated fasting response in the mouse. Gastroenterology 2003, 124, 160–171. [Google Scholar] [CrossRef]
- Lee, J.M.; Wagner, M.; Xiao, R.; Kim, K.H.; Feng, D.; Lazar, M.A.; Moore, D.D. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 2014, 516, 112–115. [Google Scholar] [CrossRef]
- Iershov, A.; Nemazanyy, I.; Alkhoury, C.; Girard, M.; Barth, E.; Cagnard, N.; Montagner, A.; Chretien, D.; Rugarli, E.I.; Guillou, H.; et al. The class 3 PI3K coordinates autophagy and mitochondrial lipid catabolism by controlling nuclear receptor PPARalpha. Nat. Commun. 2019, 10, 1566. [Google Scholar] [CrossRef]
- Saito, T.; Kuma, A.; Sugiura, Y.; Ichimura, Y.; Obata, M.; Kitamura, H.; Okuda, S.; Lee, H.C.; Ikeda, K.; Kanegae, Y.; et al. Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat. Commun. 2019, 10, 1567. [Google Scholar] [CrossRef] [PubMed]
- Khambu, B.; Li, T.; Yan, S.; Yu, C.; Chen, X.; Goheen, M.; Li, Y.; Lin, J.; Cummings, O.W.; Lee, Y.A.; et al. Hepatic Autophagy Deficiency Compromises Farnesoid X Receptor Functionality and Causes Cholestatic Injury. Hepatology 2019, 69, 2196–2213. [Google Scholar] [CrossRef]
- Kim, E.Y.; Lee, J.M. Basal Autophagy Is Necessary for A Pharmacologic PPARalpha Transactivation. Cells 2022, 11, 754. [Google Scholar] [CrossRef]
- Kim, E.Y.; Lee, J.M. NRF2 Activation in Autophagy Defects Suppresses a Pharmacological Transactivation of the Nuclear Receptor FXR. Antioxidants 2022, 11, 370. [Google Scholar] [CrossRef]
- Takahashi, S.S.; Sou, Y.S.; Saito, T.; Kuma, A.; Yabe, T.; Sugiura, Y.; Lee, H.C.; Suematsu, M.; Yokomizo, T.; Koike, M.; et al. Loss of autophagy impairs physiological steatosis by accumulation of NCoR1. Life Sci. Alliance 2020, 3, e201900513. [Google Scholar] [CrossRef]
- Evans, R.M. The steroid and thyroid hormone receptor superfamily. Science 1988, 240, 889–895. [Google Scholar] [CrossRef]
- Evans, R.M.; Mangelsdorf, D.J. Nuclear Receptors, RXR, and the Big Bang. Cell 2014, 157, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Artandi, S.E.; Cooper, J.P. Reverse transcribing the code for chromosome stability. Mol. Cell 2009, 36, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, J.; Pei, L.; Evans, R.M. Nuclear receptors: Decoding metabolic disease. FEBS Lett. 2008, 582, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Ahmadian, M.; Yu, R.T.; Atkins, A.R.; Downes, M.; Evans, R.M. Nuclear receptors and metabolism: From feast to famine. Diabetologia 2014, 57, 860–867. [Google Scholar] [CrossRef]
- Chawla, A.; Repa, J.J.; Evans, R.M.; Mangelsdorf, D.J. Nuclear receptors and lipid physiology: Opening the X-files. Science 2001, 294, 1866–1870. [Google Scholar] [CrossRef]
- Modica, S.; Gadaleta, R.M.; Moschetta, A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl. Recept. Signal 2010, 8, e005. [Google Scholar] [CrossRef]
- Gearing, K.L.; Gottlicher, M.; Teboul, M.; Widmark, E.; Gustafsson, J.A. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc. Natl. Acad. Sci. USA 1993, 90, 1440–1444. [Google Scholar] [CrossRef]
- Miyata, K.S.; McCaw, S.E.; Marcus, S.L.; Rachubinski, R.A.; Capone, J.P. The peroxisome proliferator-activated receptor interacts with the retinoid X receptor in vivo. Gene 1994, 148, 327–330. [Google Scholar] [CrossRef]
- Bardot, O.; Aldridge, T.C.; Latruffe, N.; Green, S. PPAR-RXR heterodimer activates a peroxisome proliferator response element upstream of the bifunctional enzyme gene. Biochem. Biophys. Res. Commun. 1993, 192, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Schoonjans, K.; Peinado-Onsurbe, J.; Lefebvre, A.M.; Heyman, R.A.; Briggs, M.; Deeb, S.; Staels, B.; Auwerx, J. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996, 15, 5336–5348. [Google Scholar] [CrossRef] [PubMed]
- Watts, G.F.; Dimmitt, S.B. Fibrates, dyslipoproteinaemia and cardiovascular disease. Curr. Opin. Lipidol. 1999, 10, 561–574. [Google Scholar] [CrossRef]
- Leone, T.C.; Weinheimer, C.J.; Kelly, D.P. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: The PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. USA 1999, 96, 7473–7478. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S.; Seydoux, J.; Peters, J.M.; Gonzalez, F.J.; Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Investig. 1999, 103, 1489–1498. [Google Scholar] [CrossRef]
- Rakhshandehroo, M.; Sanderson, L.M.; Matilainen, M.; Stienstra, R.; Carlberg, C.; de Groot, P.J.; Muller, M.; Kersten, S. Comprehensive analysis of PPARalpha-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007, 2007, 26839. [Google Scholar] [CrossRef]
- Rakhshandehroo, M.; Hooiveld, G.; Muller, M.; Kersten, S. Comparative analysis of gene regulation by the transcription factor PPARalpha between mouse and human. PLoS ONE 2009, 4, e6796. [Google Scholar] [CrossRef]
- Rakhshandehroo, M.; Knoch, B.; Muller, M.; Kersten, S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010, 2010, 612089. [Google Scholar] [CrossRef]
- Lu, Y.; Boekschoten, M.V.; Wopereis, S.; Muller, M.; Kersten, S. Comparative transcriptomic and metabolomic analysis of fenofibrate and fish oil treatments in mice. Physiol. Genom. 2011, 43, 1307–1318. [Google Scholar] [CrossRef]
- Janssen, A.W.; Betzel, B.; Stoopen, G.; Berends, F.J.; Janssen, I.M.; Peijnenburg, A.A.; Kersten, S. The impact of PPARalpha activation on whole genome gene expression in human precision cut liver slices. BMC Genom. 2015, 16, 760. [Google Scholar] [CrossRef]
- Christofides, A.; Konstantinidou, E.; Jani, C.; Boussiotis, V.A. The role of peroxisome proliferator-activated receptors (PPAR) in immune responses. Metabolism 2021, 114, 154338. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, Y.; Li, P.F. PPARalpha: An emerging target of metabolic syndrome, neurodegenerative and cardiovascular diseases. Front. Endocrinol. 2022, 13, 1074911. [Google Scholar] [CrossRef]
- Lee, S.S.; Pineau, T.; Drago, J.; Lee, E.J.; Owens, J.W.; Kroetz, D.L.; Fernandez-Salguero, P.M.; Westphal, H.; Gonzalez, F.J. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell Biol. 1995, 15, 3012–3022. [Google Scholar] [CrossRef]
- Ren, B.; Thelen, A.; Jump, D.B. Peroxisome proliferator-activated receptor alpha inhibits hepatic S14 gene transcription. Evidence against the peroxisome proliferator-activated receptor alpha as the mediator of polyunsaturated fatty acid regulation of s14 gene transcription. J. Biol. Chem. 1996, 271, 17167–17173. [Google Scholar] [CrossRef]
- Staels, B.; Koenig, W.; Habib, A.; Merval, R.; Lebret, M.; Torra, I.P.; Delerive, P.; Fadel, A.; Chinetti, G.; Fruchart, J.C.; et al. Activation of human aortic smooth-muscle cells is inhibited by PPARalpha but not by PPARgamma activators. Nature 1998, 393, 790–793. [Google Scholar] [CrossRef]
- Chinetti, G.; Fruchart, J.C.; Staels, B. Peroxisome proliferator-activated receptors (PPARs): Nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm. Res. 2000, 49, 497–505. [Google Scholar] [CrossRef]
- Gervois, P.; Vu-Dac, N.; Kleemann, R.; Kockx, M.; Dubois, G.; Laine, B.; Kosykh, V.; Fruchart, J.C.; Kooistra, T.; Staels, B. Negative regulation of human fibrinogen gene expression by peroxisome proliferator-activated receptor alpha agonists via inhibition of CCAAT box/enhancer-binding protein beta. J. Biol. Chem. 2001, 276, 33471–33477. [Google Scholar] [CrossRef]
- Oka, S.; Alcendor, R.; Zhai, P.; Park, J.Y.; Shao, D.; Cho, J.; Yamamoto, T.; Tian, B.; Sadoshima, J. PPARalpha-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011, 14, 598–611. [Google Scholar] [CrossRef]
- Oka, S.; Zhai, P.; Alcendor, R.; Park, J.Y.; Tian, B.; Sadoshima, J. Suppression of ERR targets by a PPARalpha/Sirt1 complex in the failing heart. Cell Cycle 2012, 11, 856–864. [Google Scholar] [CrossRef][Green Version]
- Bougarne, N.; Paumelle, R.; Caron, S.; Hennuyer, N.; Mansouri, R.; Gervois, P.; Staels, B.; Haegeman, G.; De Bosscher, K. PPARalpha blocks glucocorticoid receptor alpha-mediated transactivation but cooperates with the activated glucocorticoid receptor alpha for transrepression on NF-kappaB. Proc. Natl. Acad. Sci. USA 2009, 106, 7397–7402. [Google Scholar] [CrossRef]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular Actions of PPARalpha in Lipid Metabolism and Inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef]
- Liang, C.P.; Tall, A.R. Transcriptional profiling reveals global defects in energy metabolism, lipoprotein, and bile acid synthesis and transport with reversal by leptin treatment in ob/ob mouse liver. J. Biol. Chem. 2001, 276, 49066–49076. [Google Scholar] [CrossRef]
- Suzuki, A.; Okamoto, S.; Lee, S.; Saito, K.; Shiuchi, T.; Minokoshi, Y. Leptin stimulates fatty acid oxidation and peroxisome proliferator-activated receptor alpha gene expression in mouse C2C12 myoblasts by changing the subcellular localization of the alpha2 form of AMP-activated protein kinase. Mol. Cell Biol. 2007, 27, 4317–4327. [Google Scholar] [CrossRef]
- Zhou, Y.T.; Shimabukuro, M.; Wang, M.Y.; Lee, Y.; Higa, M.; Milburn, J.L.; Newgard, C.B.; Unger, R.H. Role of peroxisome proliferator-activated receptor alpha in disease of pancreatic beta cells. Proc. Natl. Acad. Sci. USA 1998, 95, 8898–8903. [Google Scholar] [CrossRef]
- You, M.; Considine, R.V.; Leone, T.C.; Kelly, D.P.; Crabb, D.W. Role of adiponectin in the protective action of dietary saturated fat against alcoholic fatty liver in mice. Hepatology 2005, 42, 568–577. [Google Scholar] [CrossRef]
- Carlsson, L.; Linden, D.; Jalouli, M.; Oscarsson, J. Effects of fatty acids and growth hormone on liver fatty acid binding protein and PPARalpha in rat liver. Am. J. Physiol. Endocrinol. Metab. 2001, 281, E772–E781. [Google Scholar] [CrossRef]
- Yamada, J.; Sugiyama, H.; Watanabe, T.; Suga, T. Suppressive effect of growth hormone on the expression of peroxisome proliferator-activated receptor in cultured rat hepatocytes. Res. Commun. Mol. Pathol. Pharmacol. 1995, 90, 173–176. [Google Scholar]
- Jalouli, M.; Carlsson, L.; Ameen, C.; Linden, D.; Ljungberg, A.; Michalik, L.; Eden, S.; Wahli, W.; Oscarsson, J. Sex difference in hepatic peroxisome proliferator-activated receptor alpha expression: Influence of pituitary and gonadal hormones. Endocrinology 2003, 144, 101–109. [Google Scholar] [CrossRef]
- Zhou, Y.C.; Waxman, D.J. Cross-talk between janus kinase-signal transducer and activator of transcription (JAK-STAT) and peroxisome proliferator-activated receptor-alpha (PPARalpha) signaling pathways. Growth hormone inhibition of pparalpha transcriptional activity mediated by stat5b. J. Biol. Chem. 1999, 274, 2672–2681. [Google Scholar]
- Ljungberg, A.; Linden, D.; Ameen, C.; Bergstrom, G.; Oscarsson, J. Importance of PPAR alpha for the effects of growth hormone on hepatic lipid and lipoprotein metabolism. Growth Horm. IGF Res. 2007, 17, 154–164. [Google Scholar] [CrossRef]
- Steineger, H.H.; Sorensen, H.N.; Tugwood, J.D.; Skrede, S.; Spydevold, O.; Gautvik, K.M. Dexamethasone and insulin demonstrate marked and opposite regulation of the steady-state mRNA level of the peroxisomal proliferator-activated receptor (PPAR) in hepatic cells. Hormonal modulation of fatty-acid-induced transcription. Eur. J. Biochem. 1994, 225, 967–974. [Google Scholar] [CrossRef]
- Yang, X.; Downes, M.; Yu, R.T.; Bookout, A.L.; He, W.; Straume, M.; Mangelsdorf, D.J.; Evans, R.M. Nuclear receptor expression links the circadian clock to metabolism. Cell 2006, 126, 801–810. [Google Scholar] [CrossRef]
- Lemberger, T.; Staels, B.; Saladin, R.; Desvergne, B.; Auwerx, J.; Wahli, W. Regulation of the peroxisome proliferator-activated receptor alpha gene by glucocorticoids. J. Biol. Chem. 1994, 269, 24527–24530. [Google Scholar] [CrossRef]
- Lemberger, T.; Saladin, R.; Vazquez, M.; Assimacopoulos, F.; Staels, B.; Desvergne, B.; Wahli, W.; Auwerx, J. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J. Biol. Chem. 1996, 271, 1764–1769. [Google Scholar] [CrossRef]
- Oishi, K.; Shirai, H.; Ishida, N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem. J. 2005, 386, 575–581. [Google Scholar] [CrossRef]
- Gachon, F.; Leuenberger, N.; Claudel, T.; Gos, P.; Jouffe, C.; Fleury Olela, F.; de Mollerat du Jeu, X.; Wahli, W.; Schibler, U. Proline- and acidic amino acid-rich basic leucine zipper proteins modulate peroxisome proliferator-activated receptor alpha (PPARalpha) activity. Proc. Natl. Acad. Sci. USA 2011, 108, 4794–4799. [Google Scholar] [CrossRef]
- Poynter, M.E.; Daynes, R.A. Peroxisome proliferator-activated receptor alpha activation modulates cellular redox status, represses nuclear factor-kappaB signaling, and reduces inflammatory cytokine production in aging. J. Biol. Chem. 1998, 273, 32833–32841. [Google Scholar] [CrossRef]
- Hostetler, H.A.; Huang, H.; Kier, A.B.; Schroeder, F. Glucose directly links to lipid metabolism through high affinity interaction with peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 2008, 283, 2246–2254. [Google Scholar] [CrossRef]
- Sartippour, M.R.; Renier, G. Differential regulation of macrophage peroxisome proliferator-activated receptor expression by glucose: Role of peroxisome proliferator-activated receptors in lipoprotein lipase gene expression. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 104–110. [Google Scholar] [CrossRef]
- Roduit, R.; Morin, J.; Masse, F.; Segall, L.; Roche, E.; Newgard, C.B.; Assimacopoulos-Jeannet, F.; Prentki, M. Glucose down-regulates the expression of the peroxisome proliferator-activated receptor-alpha gene in the pancreatic beta -cell. J. Biol. Chem. 2000, 275, 35799–35806. [Google Scholar] [CrossRef]
- Beier, K.; Volkl, A.; Fahimi, H.D. TNF-alpha downregulates the peroxisome proliferator activated receptor-alpha and the mRNAs encoding peroxisomal proteins in rat liver. FEBS Lett. 1997, 412, 385–387. [Google Scholar] [CrossRef]
- Parmentier, J.H.; Schohn, H.; Bronner, M.; Ferrari, L.; Batt, A.M.; Dauca, M.; Kremers, P. Regulation of CYP4A1 and peroxisome proliferator-activated receptor alpha expression by interleukin-1beta, interleukin-6, and dexamethasone in cultured fetal rat hepatocytes. Biochem. Pharmacol. 1997, 54, 889–898. [Google Scholar] [CrossRef]
- Bechmann, L.P.; Vetter, D.; Ishida, J.; Hannivoort, R.A.; Lang, U.E.; Kocabayoglu, P.; Fiel, M.I.; Munoz, U.; Patman, G.L.; Ge, F.; et al. Post-transcriptional activation of PPAR alpha by KLF6 in hepatic steatosis. J. Hepatol. 2013, 58, 1000–1006. [Google Scholar] [CrossRef]
- Pineda Torra, I.; Jamshidi, Y.; Flavell, D.M.; Fruchart, J.C.; Staels, B. Characterization of the human PPARalpha promoter: Identification of a functional nuclear receptor response element. Mol. Endocrinol. 2002, 16, 1013–1028. [Google Scholar]
- Hayhurst, G.P.; Lee, Y.H.; Lambert, G.; Ward, J.M.; Gonzalez, F.J. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell Biol. 2001, 21, 1393–1403. [Google Scholar] [CrossRef]
- Inoue, J.; Satoh, S.; Kita, M.; Nakahara, M.; Hachimura, S.; Miyata, M.; Nishimaki-Mogami, T.; Sato, R. PPARalpha gene expression is up-regulated by LXR and PXR activators in the small intestine. Biochem. Biophys. Res. Commun. 2008, 371, 675–678. [Google Scholar] [CrossRef]
- Roszer, T.; Menendez-Gutierrez, M.P.; Cedenilla, M.; Ricote, M. Retinoid X receptors in macrophage biology. Trends Endocrinol. Metab. 2013, 24, 460–468. [Google Scholar] [CrossRef]
- Blanquart, C.; Barbier, O.; Fruchart, J.C.; Staels, B.; Glineur, C. Peroxisome proliferator-activated receptor alpha (PPARalpha ) turnover by the ubiquitin-proteasome system controls the ligand-induced expression level of its target genes. J. Biol. Chem. 2002, 277, 37254–37259. [Google Scholar] [CrossRef]
- Kersten, S.; Desvergne, B.; Wahli, W. Roles of PPARs in health and disease. Nature 2000, 405, 421–424. [Google Scholar] [CrossRef]
- Devchand, P.R.; Keller, H.; Peters, J.M.; Vazquez, M.; Gonzalez, F.J.; Wahli, W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature 1996, 384, 39–43. [Google Scholar] [CrossRef]
- Hostetler, H.A.; Petrescu, A.D.; Kier, A.B.; Schroeder, F. Peroxisome proliferator-activated receptor alpha interacts with high affinity and is conformationally responsive to endogenous ligands. J. Biol. Chem. 2005, 280, 18667–18682. [Google Scholar] [CrossRef]
- Schoonjans, K.; Staels, B.; Auwerx, J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J. Lipid Res. 1996, 37, 907–925. [Google Scholar] [CrossRef]
- Ellinghaus, P.; Wolfrum, C.; Assmann, G.; Spener, F.; Seedorf, U. Phytanic acid activates the peroxisome proliferator-activated receptor alpha (PPARalpha) in sterol carrier protein 2-/ sterol carrier protein x-deficient mice. J. Biol. Chem. 1999, 274, 2766–2772. [Google Scholar] [CrossRef]
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef]
- Yu, K.; Bayona, W.; Kallen, C.B.; Harding, H.P.; Ravera, C.P.; McMahon, G.; Brown, M.; Lazar, M.A. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J. Biol. Chem. 1995, 270, 23975–23983. [Google Scholar] [CrossRef]
- Ng, V.Y.; Huang, Y.; Reddy, L.M.; Falck, J.R.; Lin, E.T.; Kroetz, D.L. Cytochrome P450 eicosanoids are activators of peroxisome proliferator-activated receptor alpha. Drug Metab. Dispos. 2007, 35, 1126–1134. [Google Scholar] [CrossRef]
- Lin, Q.; Ruuska, S.E.; Shaw, N.S.; Dong, D.; Noy, N. Ligand selectivity of the peroxisome proliferator-activated receptor alpha. Biochemistry 1999, 38, 185–190. [Google Scholar] [CrossRef]
- Narala, V.R.; Adapala, R.K.; Suresh, M.V.; Brock, T.G.; Peters-Golden, M.; Reddy, R.C. Leukotriene B4 is a physiologically relevant endogenous peroxisome proliferator-activated receptor-alpha agonist. J. Biol. Chem. 2010, 285, 22067–22074. [Google Scholar] [CrossRef]
- Delerive, P.; Furman, C.; Teissier, E.; Fruchart, J.; Duriez, P.; Staels, B. Oxidized phospholipids activate PPARalpha in a phospholipase A2-dependent manner. FEBS Lett. 2000, 471, 34–38. [Google Scholar] [CrossRef]
- Chakravarthy, M.V.; Lodhi, I.J.; Yin, L.; Malapaka, R.R.; Xu, H.E.; Turk, J.; Semenkovich, C.F. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell 2009, 138, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Fougerat, A.; Schoiswohl, G.; Polizzi, A.; Regnier, M.; Wagner, C.; Smati, S.; Fougeray, T.; Lippi, Y.; Lasserre, F.; Raho, I.; et al. ATGL-dependent white adipose tissue lipolysis controls hepatocyte PPARalpha activity. Cell Rep. 2022, 39, 110910. [Google Scholar] [CrossRef] [PubMed]
- Polizzi, A.; Fouche, E.; Ducheix, S.; Lasserre, F.; Marmugi, A.P.; Mselli-Lakhal, L.; Loiseau, N.; Wahli, W.; Guillou, H.; Montagner, A. Hepatic Fasting-Induced PPARalpha Activity Does Not Depend on Essential Fatty Acids. Int. J. Mol. Sci. 2016, 17, 1624. [Google Scholar] [CrossRef]
- Badman, M.K.; Pissios, P.; Kennedy, A.R.; Koukos, G.; Flier, J.S.; Maratos-Flier, E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007, 5, 426–437. [Google Scholar] [CrossRef]
- Chakravarthy, M.V.; Pan, Z.; Zhu, Y.; Tordjman, K.; Schneider, J.G.; Coleman, T.; Turk, J.; Semenkovich, C.F. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005, 1, 309–322. [Google Scholar] [CrossRef]
- Lodhi, I.J.; Wei, X.; Semenkovich, C.F. Lipoexpediency: De novo lipogenesis as a metabolic signal transmitter. Trends Endocrinol. Metab. 2011, 22, 1–8. [Google Scholar] [CrossRef]
- Fu, J.; Gaetani, S.; Oveisi, F.; Lo Verme, J.; Serrano, A.; Rodriguez De Fonseca, F.; Rosengarth, A.; Luecke, H.; Di Giacomo, B.; Tarzia, G.; et al. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature 2003, 425, 90–93. [Google Scholar] [CrossRef]
- Issemann, I.; Green, S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 1990, 347, 645–650. [Google Scholar] [CrossRef]
- Morimura, K.; Cheung, C.; Ward, J.M.; Reddy, J.K.; Gonzalez, F.J. Differential susceptibility of mice humanized for peroxisome proliferator-activated receptor alpha to Wy-14,643-induced liver tumorigenesis. Carcinogenesis 2006, 27, 1074–1080. [Google Scholar] [CrossRef]
- De La Iglesia, F.A.; Lewis, J.E.; Buchanan, R.A.; Marcus, E.L.; McMahon, G. Light and electron microscopy of liver in hyperlipoproteinemic patients under long-term gemfibrozil treatment. Atherosclerosis 1982, 43, 19–37. [Google Scholar] [CrossRef]
- Blumcke, S.; Schwartzkopff, W.; Lobeck, H.; Edmondson, N.A.; Prentice, D.E.; Blane, G.F. Influence of fenofibrate on cellular and subcellular liver structure in hyperlipidemic patients. Atherosclerosis 1983, 46, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Feige, J.N.; Gelman, L.; Michalik, L.; Desvergne, B.; Wahli, W. From molecular action to physiological outputs: Peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog. Lipid Res. 2006, 45, 120–159. [Google Scholar] [CrossRef] [PubMed]
- White, J.H.; Fernandes, I.; Mader, S.; Yang, X.J. Corepressor recruitment by agonist-bound nuclear receptors. Vitam. Horm. 2004, 68, 123–143. [Google Scholar] [PubMed]
- Leonardsson, G.; Steel, J.H.; Christian, M.; Pocock, V.; Milligan, S.; Bell, J.; So, P.W.; Medina-Gomez, G.; Vidal-Puig, A.; White, R.; et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. USA 2004, 101, 8437–8442. [Google Scholar] [CrossRef]
- Viswakarma, N.; Jia, Y.; Bai, L.; Vluggens, A.; Borensztajn, J.; Xu, J.; Reddy, J.K. Coactivators in PPAR-Regulated Gene Expression. PPAR Res. 2010, 2010, 250126. [Google Scholar] [CrossRef]
- Surapureddi, S.; Yu, S.; Bu, H.; Hashimoto, T.; Yeldandi, A.V.; Kashireddy, P.; Cherkaoui-Malki, M.; Qi, C.; Zhu, Y.J.; Rao, M.S.; et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc. Natl. Acad. Sci. USA 2002, 99, 11836–11841. [Google Scholar] [CrossRef]
- Matsumoto, K.; Yu, S.; Jia, Y.; Ahmed, M.R.; Viswakarma, N.; Sarkar, J.; Kashireddy, P.V.; Rao, M.S.; Karpus, W.; Gonzalez, F.J.; et al. Critical role for transcription coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein/TRAP220 in liver regeneration and PPARalpha ligand-induced liver tumor development. J. Biol. Chem. 2007, 282, 17053–17060. [Google Scholar] [CrossRef]
- Dasgupta, S.; Lonard, D.M.; O′Malley, B.W. Nuclear receptor coactivators: Master regulators of human health and disease. Annu. Rev. Med. 2014, 65, 279–292. [Google Scholar] [CrossRef]
- Ogryzko, V.V.; Schiltz, R.L.; Russanova, V.; Howard, B.H.; Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 1996, 87, 953–959. [Google Scholar] [CrossRef]
- Spencer, T.E.; Jenster, G.; Burcin, M.M.; Allis, C.D.; Zhou, J.; Mizzen, C.A.; McKenna, N.J.; Onate, S.A.; Tsai, S.Y.; Tsai, M.J.; et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 1997, 389, 194–198. [Google Scholar] [CrossRef]
- Pourcet, B.; Pineda-Torra, I.; Derudas, B.; Staels, B.; Glineur, C. SUMOylation of human peroxisome proliferator-activated receptor alpha inhibits its trans-activity through the recruitment of the nuclear corepressor NCoR. J. Biol. Chem. 2010, 285, 5983–5992. [Google Scholar] [CrossRef] [PubMed]
- Shalev, A.; Siegrist-Kaiser, C.A.; Yen, P.M.; Wahli, W.; Burger, A.G.; Chin, W.W.; Meier, C.A. The peroxisome proliferator-activated receptor alpha is a phosphoprotein: Regulation by insulin. Endocrinology 1996, 137, 4499–4502. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.A.; Vanden Heuvel, J.P. Modulation of PPAR activity via phosphorylation. Biochim. Biophys. Acta 2007, 1771, 952–960. [Google Scholar] [CrossRef]
- Blanquart, C.; Mansouri, R.; Paumelle, R.; Fruchart, J.C.; Staels, B.; Glineur, C. The protein kinase C signaling pathway regulates a molecular switch between transactivation and transrepression activity of the peroxisome proliferator-activated receptor alpha. Mol. Endocrinol. 2004, 18, 1906–1918. [Google Scholar] [CrossRef]
- Juge-Aubry, C.E.; Hammar, E.; Siegrist-Kaiser, C.; Pernin, A.; Takeshita, A.; Chin, W.W.; Burger, A.G.; Meier, C.A. Regulation of the transcriptional activity of the peroxisome proliferator-activated receptor alpha by phosphorylation of a ligand-independent trans-activating domain. J. Biol. Chem. 1999, 274, 10505–10510. [Google Scholar] [CrossRef]
- Barger, P.M.; Browning, A.C.; Garner, A.N.; Kelly, D.P. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: A potential role in the cardiac metabolic stress response. J. Biol. Chem. 2001, 276, 44495–44501. [Google Scholar] [CrossRef]
- Lazennec, G.; Canaple, L.; Saugy, D.; Wahli, W. Activation of peroxisome proliferator-activated receptors (PPARs) by their ligands and protein kinase A activators. Mol. Endocrinol. 2000, 14, 1962–1975. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Kim, M.; Park, H.S.; Kim, H.S.; Jeon, M.J.; Oh, K.S.; Koh, E.H.; Won, J.C.; Kim, M.S.; Oh, G.T.; et al. AMPK activation increases fatty acid oxidation in skeletal muscle by activating PPARalpha and PGC-1. Biochem. Biophys. Res. Commun. 2006, 340, 291–295. [Google Scholar] [CrossRef]
- Yoon, M.J.; Lee, G.Y.; Chung, J.J.; Ahn, Y.H.; Hong, S.H.; Kim, J.B. Adiponectin increases fatty acid oxidation in skeletal muscle cells by sequential activation of AMP-activated protein kinase, p38 mitogen-activated protein kinase, and peroxisome proliferator-activated receptor alpha. Diabetes 2006, 55, 2562–2570. [Google Scholar] [CrossRef]
- Hinds, T.D., Jr.; Burns, K.A.; Hosick, P.A.; McBeth, L.; Nestor-Kalinoski, A.; Drummond, H.A.; AlAmodi, A.A.; Hankins, M.W.; Vanden Heuvel, J.P.; Stec, D.E. Biliverdin Reductase A Attenuates Hepatic Steatosis by Inhibition of Glycogen Synthase Kinase (GSK) 3beta Phosphorylation of Serine 73 of Peroxisome Proliferator-activated Receptor (PPAR) alpha. J. Biol. Chem. 2016, 291, 25179–25191. [Google Scholar] [CrossRef]
- Sengupta, S.; Peterson, T.R.; Laplante, M.; Oh, S.; Sabatini, D.M. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 2010, 468, 1100–1104. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Pyo, S.; Um, S.H. S6 kinase 2 deficiency enhances ketone body production and increases peroxisome proliferator-activated receptor alpha activity in the liver. Hepatology 2012, 55, 1727–1737. [Google Scholar] [CrossRef]
- Quinn, W.J., 3rd; Wan, M.; Shewale, S.V.; Gelfer, R.; Rader, D.J.; Birnbaum, M.J.; Titchenell, P.M. mTORC1 stimulates phosphatidylcholine synthesis to promote triglyceride secretion. J. Clin. Investig. 2017, 127, 4207–4215. [Google Scholar] [CrossRef] [PubMed]
- Selen, E.S.; Wolfgang, M.J. mTORC1 activation is not sufficient to suppress hepatic PPARalpha signaling or ketogenesis. J. Biol. Chem. 2021, 297, 100884. [Google Scholar] [CrossRef]
- Bronner, M.; Hertz, R.; Bar-Tana, J. Kinase-independent transcriptional co-activation of peroxisome proliferator-activated receptor alpha by AMP-activated protein kinase. Biochem. J. 2004, 384, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Joly, E.; Roduit, R.; Peyot, M.L.; Habinowski, S.A.; Ruderman, N.B.; Witters, L.A.; Prentki, M. Glucose represses PPARalpha gene expression via AMP-activated protein kinase but not via p38 mitogen-activated protein kinase in the pancreatic beta-cell. J. Diabetes 2009, 1, 263–272. [Google Scholar] [CrossRef]
- Ravnskjaer, K.; Boergesen, M.; Dalgaard, L.T.; Mandrup, S. Glucose-induced repression of PPARalpha gene expression in pancreatic beta-cells involves PP2A activation and AMPK inactivation. J. Mol. Endocrinol. 2006, 36, 289–299. [Google Scholar] [CrossRef]
- Yamauchi, T.; Nio, Y.; Maki, T.; Kobayashi, M.; Takazawa, T.; Iwabu, M.; Okada-Iwabu, M.; Kawamoto, S.; Kubota, N.; Kubota, T.; et al. Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat. Med. 2007, 13, 332–339. [Google Scholar] [CrossRef]
- Seol, W.; Choi, H.S.; Moore, D.D. Isolation of proteins that interact specifically with the retinoid X receptor: Two novel orphan receptors. Mol. Endocrinol. 1995, 9, 72–85. [Google Scholar]
- Forman, B.M.; Goode, E.; Chen, J.; Oro, A.E.; Bradley, D.J.; Perlmann, T.; Noonan, D.J.; Burka, L.T.; McMorris, T.; Lamph, W.W.; et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 1995, 81, 687–693. [Google Scholar] [CrossRef]
- Ramos Pittol, J.M.; Milona, A.; Morris, I.; Willemsen, E.C.L.; van der Veen, S.W.; Kalkhoven, E.; van Mil, S.W.C. FXR Isoforms Control Different Metabolic Functions in Liver Cells via Binding to Specific DNA Motifs. Gastroenterology 2020, 159, 1853–1865.e10. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Hart, S.N.; Kong, B.; Fang, J.; Zhong, X.B.; Guo, G.L. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology 2010, 51, 1410–1419. [Google Scholar] [CrossRef] [PubMed]
- van der Veen, S.W.; Dijkstra, J.J.; Willemsen, E.C.L.; Houtman, R.; Milona, A.; Marchet, N.; Spit, M.; Hollman, D.; Zwartkruis, F.J.T.; Vermeulen, M.; et al. The farnesoid X receptor activates transcription independently of RXR at non-canonical response elements. Nucleic Acids Res. 2025, 53, gkae1214. [Google Scholar] [CrossRef]
- Ma, K.; Saha, P.K.; Chan, L.; Moore, D.D. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Investig. 2006, 116, 1102–1109. [Google Scholar] [CrossRef]
- Massafra, V.; Milona, A.; Vos, H.R.; Ramos, R.J.J.; Gerrits, J.; Willemsen, E.C.L.; Ramos Pittol, J.M.; Ijssennagger, N.; Houweling, M.; Prinsen, H.; et al. Farnesoid X Receptor Activation Promotes Hepatic Amino Acid Catabolism and Ammonium Clearance in Mice. Gastroenterology 2017, 152, 1462–1476.e10. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Cai, J.; Gonzalez, F.J. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 335–347. [Google Scholar] [CrossRef]
- Fuchs, C.D.; Trauner, M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 432–450. [Google Scholar] [CrossRef]
- Yang, J.; Pontoglio, M.; Terzi, F. Bile Acids and Farnesoid X Receptor in Renal Pathophysiology. Nephron 2024, 148, 618–630. [Google Scholar] [CrossRef]
- Sinal, C.J.; Tohkin, M.; Miyata, M.; Ward, J.M.; Lambert, G.; Gonzalez, F.J. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 2000, 102, 731–744. [Google Scholar] [CrossRef]
- De Magalhaes Filho, C.D.; Downes, M.; Evans, R.M. Farnesoid X Receptor an Emerging Target to Combat Obesity. Dig. Dis. 2017, 35, 185–190. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Mangelsdorf, D.J. Bile Acids as Hormones: The FXR-FGF15/19 Pathway. Dig. Dis. 2015, 33, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Makishima, M.; Okamoto, A.Y.; Repa, J.J.; Tu, H.; Learned, R.M.; Luk, A.; Hull, M.V.; Lustig, K.D.; Mangelsdorf, D.J.; Shan, B. Identification of a nuclear receptor for bile acids. Science 1999, 284, 1362–1365. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.J.; Blanchard, S.G.; Bledsoe, R.K.; Chandra, G.; Consler, T.G.; Kliewer, S.A.; Stimmel, J.B.; Willson, T.M.; Zavacki, A.M.; Moore, D.D.; et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science 1999, 284, 1365–1368. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Hollister, K.; Sowers, L.C.; Forman, B.M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 1999, 3, 543–553. [Google Scholar] [CrossRef]
- Li, F.; Jiang, C.; Krausz, K.W.; Li, Y.; Albert, I.; Hao, H.; Fabre, K.M.; Mitchell, J.B.; Patterson, A.D.; Gonzalez, F.J. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 2013, 4, 2384. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Won, T.H.; Arifuzzaman, M.; Parkhurst, C.N.; Miranda, I.C.; Zhang, B.; Hu, E.; Kashyap, S.; Letourneau, J.; Jin, W.B.; Fu, Y.; et al. Host metabolism balances microbial regulation of bile acid signalling. Nature 2025, 638, 216–224. [Google Scholar] [CrossRef]
- Mueller, M.; Thorell, A.; Claudel, T.; Jha, P.; Koefeler, H.; Lackner, C.; Hoesel, B.; Fauler, G.; Stojakovic, T.; Einarsson, C.; et al. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J. Hepatol. 2015, 62, 1398–1404. [Google Scholar] [CrossRef]
- Urizar, N.L.; Liverman, A.B.; Dodds, D.T.; Silva, F.V.; Ordentlich, P.; Yan, Y.; Gonzalez, F.J.; Heyman, R.A.; Mangelsdorf, D.J.; Moore, D.D. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science 2002, 296, 1703–1706. [Google Scholar] [CrossRef]
- Brevini, T.; Maes, M.; Webb, G.J.; John, B.V.; Fuchs, C.D.; Buescher, G.; Wang, L.; Griffiths, C.; Brown, M.L.; Scott, W.E., 3rd; et al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature 2023, 615, 134–142. [Google Scholar] [CrossRef]
- Maloney, P.R.; Parks, D.J.; Haffner, C.D.; Fivush, A.M.; Chandra, G.; Plunket, K.D.; Creech, K.L.; Moore, L.B.; Wilson, J.G.; Lewis, M.C.; et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 2000, 43, 2971–2974. [Google Scholar] [CrossRef] [PubMed]
- Cariou, B.; van Harmelen, K.; Duran-Sandoval, D.; van Dijk, T.H.; Grefhorst, A.; Abdelkarim, M.; Caron, S.; Torpier, G.; Fruchart, J.C.; Gonzalez, F.J.; et al. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J. Biol. Chem. 2006, 281, 11039–11049. [Google Scholar] [CrossRef]
- Ma, Y.; Huang, Y.; Yan, L.; Gao, M.; Liu, D. Synthetic FXR agonist GW4064 prevents diet-induced hepatic steatosis and insulin resistance. Pharm. Res. 2013, 30, 1447–1457. [Google Scholar] [CrossRef]
- Downes, M.; Verdecia, M.A.; Roecker, A.J.; Hughes, R.; Hogenesch, J.B.; Kast-Woelbern, H.R.; Bowman, M.E.; Ferrer, J.L.; Anisfeld, A.M.; Edwards, P.A.; et al. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell 2003, 11, 1079–1092. [Google Scholar] [CrossRef]
- Fang, S.; Suh, J.M.; Reilly, S.M.; Yu, E.; Osborn, O.; Lackey, D.; Yoshihara, E.; Perino, A.; Jacinto, S.; Lukasheva, Y.; et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat. Med. 2015, 21, 159–165. [Google Scholar] [CrossRef]
- Pellicciari, R.; Fiorucci, S.; Camaioni, E.; Clerici, C.; Costantino, G.; Maloney, P.R.; Morelli, A.; Parks, D.J.; Willson, T.M. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 2002, 45, 3569–3572. [Google Scholar] [CrossRef]
- Nevens, F.; Andreone, P.; Mazzella, G.; Strasser, S.I.; Bowlus, C.; Invernizzi, P.; Drenth, J.P.; Pockros, P.J.; Regula, J.; Beuers, U.; et al. A Placebo-Controlled Trial of Obeticholic Acid in Primary Biliary Cholangitis. N. Engl. J. Med. 2016, 375, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Hirschfield, G.M.; Mason, A.; Luketic, V.; Lindor, K.; Gordon, S.C.; Mayo, M.; Kowdley, K.V.; Vincent, C.; Bodhenheimer, H.C., Jr.; Pares, A.; et al. Efficacy of obeticholic acid in patients with primary biliary cirrhosis and inadequate response to ursodeoxycholic acid. Gastroenterology 2015, 148, 751–761.e8. [Google Scholar] [CrossRef] [PubMed]
- Trauner, M.; Nevens, F.; Shiffman, M.L.; Drenth, J.P.H.; Bowlus, C.L.; Vargas, V.; Andreone, P.; Hirschfield, G.M.; Pencek, R.; Malecha, E.S.; et al. Long-term efficacy and safety of obeticholic acid for patients with primary biliary cholangitis: 3-year results of an international open-label extension study. Lancet Gastroenterol. Hepatol. 2019, 4, 445–453. [Google Scholar] [CrossRef]
- Kowdley, K.V.; Luketic, V.; Chapman, R.; Hirschfield, G.M.; Poupon, R.; Schramm, C.; Vincent, C.; Rust, C.; Pares, A.; Mason, A.; et al. A randomized trial of obeticholic acid monotherapy in patients with primary biliary cholangitis. Hepatology 2018, 67, 1890–1902. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Y.; Wang, G.; Hao, H.; Wang, H. FXR agonists for MASH therapy: Lessons and perspectives from obeticholic acid. Med. Res. Rev. 2024, 44, 568–586. [Google Scholar] [CrossRef] [PubMed]
- Massafra, V.; Pellicciari, R.; Gioiello, A.; van Mil, S.W.C. Progress and challenges of selective Farnesoid X Receptor modulation. Pharmacol. Ther. 2018, 191, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Kok, T.; Hulzebos, C.V.; Wolters, H.; Havinga, R.; Agellon, L.B.; Stellaard, F.; Shan, B.; Schwarz, M.; Kuipers, F. Enterohepatic circulation of bile salts in farnesoid X receptor-deficient mice: Efficient intestinal bile salt absorption in the absence of ileal bile acid-binding protein. J. Biol. Chem. 2003, 278, 41930–41937. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Morimura, K.; Shah, Y.; Yang, Q.; Ward, J.M.; Gonzalez, F.J. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 2007, 28, 940–946. [Google Scholar] [CrossRef]
- Bjursell, M.; Wedin, M.; Admyre, T.; Hermansson, M.; Bottcher, G.; Goransson, M.; Linden, D.; Bamberg, K.; Oscarsson, J.; Bohlooly, Y.M. Ageing Fxr deficient mice develop increased energy expenditure, improved glucose control and liver damage resembling NASH. PLoS ONE 2013, 8, e64721. [Google Scholar] [CrossRef]
- Yang, F.; Huang, X.; Yi, T.; Yen, Y.; Moore, D.D.; Huang, W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007, 67, 863–867. [Google Scholar] [CrossRef]
- Huang, W.; Ma, K.; Zhang, J.; Qatanani, M.; Cuvillier, J.; Liu, J.; Dong, B.; Huang, X.; Moore, D.D. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 2006, 312, 233–236. [Google Scholar] [CrossRef]
- Ryan, K.K.; Tremaroli, V.; Clemmensen, C.; Kovatcheva-Datchary, P.; Myronovych, A.; Karns, R.; Wilson-Perez, H.E.; Sandoval, D.A.; Kohli, R.; Backhed, F.; et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 2014, 509, 183–188. [Google Scholar] [CrossRef]
- Fu, T.; Coulter, S.; Yoshihara, E.; Oh, T.G.; Fang, S.; Cayabyab, F.; Zhu, Q.; Zhang, T.; Leblanc, M.; Liu, S.; et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 2019, 176, 1098–1112.e18. [Google Scholar] [CrossRef]
- Berrabah, W.; Aumercier, P.; Gheeraert, C.; Dehondt, H.; Bouchaert, E.; Alexandre, J.; Ploton, M.; Mazuy, C.; Caron, S.; Tailleux, A.; et al. Glucose sensing O-GlcNAcylation pathway regulates the nuclear bile acid receptor farnesoid X receptor (FXR). Hepatology 2014, 59, 2022–2033. [Google Scholar] [CrossRef]
- Kir, S.; Beddow, S.A.; Samuel, V.T.; Miller, P.; Previs, S.F.; Suino-Powell, K.; Xu, H.E.; Shulman, G.I.; Kliewer, S.A.; Mangelsdorf, D.J. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 2011, 331, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Patsouris, D.; Mandard, S.; Voshol, P.J.; Escher, P.; Tan, N.S.; Havekes, L.M.; Koenig, W.; Marz, W.; Tafuri, S.; Wahli, W.; et al. PPARalpha governs glycerol metabolism. J. Clin. Investig. 2004, 114, 94–103. [Google Scholar] [CrossRef]
- Xu, J.; Xiao, G.; Trujillo, C.; Chang, V.; Blanco, L.; Joseph, S.B.; Bassilian, S.; Saad, M.F.; Tontonoz, P.; Lee, W.N.; et al. Peroxisome proliferator-activated receptor alpha (PPARalpha) influences substrate utilization for hepatic glucose production. J. Biol. Chem. 2002, 277, 50237–50244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lee, F.Y.; Barrera, G.; Lee, H.; Vales, C.; Gonzalez, F.J.; Willson, T.M.; Edwards, P.A. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc. Natl. Acad. Sci. USA 2006, 103, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Stayrook, K.R.; Bramlett, K.S.; Savkur, R.S.; Ficorilli, J.; Cook, T.; Christe, M.E.; Michael, L.F.; Burris, T.P. Regulation of carbohydrate metabolism by the farnesoid X receptor. Endocrinology 2005, 146, 984–991. [Google Scholar] [CrossRef]
- Preidis, G.A.; Kim, K.H.; Moore, D.D. Nutrient-sensing nuclear receptors PPARalpha and FXR control liver energy balance. J. Clin. Investig. 2017, 127, 1193–1201. [Google Scholar] [CrossRef]
- Kim, K.H.; Moore, D.D. Regulation of Liver Energy Balance by the Nuclear Receptors Farnesoid X Receptor and Peroxisome Proliferator Activated Receptor alpha. Dig. Dis. 2017, 35, 203–209. [Google Scholar] [CrossRef]
- Oosterveer, M.H.; Grefhorst, A.; van Dijk, T.H.; Havinga, R.; Staels, B.; Kuipers, F.; Groen, A.K.; Reijngoud, D.J. Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. J. Biol. Chem. 2009, 284, 34036–34044. [Google Scholar] [CrossRef]
- Motojima, K.; Seto, K. Fibrates and statins rapidly and synergistically induce pyruvate dehydrogenase kinase 4 mRNA in the liver and muscles of mice. Biol. Pharm. Bull. 2003, 26, 954–958. [Google Scholar] [CrossRef]
- Duran-Sandoval, D.; Cariou, B.; Percevault, F.; Hennuyer, N.; Grefhorst, A.; van Dijk, T.H.; Gonzalez, F.J.; Fruchart, J.C.; Kuipers, F.; Staels, B. The farnesoid X receptor modulates hepatic carbohydrate metabolism during the fasting-refeeding transition. J. Biol. Chem. 2005, 280, 29971–29979. [Google Scholar] [CrossRef]
- Savkur, R.S.; Bramlett, K.S.; Michael, L.F.; Burris, T.P. Regulation of pyruvate dehydrogenase kinase expression by the farnesoid X receptor. Biochem. Biophys. Res. Commun. 2005, 329, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Bandsma, R.H.; Van Dijk, T.H.; Harmsel At, A.; Kok, T.; Reijngoud, D.J.; Staels, B.; Kuipers, F. Hepatic de novo synthesis of glucose 6-phosphate is not affected in peroxisome proliferator-activated receptor alpha-deficient mice but is preferentially directed toward hepatic glycogen stores after a short term fast. J. Biol. Chem. 2004, 279, 8930–8937. [Google Scholar] [CrossRef] [PubMed]
- Minassian, C.; Montano, S.; Mithieux, G. Regulatory role of glucose-6 phosphatase in the repletion of liver glycogen during refeeding in fasted rats. Biochim. Biophys. Acta 1999, 1452, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Mandard, S.; Stienstra, R.; Escher, P.; Tan, N.S.; Kim, I.; Gonzalez, F.J.; Wahli, W.; Desvergne, B.; Muller, M.; Kersten, S. Glycogen synthase 2 is a novel target gene of peroxisome proliferator-activated receptors. Cell Mol. Life Sci. 2007, 64, 1145–1157. [Google Scholar] [CrossRef]
- Hashimoto, T.; Cook, W.S.; Qi, C.; Yeldandi, A.V.; Reddy, J.K.; Rao, M.S. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 2000, 275, 28918–28928. [Google Scholar] [CrossRef]
- Cariou, B.; van Harmelen, K.; Duran-Sandoval, D.; van Dijk, T.; Grefhorst, A.; Bouchaert, E.; Fruchart, J.C.; Gonzalez, F.J.; Kuipers, F.; Staels, B. Transient impairment of the adaptive response to fasting in FXR-deficient mice. FEBS Lett. 2005, 579, 4076–4080. [Google Scholar] [CrossRef]
- Inagaki, T.; Dutchak, P.; Zhao, G.; Ding, X.; Gautron, L.; Parameswara, V.; Li, Y.; Goetz, R.; Mohammadi, M.; Esser, V.; et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007, 5, 415–425. [Google Scholar] [CrossRef]
- Sinal, C.J.; Yoon, M.; Gonzalez, F.J. Antagonism of the actions of peroxisome proliferator-activated receptor-alpha by bile acids. J. Biol. Chem. 2001, 276, 47154–47162. [Google Scholar] [CrossRef]
- Srivastava, R.A.; Jahagirdar, R.; Azhar, S.; Sharma, S.; Bisgaier, C.L. Peroxisome proliferator-activated receptor-alpha selective ligand reduces adiposity, improves insulin sensitivity and inhibits atherosclerosis in LDL receptor-deficient mice. Mol. Cell Biochem. 2006, 285, 35–50. [Google Scholar] [CrossRef]
- Rizvi, F.; Puri, A.; Bhatia, G.; Khanna, A.K.; Wulff, E.M.; Rastogi, A.K.; Chander, R. Antidyslipidemic action of fenofibrate in dyslipidemic-diabetic hamster model. Biochem. Biophys. Res. Commun. 2003, 305, 215–222. [Google Scholar] [CrossRef]
- Forcheron, F.; Cachefo, A.; Thevenon, S.; Pinteur, C.; Beylot, M. Mechanisms of the triglyceride- and cholesterol-lowering effect of fenofibrate in hyperlipidemic type 2 diabetic patients. Diabetes 2002, 51, 3486–3491. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Houten, S.M.; Wang, L.; Moschetta, A.; Mangelsdorf, D.J.; Heyman, R.A.; Moore, D.D.; Auwerx, J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Investig. 2004, 113, 1408–1418. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A.; Loomba, R.; Sanyal, A.J.; Lavine, J.E.; Van Natta, M.L.; Abdelmalek, M.F.; Chalasani, N.; Dasarathy, S.; Diehl, A.M.; Hameed, B.; et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet 2015, 385, 956–965. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Klionsky, D.J. Autophagy in health and disease: A double-edged sword. Science 2004, 306, 990–995. [Google Scholar] [CrossRef]
- Seglen, P.O.; Gordon, P.B.; Holen, I. Non-selective autophagy. Semin. Cell Biol. 1990, 1, 441–448. [Google Scholar]
- Lamark, T.; Johansen, T. Mechanisms of Selective Autophagy. Annu. Rev. Cell Dev. Biol. 2021, 37, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.N.S.; Hamasaki, M.; Kawabata, T.; Youle, R.J.; Yoshimori, T. The mechanisms and roles of selective autophagy in mammals. Nat. Rev. Mol. Cell Biol. 2023, 24, 167–185. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 2008, 19, 5360–5372. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; White, E. Autophagy and metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef]
- Seok, S.; Fu, T.; Choi, S.E.; Li, Y.; Zhu, R.; Kumar, S.; Sun, X.; Yoon, G.; Kang, Y.; Zhong, W.; et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 2014, 516, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Panzitt, K.; Jungwirth, E.; Krones, E.; Lee, J.M.; Pollheimer, M.; Thallinger, G.G.; Kolb-Lenz, D.; Xiao, R.; Thorell, A.; Trauner, M.; et al. FXR-dependent Rubicon induction impairs autophagy in models of human cholestasis. J. Hepatol. 2020, 72, 1122–1131. [Google Scholar] [CrossRef]
- Claudel, T.; Inoue, Y.; Barbier, O.; Duran-Sandoval, D.; Kosykh, V.; Fruchart, J.; Fruchart, J.C.; Gonzalez, F.J.; Staels, B. Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology 2003, 125, 544–555. [Google Scholar] [CrossRef]
- Chennamsetty, I.; Claudel, T.; Kostner, K.M.; Baghdasaryan, A.; Kratky, D.; Levak-Frank, S.; Frank, S.; Gonzalez, F.J.; Trauner, M.; Kostner, G.M. Farnesoid X receptor represses hepatic human APOA gene expression. J. Clin. Investig. 2011, 121, 3724–3734. [Google Scholar] [CrossRef]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef]
- Ohsumi, Y.; Mizushima, N. Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 2004, 15, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef]
- Xie, Z.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Mizushima, N. Autophagy and human diseases. Cell Res. 2014, 24, 69–79. [Google Scholar] [CrossRef]
- Mizushima, N.; Levine, B. Autophagy in Human Diseases. N. Engl. J. Med. 2020, 383, 1564–1576. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef]
- Kitada, M.; Koya, D. Autophagy in metabolic disease and ageing. Nat. Rev. Endocrinol. 2021, 17, 647–661. [Google Scholar] [CrossRef]
- Sun, Y.; Li, T.Y.; Song, L.; Zhang, C.; Li, J.; Lin, Z.Z.; Lin, S.C.; Lin, S.Y. Liver-specific deficiency of unc-51 like kinase 1 and 2 protects mice from acetaminophen-induced liver injury. Hepatology 2018, 67, 2397–2413. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Molusky, M.M.; Song, J.; Hu, C.R.; Fang, F.; Rui, C.; Mathew, A.V.; Pennathur, S.; Liu, F.; Cheng, J.X.; et al. Autophagy deficiency by hepatic FIP200 deletion uncouples steatosis from liver injury in NAFLD. Mol. Endocrinol. 2013, 27, 1643–1654. [Google Scholar] [CrossRef]
- Jaber, N.; Dou, Z.; Chen, J.S.; Catanzaro, J.; Jiang, Y.P.; Ballou, L.M.; Selinger, E.; Ouyang, X.; Lin, R.Z.; Zhang, J.; et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. USA 2012, 109, 2003–2008. [Google Scholar] [CrossRef]
- Nemazanyy, I.; Montagnac, G.; Russell, R.C.; Morzyglod, L.; Burnol, A.F.; Guan, K.L.; Pende, M.; Panasyuk, G. Class III PI3K regulates organismal glucose homeostasis by providing negative feedback on hepatic insulin signalling. Nat. Commun. 2015, 6, 8283. [Google Scholar] [CrossRef]
- Lu, J.; He, L.; Behrends, C.; Araki, M.; Araki, K.; Jun Wang, Q.; Catanzaro, J.M.; Friedman, S.L.; Zong, W.X.; Fiel, M.I.; et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat. Commun. 2014, 5, 3920. [Google Scholar] [CrossRef]
- Osawa, T.; Kotani, T.; Kawaoka, T.; Hirata, E.; Suzuki, K.; Nakatogawa, H.; Ohsumi, Y.; Noda, N.N. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019, 26, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef] [PubMed]
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020, 27, 1185–1193. [Google Scholar] [CrossRef]
- da Silva Lima, N.; Fondevila, M.F.; Novoa, E.; Buque, X.; Mercado-Gomez, M.; Gallet, S.; Gonzalez-Rellan, M.J.; Fernandez, U.; Loyens, A.; Garcia-Vence, M.; et al. Inhibition of ATG3 ameliorates liver steatosis by increasing mitochondrial function. J. Hepatol. 2022, 76, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-deficient mice develop multiple liver tumors. Genes. Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef]
- Ni, H.M.; Woolbright, B.L.; Williams, J.; Copple, B.; Cui, W.; Luyendyk, J.P.; Jaeschke, H.; Ding, W.X. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J. Hepatol. 2014, 61, 617–625. [Google Scholar] [CrossRef]
- Takagi, A.; Kume, S.; Kondo, M.; Nakazawa, J.; Chin-Kanasaki, M.; Araki, H.; Araki, S.; Koya, D.; Haneda, M.; Chano, T.; et al. Mammalian autophagy is essential for hepatic and renal ketogenesis during starvation. Sci. Rep. 2016, 6, 18944. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chao, X.; Yang, L.; Lu, Q.; Li, T.; Ding, W.X.; Ni, H.M. Impaired Fasting-Induced Adaptive Lipid Droplet Biogenesis in Liver-Specific Atg5-Deficient Mouse Liver Is Mediated by Persistent Nuclear Factor-Like 2 Activation. Am. J. Pathol. 2018, 188, 1833–1846. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Ueno, T.; Iwata, J.; Murata, S.; Tanida, I.; Ezaki, J.; Mizushima, N.; Ohsumi, Y.; Uchiyama, Y.; et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005, 169, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef]
- Shibata, M.; Yoshimura, K.; Furuya, N.; Koike, M.; Ueno, T.; Komatsu, M.; Arai, H.; Tanaka, K.; Kominami, E.; Uchiyama, Y. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem. Biophys. Res. Commun. 2009, 382, 419–423. [Google Scholar] [CrossRef]
- Kim, K.H.; Jeong, Y.T.; Oh, H.; Kim, S.H.; Cho, J.M.; Kim, Y.N.; Kim, S.S.; Kim, D.H.; Hur, K.Y.; Kim, H.K.; et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 2013, 19, 83–92. [Google Scholar] [CrossRef]
- Amir, M.; Zhao, E.; Fontana, L.; Rosenberg, H.; Tanaka, K.; Gao, G.; Czaja, M.J. Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ. 2013, 20, 878–887. [Google Scholar] [CrossRef]
- Kuhn, R.; Schwenk, F.; Aguet, M.; Rajewsky, K. Inducible gene targeting in mice. Science 1995, 269, 1427–1429. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Ma, X.; Zhu, Q.; Song, D.; Ding, X.; Li, L.; Jiang, X.; Wang, X.; Tian, R.; Su, H.; et al. Pacer Is a Mediator of mTORC1 and GSK3-TIP60 Signaling in Regulation of Autophagosome Maturation and Lipid Metabolism. Mol. Cell 2019, 73, 788–802.e7. [Google Scholar] [CrossRef]
- Tanaka, S.; Hikita, H.; Tatsumi, T.; Sakamori, R.; Nozaki, Y.; Sakane, S.; Shiode, Y.; Nakabori, T.; Saito, Y.; Hiramatsu, N.; et al. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology 2016, 64, 1994–2014. [Google Scholar] [CrossRef]
- Riley, B.E.; Kaiser, S.E.; Shaler, T.A.; Ng, A.C.; Hara, T.; Hipp, M.S.; Lage, K.; Xavier, R.J.; Ryu, K.Y.; Taguchi, K.; et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: A potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 2010, 191, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.A.; Singh, B.K.; Zhou, J.; Xie, S.; Farah, B.L.; Lesmana, R.; Ohba, K.; Tripathi, M.; Ghosh, S.; Hollenberg, A.N.; et al. Loss of ULK1 increases RPS6KB1-NCOR1 repression of NR1H/LXR-mediated Scd1 transcription and augments lipotoxicity in hepatic cells. Autophagy 2017, 13, 169–186. [Google Scholar] [CrossRef]
- Choi, Y.J.; Yun, S.H.; Yu, J.; Mun, Y.; Lee, W.; Park, C.J.; Han, B.W.; Lee, B.H. Chaperone-mediated autophagy dysregulation during aging impairs hepatic fatty acid oxidation via accumulation of NCoR1. Mol. Metab. 2023, 76, 101784. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Inami, Y.; Waguri, S.; Sakamoto, A.; Kouno, T.; Nakada, K.; Hino, O.; Watanabe, S.; Ando, J.; Iwadate, M.; Yamamoto, M.; et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011, 193, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Ospina, N.; Potter, C.J.; Xiao, R.; Manickam, K.; Kim, M.S.; Kim, K.H.; Shneider, B.L.; Picarsic, J.L.; Jacobson, T.A.; Zhang, J.; et al. Mutations in the nuclear bile acid receptor FXR cause progressive familial intrahepatic cholestasis. Nat. Commun. 2016, 7, 10713. [Google Scholar] [CrossRef]
- Amirneni, S.; Haep, N.; Gad, M.A.; Soto-Gutierrez, A.; Squires, J.E.; Florentino, R.M. Molecular overview of progressive familial intrahepatic cholestasis. World J. Gastroenterol. 2020, 26, 7470–7484. [Google Scholar] [CrossRef]
- Kim, I.; Ahn, S.H.; Inagaki, T.; Choi, M.; Ito, S.; Guo, G.L.; Kliewer, S.A.; Gonzalez, F.J. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 2007, 48, 2664–2672. [Google Scholar] [CrossRef]
- Namani, A.; Li, Y.; Wang, X.J.; Tang, X. Modulation of NRF2 signaling pathway by nuclear receptors: Implications for cancer. Biochim. Biophys. Acta 2014, 1843, 1875–1885. [Google Scholar] [CrossRef]
- McKenna, N.J.; Lanz, R.B.; O′Malley, B.W. Nuclear receptor coregulators: Cellular and molecular biology. Endocr. Rev. 1999, 20, 321–344. [Google Scholar]
- Sun, Z.; Chin, Y.E.; Zhang, D.D. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol. Cell Biol. 2009, 29, 2658–2672. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Garduno, L.; Theodore, M.; Yang, J.; Arinze, I.J. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J. Biol. Chem. 2011, 286, 7629–7640. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.D.; et al. RXRalpha inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013, 73, 3097–3108. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Halder, G. The two faces of Hippo: Targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 2014, 13, 63–79. [Google Scholar] [CrossRef]
- Moya, I.M.; Halder, G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019, 20, 211–226. [Google Scholar] [CrossRef]
- Lee, Y.A.; Noon, L.A.; Akat, K.M.; Ybanez, M.D.; Lee, T.F.; Berres, M.L.; Fujiwara, N.; Goossens, N.; Chou, H.I.; Parvin-Nejad, F.P.; et al. Autophagy is a gatekeeper of hepatic differentiation and carcinogenesis by controlling the degradation of Yap. Nat. Commun. 2018, 9, 4962. [Google Scholar] [CrossRef]

| Gene | Model | Phenotype | Reference | |
|---|---|---|---|---|
| Autophagy initiation | Ulk1/2 | Alb-Cre; Ulk1F/F; Ulk2−/− | Normal autophagy activity, mild hepatomegaly, protection from APAP-induced liver injury, unaltered hepatic TG/cholesterol levels | [208] |
| Fip200 | Alb-Cre; Fip200F/F | Severe hepatomegaly, enlarged hepatocytes, ubiquitin-positive aggregates, p62 accumulation, lower serum and hepatic TG levels (fasting/HFD), impaired LXRα activity, increased susceptibility to endotoxin-induced liver injury | [209] | |
| Atg13, Atg17, Atg101 | No liver-specific knockout reported | N/A | N/A | |
| Gene | Model | Phenotype | Reference | |
|---|---|---|---|---|
| Vesicle nucleation | Vps34 | Alb-Cre; Vps34F/F | Hepatomegaly, hepatic steatosis, decreased protein turnover, impaired AP production during fasting, blunted amino acid-mediated mTOR signaling | [210] |
| Vps15 | Vps15F/F injected with Adeno-Cre (IV) | Liver enlargement, decreased autophagy activity, increased size and number of hepatocytes, p62 accumulation, vacuolization in hepatocytes, altered LC3 lipidation | [211] | |
| Alb-Cre; Vps15F/F | Mitochondrial depletion, impaired FAO/ketogenesis, and compromised PPARα activation due to NCoR and HDAC3 accumulation | [7] | ||
| Nrbf2 | Nrfb2−/− | Impaired ATG14L-linked VPS34 activity, decreased vesicle nucleation, enhanced ER stress-mediated cytotoxicity, focal liver necrosis, ductular reaction | [212] | |
| Becn1, Atg14L, Ambra1, p115 | No liver-specific knockout reported | N/A | N/A | |
| Gene | Model | Phenotype | Reference | |
|---|---|---|---|---|
| Vesicle elongation | Atg3 | Atg3 knockdown in hepatocytes | Lipid accumulation (CDHFD), enhanced fatty acid catabolism, elevated mitochondrial activity, SIRT1-mediated deacetylation, CPT1α-driven fatty acid transport to mitochondria | [216] |
| Atg5 | CAG-Cre; Atg5F/F (ubiquitous expression of CRE) | Mosaic deletion of Atg5 in hepatocytes, accumulation of cytoplasmic ubiquitinated proteins and p62, benign liver adenomas, mitochondrial swelling, oxidative stress and DNA damage | [217,218] | |
| Mx1-Cre; Atg5F/F (inducible expression of CRE) | Inducible Atg5 deletion (via pIpC injection), time-dependent progression of protein aggregation, initial diffuse ubiquitinated proteins, large inclusion bodies by day 16 post-injection, disrupted proteostasis precedes visible aggregate formation | [219] | ||
| Alb-Cre; Atg5F/F | Liver pathologies (apoptosis, inflammation, fibrosis, HCC), reduced hepatic lipid accumulation during fasting, impaired ketogenesis, defective starvation-induced LD formation, pathologies attenuated by co-deletion of NRF2, persistent NRF2 activation disrupts fasting-induced lipid mobilization | [217,220,221,222] | ||
| Atg7 | Mx1-Cre; Atg7F/F (inducible expression of CRE) | Complete deletion of Atg7 in liver and spleen; partial deletion of Atg7 in kidney and heart, hepatomegaly, disorganized hepatic lobules, swollen/vacuolated hepatocytes with cell death, increased peroxisome biogenesis, elevated ubiquitin-positive aggregates, increased ALT, AST, and ALP levels indicating severe liver injury | [223] | |
| Alb-Cre; Atg7F/F (Singh et al.) | Elevated hepatic TG/cholesterol levels (fed and fasted), hepatomegaly, hepatic steatosis, reduced TG secretion due to impaired LD lipolysis, autophagy necessary for LD breakdown, TG release, and FAO | [224] | ||
| Alb-Cre; Atg7F/F (Other studies) | Reduced TG levels during fasting or hepatectomy, decreased LD size and number; autophagy is essential for fasting-induced LD biogenesis | [12,225] | ||
| Alb-Cre; Atg7F/F (Kim et al.) | Hepatomegaly, irregular hepatic lobules, decreased hepatic lipid levels and TG secretion during fasting, reduced gene expression involved in fatty acid synthesis, TG production, and FAO, improved glucose homeostasis via elevated hepatic FGF21 production through ATF4 activation, decreased BW, fat mass, and hepatic steatosis under chow or HFD, diminished fasting-induced ketogenesis due to impaired PPARα activity | [8,226] | ||
| ERt-Alb-Cre; Atg7F/F | Severe liver damage induced by GalN/LPS treatment, increased apoptosis via caspase-8 activation and mitochondrial cell death pathway, autophagy protects against TNFα-mediated tissue damage by alleviating apoptotic signaling pathways | [227] | ||
| Atg2, Atg4, Atg8, Atg9, Atg10, Atg12, Atg16l | No liver-specific knockout reported | N/A | N/A | |
| Gene | Model | Phenotype | Reference | |
|---|---|---|---|---|
| Docking and fusion of AP-lysosome | Pacer | Alb-Cre; PacerF/F | Impaired autophagy and metabolic fluxes, TG and glycogen accumulation, reduced ketogenesis, early-onset fibrosis (increased collagen deposition) and liver injury, no hepatomegaly and HCC | [229] |
| Rubcn | Alb-Cre; RubcnF/F | Enhanced autophagy, protection against HFD-induced hepatic steatosis, liver damage and ER stress, no abnormalities on NCD | [230] | |
| Becn1, Uvrag, Stx17 | No liver-specific knockout reported | N/A | N/A | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.Y.; Lee, J.M. Liver Metabolism at the Crossroads: The Reciprocal Control of Nutrient-Sensing Nuclear Receptors and Autophagy. Int. J. Mol. Sci. 2025, 26, 5825. https://doi.org/10.3390/ijms26125825
Kim EY, Lee JM. Liver Metabolism at the Crossroads: The Reciprocal Control of Nutrient-Sensing Nuclear Receptors and Autophagy. International Journal of Molecular Sciences. 2025; 26(12):5825. https://doi.org/10.3390/ijms26125825
Chicago/Turabian StyleKim, Eun Young, and Jae Man Lee. 2025. "Liver Metabolism at the Crossroads: The Reciprocal Control of Nutrient-Sensing Nuclear Receptors and Autophagy" International Journal of Molecular Sciences 26, no. 12: 5825. https://doi.org/10.3390/ijms26125825
APA StyleKim, E. Y., & Lee, J. M. (2025). Liver Metabolism at the Crossroads: The Reciprocal Control of Nutrient-Sensing Nuclear Receptors and Autophagy. International Journal of Molecular Sciences, 26(12), 5825. https://doi.org/10.3390/ijms26125825






