Synthesis of Lipopeptides Using Vegetable Oils by Newly Isolated Strain of Serratia marcescens G8-1: Genomic Characterization and Process Performance
Abstract
1. Introduction
2. Results
2.1. Surface Tension Analysis
2.2. Determination of Biomass
2.3. Ultra-Performance Liquid Chromatography Coupled to Mass Spectrometry
2.4. Genomic Characterization
3. Discussion
4. Materials and Methods
4.1. Strain
4.2. Cultivation
4.3. Surface Tension Measurements
4.4. Ultra-Performance Liquid Chromatography Coupled to Mass Spectrometry
4.5. Genomic Characterization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bar-Ness, R.; Avrahamy, N.; Matsuyama, T.; Rosenberg, M. Increased cell surface hydrophobicity of a Serratia marcescens NS 38 mutant lacking wetting activity. J. Bacteriol. 1988, 170, 4361–4364. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Xiang, Z.; Liu, Y.; Zhao, X.; Sun, Y.; Li, Z.; Li, L.; Chang, F.; Chen, T.; Wen, X.; et al. Analysis of the genomic sequences and metabolites of Serratia surfactantfaciens sp. nov. YD25T that simultaneously produces prodigiosin and serrawettin W2. BMC Genom. 2016, 17, 865. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, K.; Gopalakrishnan, S.; Ravi, T.; Sivachidambaram, P. Biosurfactants: Properties, commercial production and application. Curr. Sci. 2008, 94, 736–747. [Google Scholar]
- Kosaric, N. Biosurfactants. In Biotechnology; Rehm, H.J., Reed, G., Puhler, A., Stadler, P., Eds.; VCH Weinheim: New York, NY, USA, 1996; pp. 659–717. [Google Scholar]
- Bidlan, R.; Deepthi, N.; Rastogi, R.K.; Manonmani, H.K. Production of Biosurfactant by Serratia marcescens DT-1P. Res. J. Microbiol. 2007, 2, 705–716. [Google Scholar] [CrossRef]
- Farias, C.B.B.; Almeida, F.C.G.; Silva, I.A.; Souza, T.C.; Meira, H.M.; de Cássia, R.; da Silva, F.S.; Luna, J.M.; Santos, V.A.; Converti, A.; et al. Production of green surfactants: Market prospects. Electron. J. Biotechnol. 2021, 51, 28–39. [Google Scholar] [CrossRef]
- Rosenberg, E. High- and low-molecular-mass microbial surfactants. Appl. Microbiol. Biotechnol. 1999, 52, 154–162. [Google Scholar] [CrossRef]
- Van Hamme, J.D.; Singh, A.; Ward, O.P. Physiological aspects: Part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol. Adv. 2006, 24, 604–620. [Google Scholar] [CrossRef]
- Carolin, C.F.; Kumar, P.S.; Chitra, B.; Jackulin, C.F.; Ramamurthy, R. Stimulation of Bacillus sp. by lipopeptide biosurfactant for the degradation of aromatic amine 4-Chloroaniline. J. Hazard. Mater. 2021, 415, 125716. [Google Scholar] [CrossRef]
- Ahimou, F.; Jacques, P.; Deleu, M. Surfactin and iturin A effects on Bacillus subtilis surface hydrophobicity. In Proceedings of the Enzyme and Microbial Technology; Elsevier Science Inc.: Amsterdam, The Netherlands, 2000; Volume 27, pp. 749–754. [Google Scholar] [CrossRef]
- Gutiérrez-Chávez, C.; Benaud, N.; Ferrari, B.C. The ecological roles of microbial lipopeptides: Where are we going? Comput. Struct. Biotechnol. J. 2021, 19, 1400–1413. [Google Scholar] [CrossRef]
- Mondal, B.; Adak, A.; Datta, P. Degradation of anionic surfactant in municipal wastewater by UV-H2O2: Process optimization using response surface methodology. J. Photochem. Photobiol. A Chem. 2019, 375, 237–243. [Google Scholar] [CrossRef]
- Banat, I.M.; Satpute, S.K.; Cameotra, S.S.; Rajenda, P.; Nyayanit, N.V. Cost effective technologies and renewable substrates production. Front. Microbiol. 2014, 5, 697. [Google Scholar] [CrossRef]
- Clements, T.; Ndlovu, T.; Khan, S.; Khan, W. Biosurfactants produced by Serratia species: Classification, biosynthesis, production for biosurfactants’ and application. Appl. Microbiol. Biotechnol. 2019, 103, 589–602. [Google Scholar] [CrossRef]
- Eras-Muñoz, E.; Farré, A.; Sánchez, A.; Font, X.; Gea, T. Microbial biosurfactants: A review of recent environmental applications. Bioengineered 2022, 13, 12365–12391. [Google Scholar] [CrossRef]
- Pilz, M.; Cavelius, P.; Qoura, F.; Awad, D. Lipopeptides development in cosmetics and pharmaceutical applications: A comprehensive review. Biotechnol Adv. 2023, 67, 108210. [Google Scholar] [CrossRef]
- Wasserman, H.H.; Keggi, J.J.; McKeon, J.E. The structure of serratamolide. J. Am. Chem. Sot. 1961, 84, 2978–2982. [Google Scholar] [CrossRef]
- Zhu, L.; Pang, C.; Chen, L.; Zhu, X. Antibacterial activity of a novel depsipeptide and prodigiosine of Serratia marcescans S823. Nat. Prod. Chem. Res. 2018, 6, 312. [Google Scholar] [CrossRef]
- Matsuyama, T.; Tanikawa, T.; Nakagawa, Y. Serrawettins and Other Surfactants Produced by Serratia. In Biosurfactants; Soberón-Chávez, G., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 93–120. [Google Scholar] [CrossRef]
- Walsh, C.T. The chemical versatility of natural-product assembly lines. Acc. Chem. Res. 2008, 41, 4–10. [Google Scholar] [CrossRef]
- Iacovelli, R.; Bovenberg, R.A.L.; Driessen, A.J.M. Nonribosomal peptide synthetases and their biotechnological potential in Penicillium rubens. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab045. [Google Scholar] [CrossRef]
- Li, H.; Tanikawa, T.; Sato, Y.; Nakagawa, Y.; Matsuyama, T. Serratia marcescens gene required for surfactant serrawettin W1 production encodes putative aminolipid synthetase belonging to nonribosomal peptide synthetase family. Microbiol. Immunol. 2005, 49, 303–310. [Google Scholar] [CrossRef]
- Araújo, H.W.C.; Andrade, R.F.S.; Montero-Rodríguez, D.; Rubio-Ribeaux, D.; Silva, C.A.A.D.; Campos-Takaki, G.M. Sustainable biosurfactant produced by Serratia marcescens UCP 1549 and its suitability for agricultural and marine bioremediation applications. Microb. Cell Fact. 2019, 18, 2. [Google Scholar] [CrossRef]
- Ferraz, C.; De Araújo, Á.A.; Pastore, G.M. The Influence of Vegetable Oils on Biosurfactant Production by Serratia marcescens. In Biotechnology for Fuels and Chemicals; Finkelstein, M., McMillan, J.D., Davison, B.H., Eds.; Humana Press: Totowa, NJ, USA, 2002. [Google Scholar]
- Satpute, S.K.; Banpurkar, A.G.; Dhakephalkar, P.K.; Banat, I.M.; Chopade, B.A. Methods for investigating biosurfactants and bioemulsifiers: A review. Crit. Rev. Biotechnol. 2010, 30, 127–144. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Hausmann, R.; Lepine, F.; Muller, M.M.; Deziel, E. Biosurfactants and Bioengineering of Production. In Biosurfactants. From Genes to Applications; Soberon-Chavez, G., Ed.; Springer: Berlin/Heidelberg, Germany; Dordrecht, The Netherlands; London, UK; New York, NY, USA, 2010; pp. 13–56. [Google Scholar]
- Chew, S.C. Cold-pressed rapeseed (Brassica napus) oil: Chemistry and functionality. Food Res. Int. 2020, 131, 108997. [Google Scholar] [CrossRef]
- Orsavova, J.; Misurcova, L.; Ambrozova, J.V.; Vicha, R.; Mlcek, J. Fatty acids composition of vegetable oils and its contribution to dietary energy intake and dependence of cardiovascular mortality on dietary intake of fatty acids. Int. J. Mol. Sci. 2015, 16, 12871. [Google Scholar] [CrossRef] [PubMed]
- Stepien, A.; Wojtkowiak, K.; Pietrzak-Fiecko, R. Nutrient content, fat yield and fatty acid profile of winter rapeseed (Brassica napus L.) grown under different agricultural production systems. Chil. J. Agric. Res. 2017, 77, 266–272. [Google Scholar] [CrossRef]
- Panadare, D.; Rathod, V.K. Application of waster cooking oil other than biodiesel: A review. Iran. J. Chem. Eng. 2015, 12, 55–76. [Google Scholar]
- Liepins, J.; Balina, K.; Soloha, R.; Berzina, I.; Lukasa, L.K.; Dace, E. Glycolipid biosurfactant production from waste cooking oils by yeast: Review of substrates, producers and products. Fermentation 2021, 7, 136. [Google Scholar] [CrossRef]
- Gautam, K.; Sharma, P.; Gaur, V.K.; Gupta, P.; Pandey, U.; Varjani, S.; Pandey, A.; Wong, J.W.C.; Chang, J.S. Oily waste to biosurfactant: A path towards carbon neutrality and environmental sustainability. Environ. Technol. Innov. 2023, 30, 103095. [Google Scholar] [CrossRef]
- Elkenawy, N.M.; Gomaa, O.M. Valorization of frying oil waste for biodetergent production using Serratia marcescens N2 and gamma irradiation assisted biorecovery. Microb. Cell Fact. 2022, 21, 151. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, H.; Zheng, G.; Li, Y.; Xie, Q.; You, S.; Zhang, C. Isolation and characterization of biosurfactant-producing Serratia marcescens ZCF25 from oil sludge and application to bioremediation. Environ. Sci. Pollut. Res. 2020, 27, 27762–27772. [Google Scholar] [CrossRef]
- Roldán-Carrillo, T.; Castorena-Cortés, G.; Álvarez-Ramírez, F.; Vázquez-Moreno, F.; Olguín-Lora, P. Lipopeptide production by Serratia marcescens SmSA using a Taguchi design and its application in enhanced heavy oil recovery. Prep. Biochem. Biotechnol. 2022, 52, 872–884. [Google Scholar] [CrossRef]
- dos Santos, R.A.; Rodríguez, D.M.; Ferreira, I.N.d.S.; de Almeida, S.M.; Takaki, G.M.d.C.; de Lima, M.A.B. Novel production of biodispersant by Serratia marcescens UCP 1549 in solid-state fermentation and application for oil spill bioremediation. Environ. Technol. 2022, 43, 2956–2967. [Google Scholar] [CrossRef]
- Awatif, I.I.; Shaker, M.A. Quality characteristics of high-oleic sunflower oil extracted from some hybrids cultivated under Egyptian conditions. Helia 2014, 37, 113–126. [Google Scholar] [CrossRef]
- Popa, V.; Gruia, A.; Raba, D.-N.; Dumbrava, D.; Moldovan, C.; Bordean, D.; Mateescu, C. Fatty acids composition and oil characteristics of linseed (Linum Usitatissimum L.) from Romania. Agroaliment Processes Technol. 2012, 18, 2. [Google Scholar]
- Eckelmann, D.; Spiteller, M.; Kusari, S. Spatial-temporal profiling of prodiginines and serratamolides produced by endophytic Serratia marcescens harbored in Maytenus serrata. Sci. Rep. 2018, 8, 5283. [Google Scholar] [CrossRef]
- Clements, T.; Rautenbach, M.; Ndlovu, T.; Khan, S.; Khan, W. A Metabolomics and Molecular Networking Approach to Elucidate the Structures of Secondary Metabolites Produced by Serratia marcescens Strains. Front. Chem. 2021, 9, 633870. [Google Scholar] [CrossRef]
- Kadouri, D.E.; Shanks, R.M.Q. Identification of a methicillin-resistant Staphylococcus aureus inhibitory compound isolated from Serratia marcescens. Res. Microbiol. 2013, 164, 821–826. [Google Scholar] [CrossRef]
- Geiger, O.; González-Silva, N.; López-Lara, I.M.; Sohlenkamp, C. Amino acid-containing membrane lipids in bacteria. Prog. Lipid Res. 2010, 49, 46–60. [Google Scholar] [CrossRef]
- Sohlenkamp, C. Ornithine Lipids and Other Amino Acid-Containing Acyloxyacyl Lipids. In Biogenesis of Fatty Acids, Lipids and Membranes; Geiger, O., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 109–122. [Google Scholar]
- Igarashi, M.; Shida, T.; Sasaki, Y.; Kinoshita, N.; Naganawa, H.; Hamada, M.; Takeuchi, T. Vinylamycin, a new depsipeptide antibiotic, from Streptomyces sp. J. Antibiot. 1999, 52, 873–879. [Google Scholar] [CrossRef]
- Hage-Hülsmann, J.; Grünberger, A.; Thies, S.; Santiago-Schübel, B.; Klein, A.S.; Pietruszka, J.; Binder, D.; Hilgers, F.; Domröse, A.; Drepper, T.; et al. Natural biocide cocktails: Combinatorial antibiotic effects of prodigiosin and biosurfactants. PLoS ONE 2018, 13, e0200940. [Google Scholar] [CrossRef]
- Wang, J.; Kuang, B.; Guo, X.; Liu, J.; Ding, Y.; Li, J.; Jiang, S.; Liu, Y.; Bai, F.; Li, L.; et al. Total Syntheses and Biological Activities of Vinylamycin Analogues. J. Med. Chem. 2017, 60, 1189–1209. [Google Scholar] [CrossRef]
- Marques-Pereira, C.; Proença, D.N.; Morais, P.V. Genome sequences of Serratia strains revealed common genes in both serratomolides gene clusters. Biology 2020, 9, 482. [Google Scholar] [CrossRef]
- Shanks, R.M.Q.; Stella, N.A.; Kalivoda, E.J.; Doe, M.R.; O’Dee, D.M.; Lathrop, K.L.; Feng, L.G.; Nau, G.J. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 2007, 189, 7262–7272. [Google Scholar] [CrossRef]
- Calvo, J.M.; Matthews, R.G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994, 58, 466–490. [Google Scholar] [CrossRef]
- Hay, N.A.; Tipper, D.J.; Gygi, D.; Hughes, C. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J Bacteriol. 1997, 179, 4741–4746. [Google Scholar] [CrossRef]
- Patrick, J.E.; Kearns, D.B. Swarming motility and the control of master regulators of flagellar biosynthesis. Mol. Microbiol. 2012, 83, 14–23. [Google Scholar] [CrossRef]
- Ciesielski, S.; Pokoj, T.; Mozejko, J.; Klimiuk, E. Molecular identification of polyhydroxyalkanoates-producing bacteria isolated from enriched microbial community. Pol. J. Microbiol. 2013, 62, 45–50. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 15, 1072–1075. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0-A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
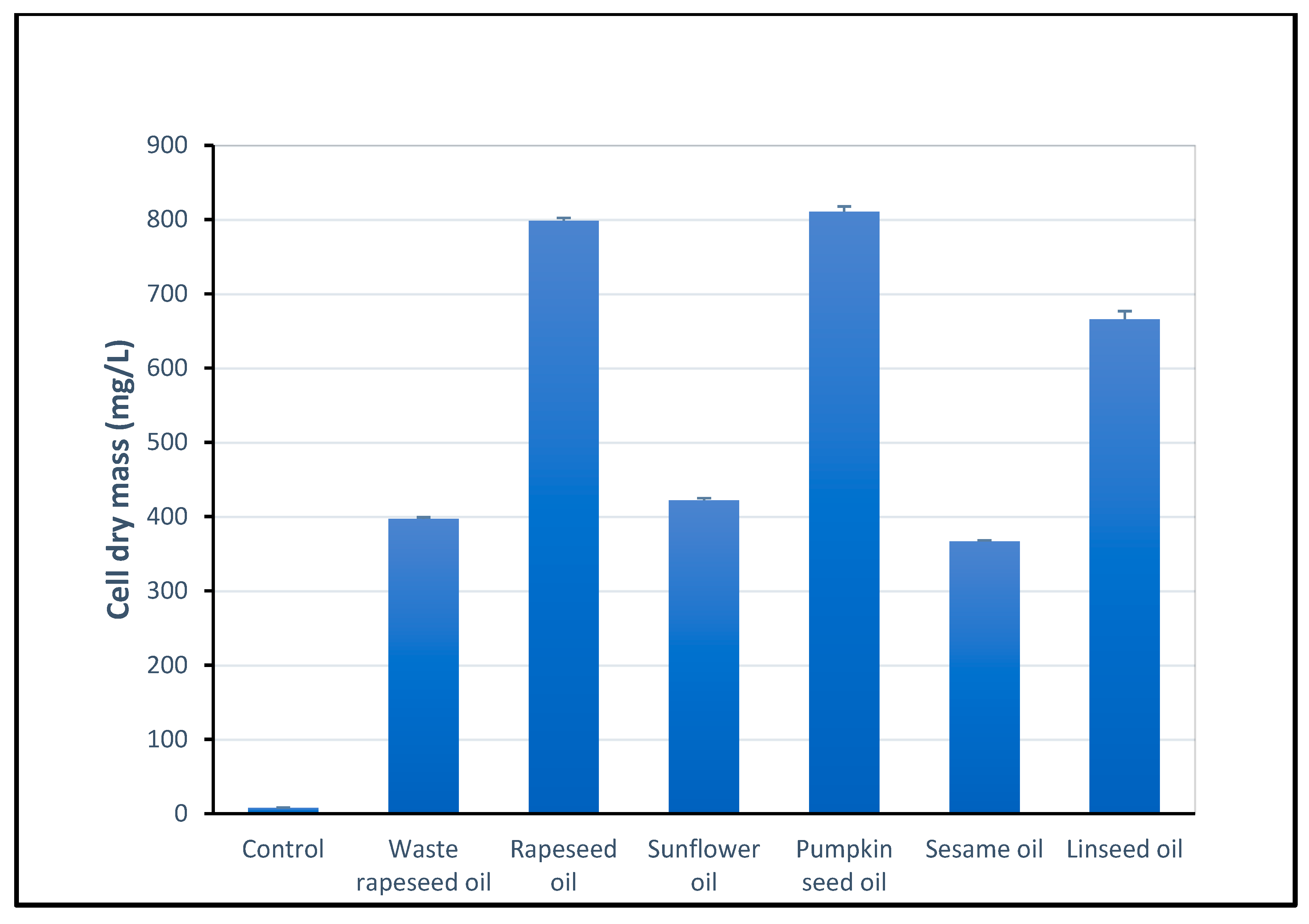

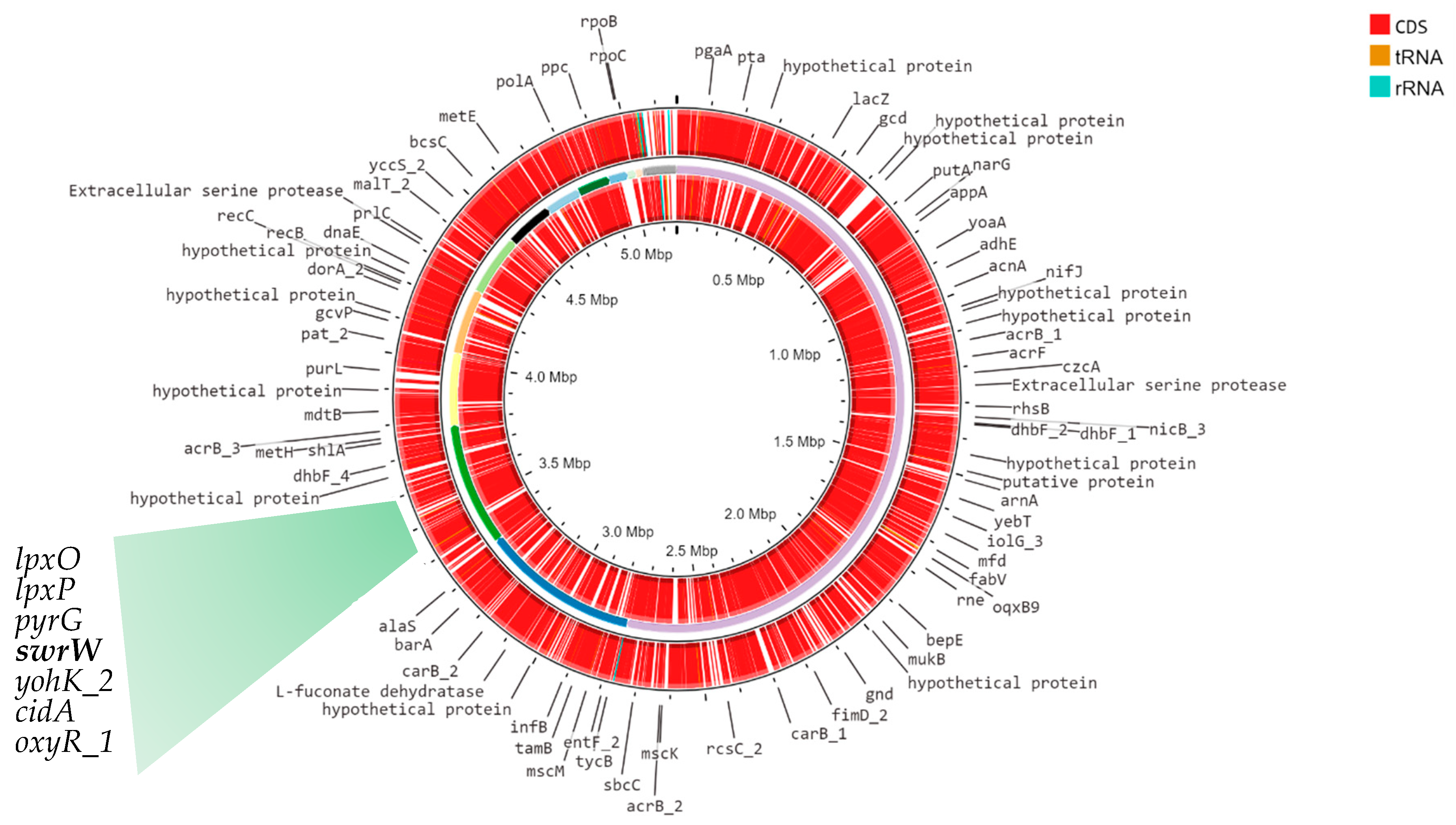
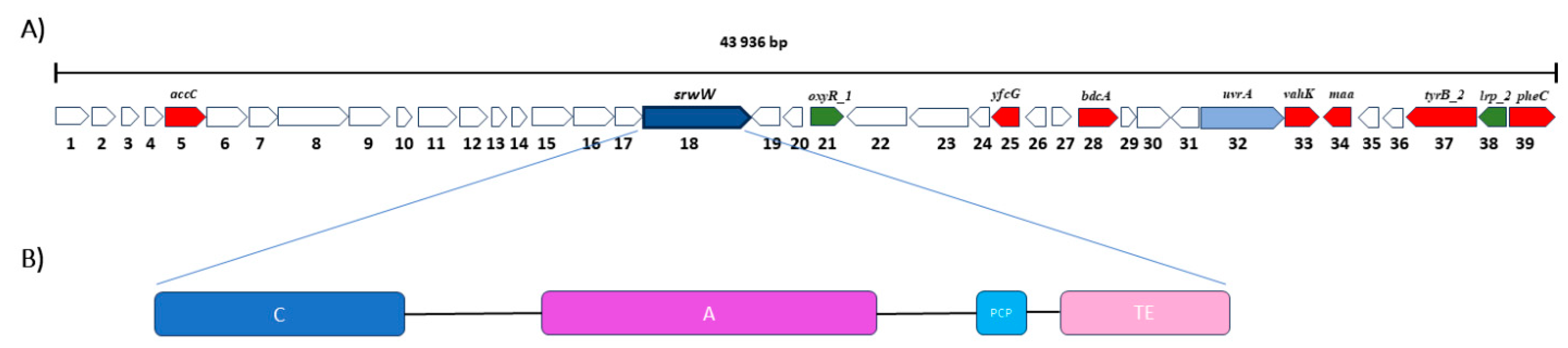
| Surface Tension (mN/m) After 120 h | Surface Tension (mN/m) After 144 h | Surface Tension (mN/m) After 168 h | Surface Tension (mN/m) Mean (for Three Measurements) | CMD−1 (mN/M) | CMD−2 (mN/M) | |
|---|---|---|---|---|---|---|
| Oil free | 29.84 ± 0.45 | 31.28 ± 0.18 | 30.985 ± 0.46 | 30.7 ± 0.76 | 48.41 ± 1.69 | 58.135 ± 2.22 |
| Fresh rapeseed oil | 42.02 ± 3.39 | 40.67 ± 1.06 | 43.19 ± 4.44 | 41.96 ± 1.26 | 52.6 ± 3.33 | 56.12 ± 1.49 |
| Waste rapeseed oil | 38.83 ± 2.53 | 40.87 ± 0.76 | 41.45 ± 0.45 | 40.38 ± 1.37 | 49.94 ± 1.13 | 52.16 ± 1.38 |
| Linseed oil | 40.5 ± 0.81 | 41.1 ± 1.07 | 41.78 ± 1.15 | 41.12 ± 0.64 | 50.53 ± 2.05 | 55.1 ± 3.32 |
| Pumpkin seed oil | 41.64 ± 0.34 | 43.1 ± 1.67 | 43.94 ± 1.10 | 42.89 ± 1.16 | 54.05 ± 1.78 | 59.9 ± 4.49 |
| Sunflower oil | 40.31 ± 1.30 | 42.03 ± 0.42 | 41.73 ± 0.39 | 41.35 ± 0.92 | 50.09 ± 2.32 | 53.11 ± 2.92 |
| Sesame oil | 40.81 ± 2.12 | 42.79 ± 2.00 | 42.97 ± 1.75 | 42.19 ± 1.19 | 52.39 ± 1.89 | 56.94 ± 3.96 |
| Compound Name | UPLC Rt | Molecular Formula | m/z [M+H]+ | m/z [M+H]+ | Oil |
|---|---|---|---|---|---|
| Theoretical | |||||
| Serratomolide A | 5.65 | C26H46N2O8 | 515.3328 | 515.3332 | rapeseed waste |
| 5.66 | sunflower; sesame; linseed | ||||
| 5.67 | rapeseed, pumpkin | ||||
| Serratomolide B | 7.01 | C28H48N2O8 | 541.3489 | 541.3489 | sunflower |
| 7.02 | sesame; linseed | ||||
| 7.03 | pumpkin | ||||
| Serratomolide C | 7.61 | C28H50N2O8 | 543.3648 | 543.3645 | rapeseed; sunflower |
| 7.62 | linseed | ||||
| Serratomolide 571 | 9.17 | C30H54N2O8 | 571.3958 | 571.3958 | rapeseed |
| 9.18 | sesame; linseed | ||||
| 9.19 | pumpkin | ||||
| Open ring serratomolide 587 | 6.72 | C30H54N2O9 | 587.3896 | 587.3908 | rapeseed waste |
| Ornithine containig lipids | 11.84 | C37H72N2O5 | 625.5518 | 625.5505 | rapeseed waste |
| 11.88 | rapeseed | ||||
| 12.58 | C40H76N2O5 | 665.5831 | 665.5832 | rapeseed | |
| 12.60 | rapeseed waste | ||||
| Vinylamicine | 7.85 | C26H43N3O6 | 494.3252 | 494.3230 | rapeseed waste |
| 7.87 | rapeseed |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciesielski, S.; Stefańska, W.; Singh, K.; Wielgus, E. Synthesis of Lipopeptides Using Vegetable Oils by Newly Isolated Strain of Serratia marcescens G8-1: Genomic Characterization and Process Performance. Int. J. Mol. Sci. 2025, 26, 5794. https://doi.org/10.3390/ijms26125794
Ciesielski S, Stefańska W, Singh K, Wielgus E. Synthesis of Lipopeptides Using Vegetable Oils by Newly Isolated Strain of Serratia marcescens G8-1: Genomic Characterization and Process Performance. International Journal of Molecular Sciences. 2025; 26(12):5794. https://doi.org/10.3390/ijms26125794
Chicago/Turabian StyleCiesielski, Slawomir, Wiktoria Stefańska, Kritika Singh, and Ewelina Wielgus. 2025. "Synthesis of Lipopeptides Using Vegetable Oils by Newly Isolated Strain of Serratia marcescens G8-1: Genomic Characterization and Process Performance" International Journal of Molecular Sciences 26, no. 12: 5794. https://doi.org/10.3390/ijms26125794
APA StyleCiesielski, S., Stefańska, W., Singh, K., & Wielgus, E. (2025). Synthesis of Lipopeptides Using Vegetable Oils by Newly Isolated Strain of Serratia marcescens G8-1: Genomic Characterization and Process Performance. International Journal of Molecular Sciences, 26(12), 5794. https://doi.org/10.3390/ijms26125794







