Binding of Transcriptional Activator to Silent Loci Causes Their Detachment from the Nuclear Lamina in Drosophila Neurons and Salivary Gland Cells
Abstract
1. Introduction
2. Results
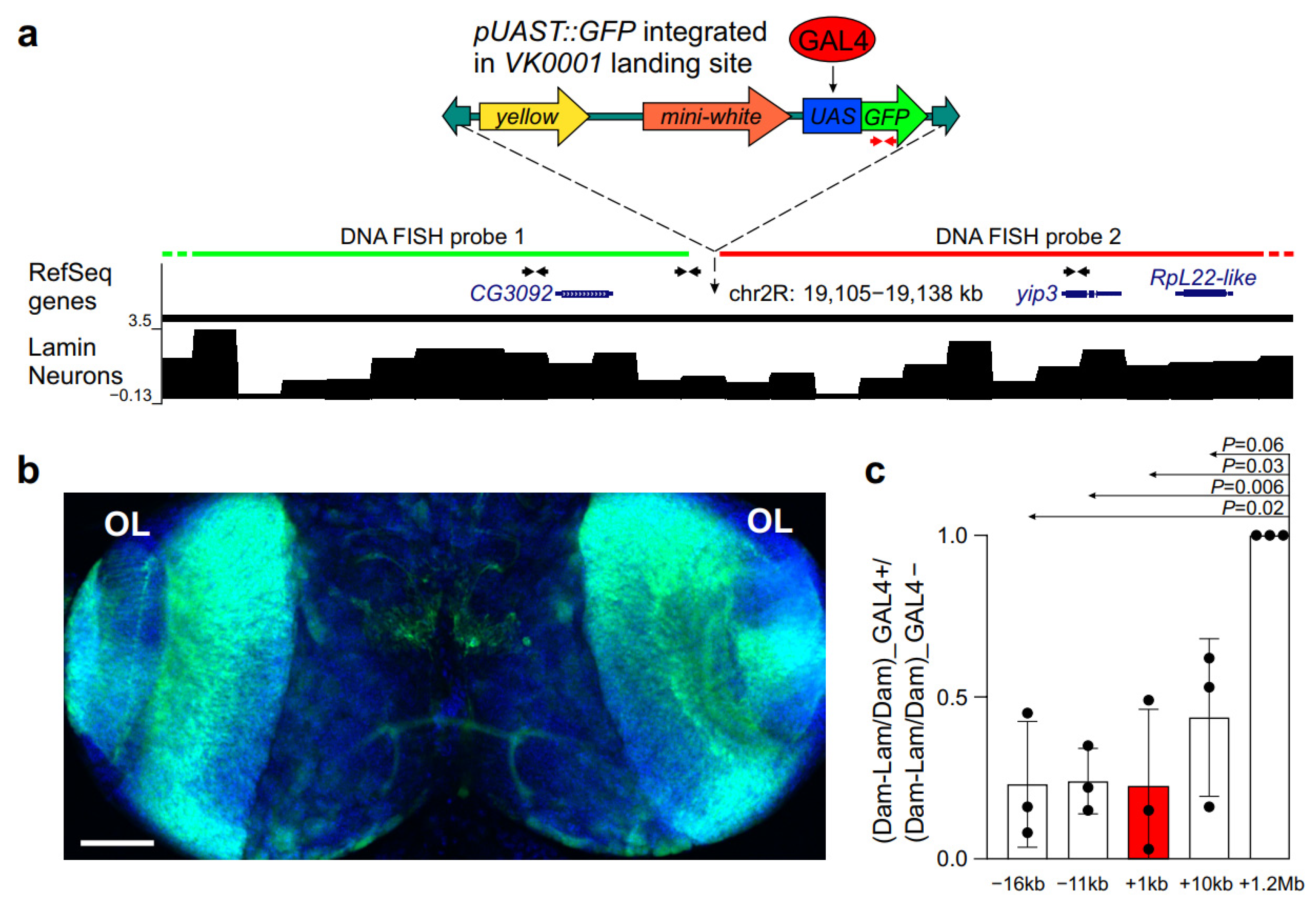
2.1. GAL4 Binding to a Silent Locus Leads to Its Repositioning from the NL to the Nuclear Interior in Salivary Gland Cells
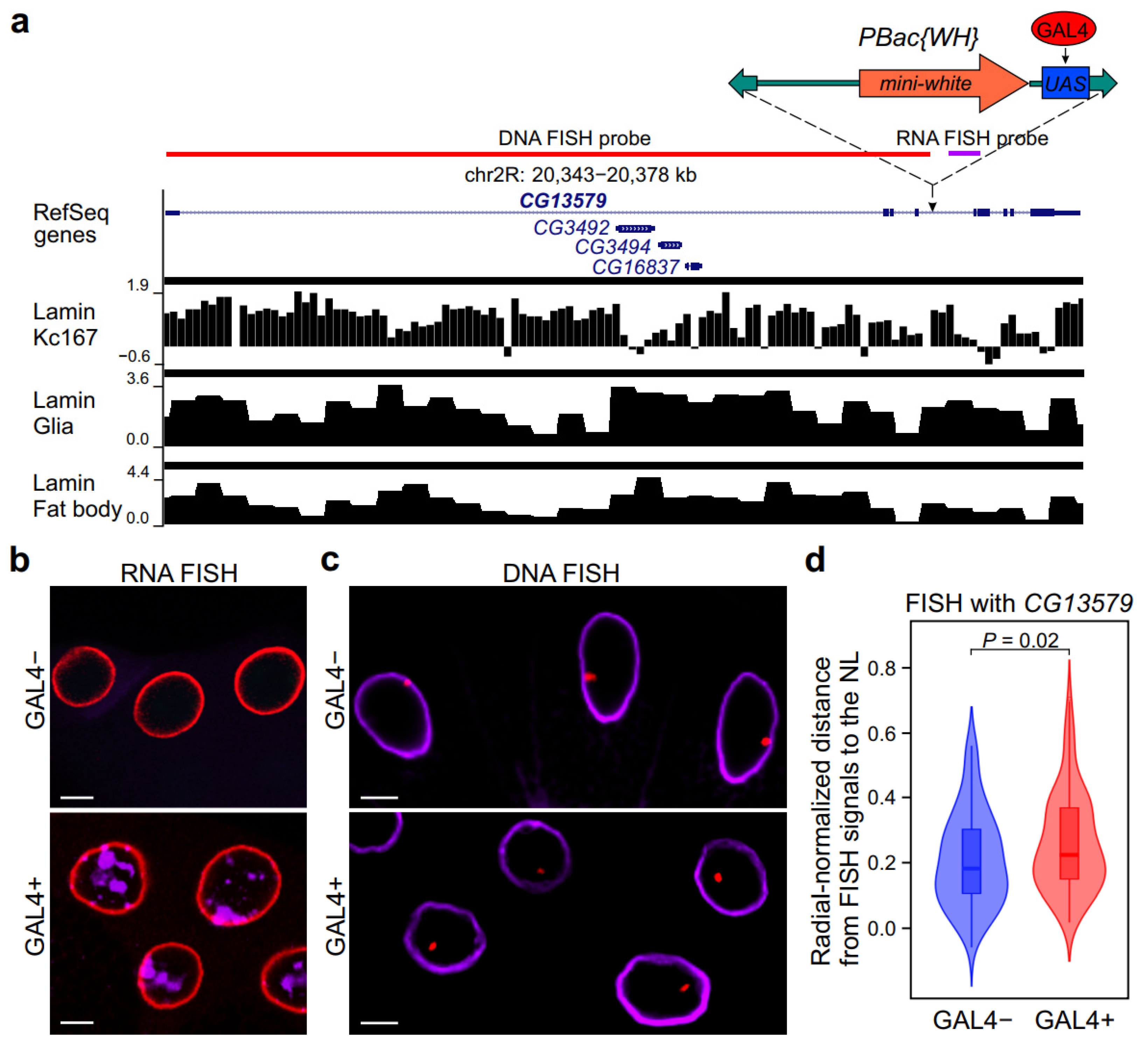
2.2. Transcriptional Activation of Neuron-Specific Gene Results in the Loss of Its Contacts with the NL and Relocalization to the Nuclear Interior
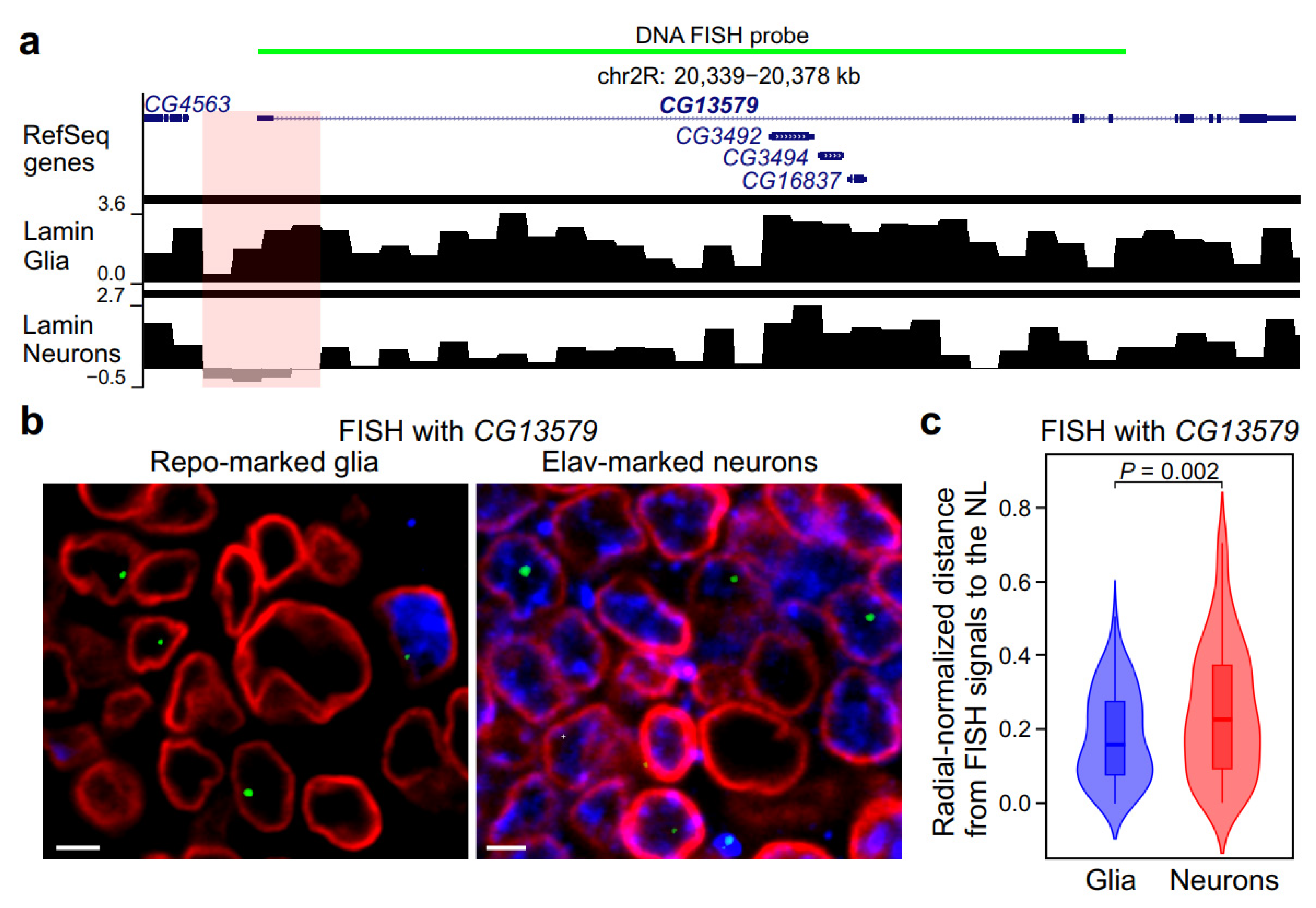
2.3. In Neurons, GAL4 Binding Within a LAD Results in Local Weakening of Interactions with the NL Around the Binding Site
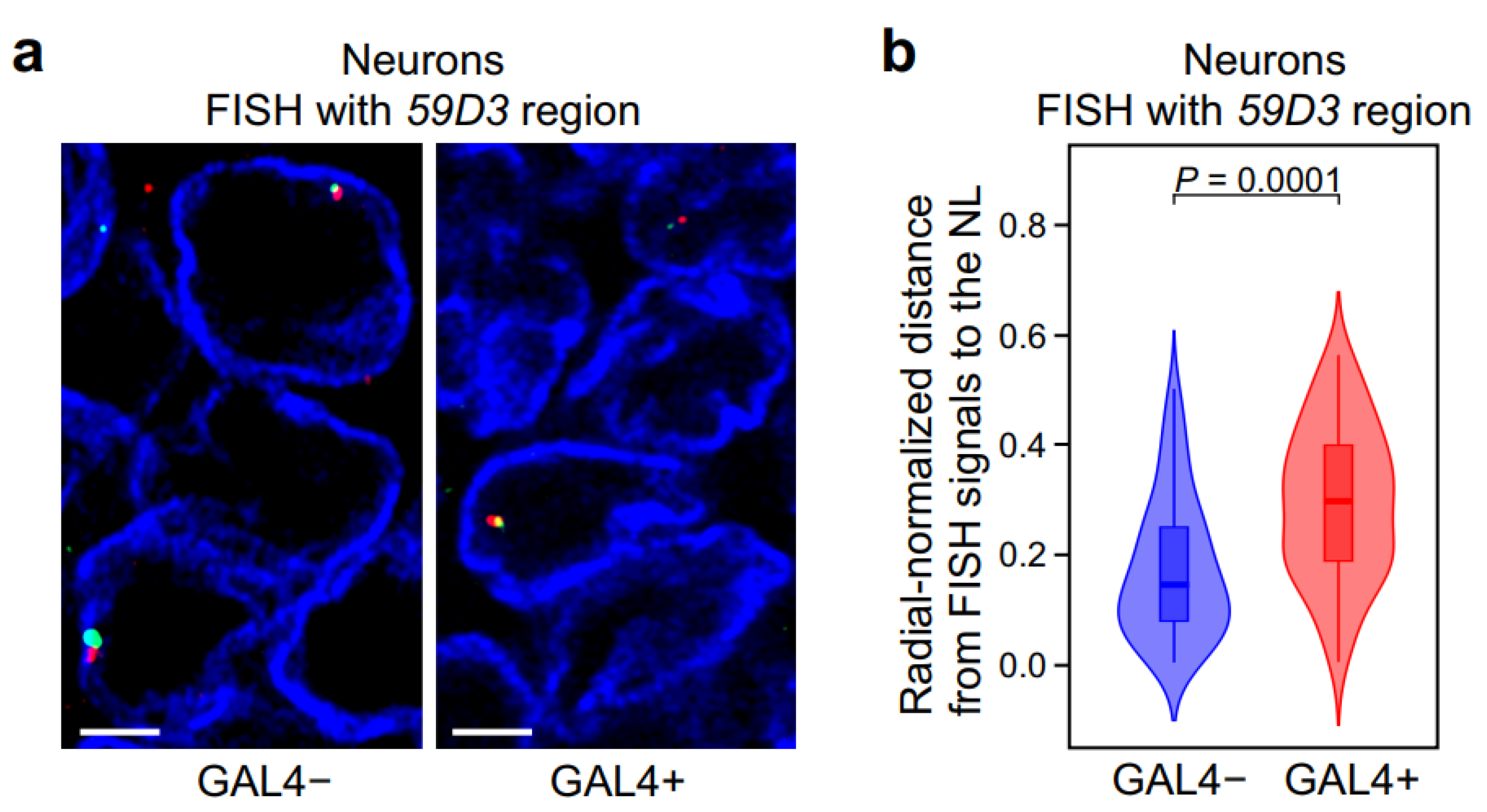
2.4. In Neurons, GAL4 Binding to a Silent Locus Leads to the Locus Repositioning from the NL to the Nuclear Interior
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Fly Stocks and Handling
4.3. Preparation of Probes for RNA and DNA FISH
4.4. RNA FISH with Immunostaining
4.5. DNA FISH with Immunostaining
4.6. Intranuclear Loci Position Quantification
4.7. DamID Procedure in Brains
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shevelyov, Y.Y.; Nurminsky, D.I. The nuclear lamina as a gene-silencing hub. Curr. Issues Mol. Biol. 2012, 14, 27–38. [Google Scholar] [PubMed]
- Kosak, S.T.; Skok, J.A.; Medina, K.L.; Riblet, R.; Le Beau, M.M.; Fisher, A.G.; Singh, H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 2002, 296, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Zink, D.; Amaral, M.D.; Englmann, A.; Lang, S.; Clarke, L.A.; Rudolph, C.; Alt, F.; Luther, K.; Braz, C.; Sadoni, N.; et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 2004, 166, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.R.; Azuara, V.; Perry, P.; Sauer, S.; Dvorkina, M.; Jørgensen, H.; Roix, J.; McQueen, P.; Misteli, T.; Merkenschlager, M.; et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 2006, 119, 132–140. [Google Scholar] [CrossRef]
- Ragoczy, T.; Bender, M.A.; Telling, A.; Byron, R.; Groudine, M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006, 20, 1447–1457. [Google Scholar] [CrossRef]
- Ballester, M.; Kress, C.; Hue-Beauvais, C.; Kiêu, K.; Lehmann, G.; Adenot, P.; Devinoy, E. The nuclear localization of WAP and CSN genes is modified by lactogenic hormones in HC11 cells. J. Cell Biochem. 2008, 105, 262–270. [Google Scholar] [CrossRef]
- Szczerbal, I.; Foster, H.A.; Bridger, J.M. The spatial repositioning of adipogenesis genes is correlated with their expression status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma 2009, 118, 647–663. [Google Scholar] [CrossRef]
- Shevelyov, Y.Y.; Lavrov, S.A.; Mikhaylova, L.M.; Nurminsky, I.D.; Kulathinal, R.J.; Egorova, K.S.; Rozovsky, Y.M.; Nurminsky, D.I. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc. Natl. Acad. Sci. USA 2009, 106, 3282–3287. [Google Scholar] [CrossRef]
- Meister, P.; Towbin, B.D.; Pike, B.L.; Ponti, A.; Gasser, S.M. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010, 24, 766–782. [Google Scholar] [CrossRef]
- van Steensel, B.; Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000, 18, 424–428. [Google Scholar] [CrossRef]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.; Solovei, I.; Brugman, W.; Gräf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Oldenburg, A.R.; Delbarre, E.; Freberg, C.T.; Duband-Goulet, I.; Eskeland, R.; Buendia, B.; Collas, P. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res. 2013, 23, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.I.; de Las Heras, J.I.; Czapiewski, R.; Lê Thành, P.; Booth, D.G.; Kelly, D.A.; Webb, S.; Kerr, A.R.W.; Schirmer, E.C. Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol. Cell 2016, 62, 834–847. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.I.; de Las Heras, J.I.; Czapiewski, R.; Sivakumar, A.; Kerr, A.R.W.; Schirmer, E.C. Constrained release of lamina-associated enhancers and genes from the nuclear envelope during T-cell activation facilitates their association in chromosome compartments. Genome Res. 2017, 27, 1126–1138. [Google Scholar] [CrossRef] [PubMed]
- Brueckner, L.; Zhao, P.A.; van Schaik, T.; Leemans, C.; Sima, J.; Peric-Hupkes, D.; Gilbert, D.M.; van Steensel, B. Local rewiring of genome-nuclear lamina interactions by transcription. EMBO J. 2020, 39, e103159. [Google Scholar] [CrossRef]
- Ilyin, A.A.; Kononkova, A.D.; Golova, A.V.; Shloma, V.V.; Olenkina, O.M.; Nenasheva, V.V.; Abramov, Y.A.; Kotov, A.A.; Maksimov, D.A.; Laktionov, P.P.; et al. Comparison of genome architecture at two stages of male germline cell differentiation in Drosophila. Nucleic Acids Res. 2022, 50, 3203–3225. [Google Scholar] [CrossRef]
- Tumbar, T.; Belmont, A.S. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 2001, 3, 134–139. [Google Scholar] [CrossRef]
- Chuang, C.H.; Carpenter, A.E.; Fuchsova, B.; Johnson, T.; de Lanerolle, P.; Belmont, A.S. Long-range directional movement of an interphase chromosome site. Curr. Biol. 2006, 16, 825–831. [Google Scholar] [CrossRef]
- Therizols, P.; Illingworth, R.S.; Courilleau, C.; Boyle, S.; Wood, A.J.; Bickmore, W.A. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 2014, 346, 1238–1242. [Google Scholar] [CrossRef]
- Kind, J.; Pagie, L.; Ortabozkoyun, H.; Boyle, S.; de Vries, S.S.; Janssen, H.; Amendola, M.; Nolen, L.D.; Bickmore, W.A.; van Steensel, B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013, 153, 178–192. [Google Scholar] [CrossRef]
- Kumaran, R.I.; Spector, D.L. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008, 180, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.L.; Zullo, J.M.; Bertolino, E.; Singh, H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008, 452, 243–247. [Google Scholar] [CrossRef]
- Shachar, S.; Voss, T.C.; Pegoraro, G.; Sciascia, N.; Misteli, T. Identification of gene positioning factors using high-throughput imaging mapping. Cell 2015, 162, 911–923. [Google Scholar] [CrossRef]
- Zhimulev, I.; Vatolina, T.; Levitsky, V.; Tsukanov, A. Developmental and housekeeping genes: Two types of genetic organization in the Drosophila genome. Int. J. Mol. Sci. 2024, 25, 4068. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef]
- Thibault, S.T.; Singer, M.A.; Miyazaki, W.Y.; Milash, B.; Dompe, N.A.; Singh, C.M.; Buchholz, R.; Demsky, M.; Fawcett, R.; Francis-Lang, H.L.; et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004, 36, 283–287. [Google Scholar] [CrossRef]
- Chintapalli, V.R.; Wang, J.; Dow, J.A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007, 39, 715–720. [Google Scholar] [CrossRef] [PubMed]
- van Bemmel, J.G.; Pagie, L.; Braunschweig, U.; Brugman, W.; Meuleman, W.; Kerkhoven, R.M.; van Steensel, B. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE 2010, 5, e15013. [Google Scholar] [CrossRef]
- Pindyurin, A.V.; Ilyin, A.A.; Ivankin, A.V.; Tselebrovsky, M.V.; Nenasheva, V.V.; Mikhaleva, E.A.; Pagie, L.; van Steensel, B.; Shevelyov, Y.Y. The large fraction of heterochromatin in Drosophila neurons is bound by both B-type lamin and HP1a. Epigenetics Chromatin 2018, 11, 65. [Google Scholar] [CrossRef]
- Yang, C.P.; Fu, C.C.; Sugino, K.; Liu, Z.; Ren, Q.; Liu, L.Y.; Yao, X.; Lee, L.P.; Lee, T. Transcriptomes of lineage-specific Drosophila neuroblasts profiled by genetic targeting and robotic sorting. Development 2016, 143, 411–421. [Google Scholar] [CrossRef]
- Venken, K.J.; He, Y.; Hoskins, R.A.; Bellen, H.J. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 2006, 314, 1747–1751. [Google Scholar] [CrossRef] [PubMed]
- Bischof, J.; Maeda, R.K.; Hediger, M.; Karch, F.; Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 2007, 104, 3312–3317. [Google Scholar] [CrossRef]
- Pindyurin, A.V.; Pagie, L.; Kozhevnikova, E.N.; van Arensbergen, J.; van Steensel, B. Inducible DamID systems for genomic mapping of chromatin proteins in Drosophila. Nucleic Acids Res. 2016, 44, 5646–5657. [Google Scholar] [CrossRef]
- Vogel, M.J.; Peric-Hupkes, D.; van Steensel, B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007, 2, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Dundr, M.; Ospina, J.K.; Sung, M.H.; John, S.; Upender, M.; Ried, T.; Hager, G.L.; Matera, A.G. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 2007, 179, 1095–1103. [Google Scholar] [CrossRef]
- Caridi, C.P.; D’Agostino, C.; Ryu, T.; Zapotoczny, G.; Delabaere, L.; Li, X.; Khodaverdian, V.Y.; Amaral, N.; Lin, E.; Rau, A.R.; et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559, 54–60. [Google Scholar] [CrossRef]
- Schrank, B.R.; Aparicio, T.; Li, Y.; Chang, W.; Chait, B.T.; Gundersen, G.G.; Gottesman, M.E.; Gautier, J. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 2018, 559, 61–66. [Google Scholar] [CrossRef]
- Hari-Gupta, Y.; Fili, N.; Dos Santos, Á.; Cook, A.W.; Gough, R.E.; Reed, H.C.W.; Wang, L.; Aaron, J.; Venit, T.; Wait, E.; et al. Myosin VI regulates the spatial organisation of mammalian transcription initiation. Nat. Commun. 2022, 13, 1346. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Kolhe, J.A.; Gioacchini, N.; Baade, I.; Brieher, W.M.; Peterson, C.L.; Freeman, B.C. Mechanism of long-range chromosome motion triggered by gene activation. Dev. Cell 2020, 52, 309–320.e5. [Google Scholar]
- Capelson, M. How genes move: Spatial repositioning of activated genes is driven by nuclear actin-based pathway. Dev. Cell 2020, 52, 252–254. [Google Scholar] [CrossRef]
- Shpiz, S.; Lavrov, S.; Kalmykova, A. Combined RNA/DNA fluorescence in situ hybridization on whole-mount Drosophila ovaries. Methods Mol. Biol. 2014, 1093, 161–169. [Google Scholar] [PubMed]
- Stuurman, N.; Maus, N.; Fisher, P.A. Interphase phosphorylation of the Drosophila nuclear lamin: Site-mapping using a monoclonal antibody. J. Cell Sci. 1995, 108, 3137–3144. [Google Scholar] [CrossRef] [PubMed]
- Milon, B.C.; Cheng, H.; Tselebrovsky, M.V.; Lavrov, S.A.; Nenasheva, V.V.; Mikhaleva, E.A.; Shevelyov, Y.Y.; Nurminsky, D.I. Role of histone deacetylases in gene regulation at nuclear lamina. PLoS ONE 2012, 7, e49692. [Google Scholar] [CrossRef] [PubMed]
- Osouda, S.; Nakamura, Y.; de Saint Phalle, B.; McConnell, M.; Horigome, T.; Sugiyama, S.; Fisher, P.A.; Furukawa, K. Null mutants of Drosophila B-type lamin Dm(0) show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev. Biol. 2005, 284, 219–232. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Doronin, S.A.; Khrameeva, E.E.; Kos, P.I.; Luzhin, A.V.; Starikov, S.S.; Galitsyna, A.A.; Nenasheva, V.V.; Ilyin, A.A.; Flyamer, I.M.; et al. Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 2019, 10, 1176. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simonov, R.A.; Olenkina, O.M.; Nenasheva, V.V.; Abramov, Y.A.; Lavrov, S.A.; Fedotova, A.A.; Shevelyov, Y.Y. Binding of Transcriptional Activator to Silent Loci Causes Their Detachment from the Nuclear Lamina in Drosophila Neurons and Salivary Gland Cells. Int. J. Mol. Sci. 2025, 26, 5793. https://doi.org/10.3390/ijms26125793
Simonov RA, Olenkina OM, Nenasheva VV, Abramov YA, Lavrov SA, Fedotova AA, Shevelyov YY. Binding of Transcriptional Activator to Silent Loci Causes Their Detachment from the Nuclear Lamina in Drosophila Neurons and Salivary Gland Cells. International Journal of Molecular Sciences. 2025; 26(12):5793. https://doi.org/10.3390/ijms26125793
Chicago/Turabian StyleSimonov, Ruslan A., Oxana M. Olenkina, Valentina V. Nenasheva, Yuri A. Abramov, Sergey A. Lavrov, Anna A. Fedotova, and Yuri Y. Shevelyov. 2025. "Binding of Transcriptional Activator to Silent Loci Causes Their Detachment from the Nuclear Lamina in Drosophila Neurons and Salivary Gland Cells" International Journal of Molecular Sciences 26, no. 12: 5793. https://doi.org/10.3390/ijms26125793
APA StyleSimonov, R. A., Olenkina, O. M., Nenasheva, V. V., Abramov, Y. A., Lavrov, S. A., Fedotova, A. A., & Shevelyov, Y. Y. (2025). Binding of Transcriptional Activator to Silent Loci Causes Their Detachment from the Nuclear Lamina in Drosophila Neurons and Salivary Gland Cells. International Journal of Molecular Sciences, 26(12), 5793. https://doi.org/10.3390/ijms26125793





