X-Ray Exposure Induces Structural Changes in Human Breast Proteins
Abstract
1. Introduction
2. Results
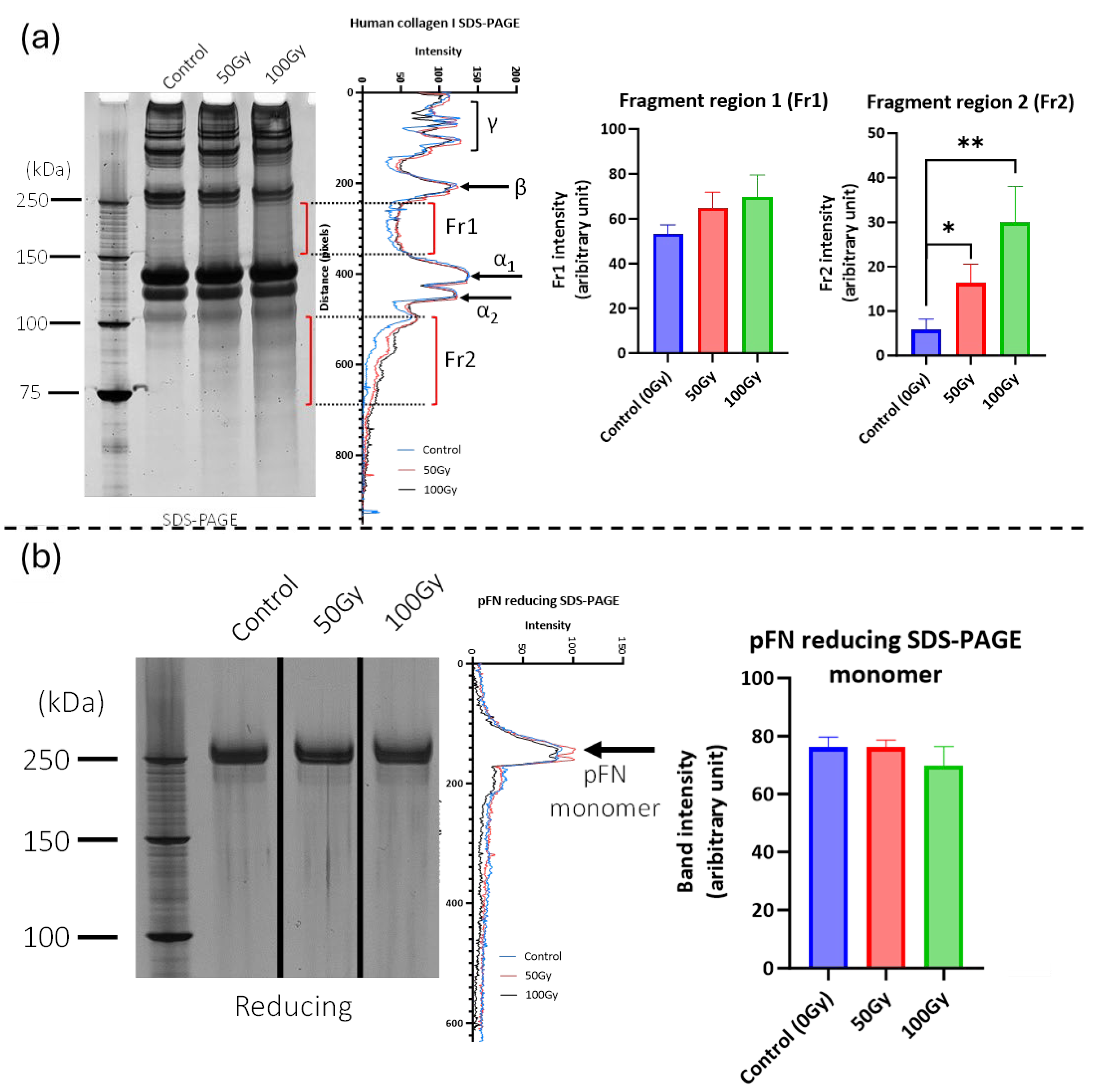
2.1. 50 Gy and 100 Gy X-Ray Exposure Affects the Structure and Binding of Purified Collagen I and Fibronectin
2.2. X-Ray Exposure Alters Structure of Extracellular Proteins in Complex Matrices
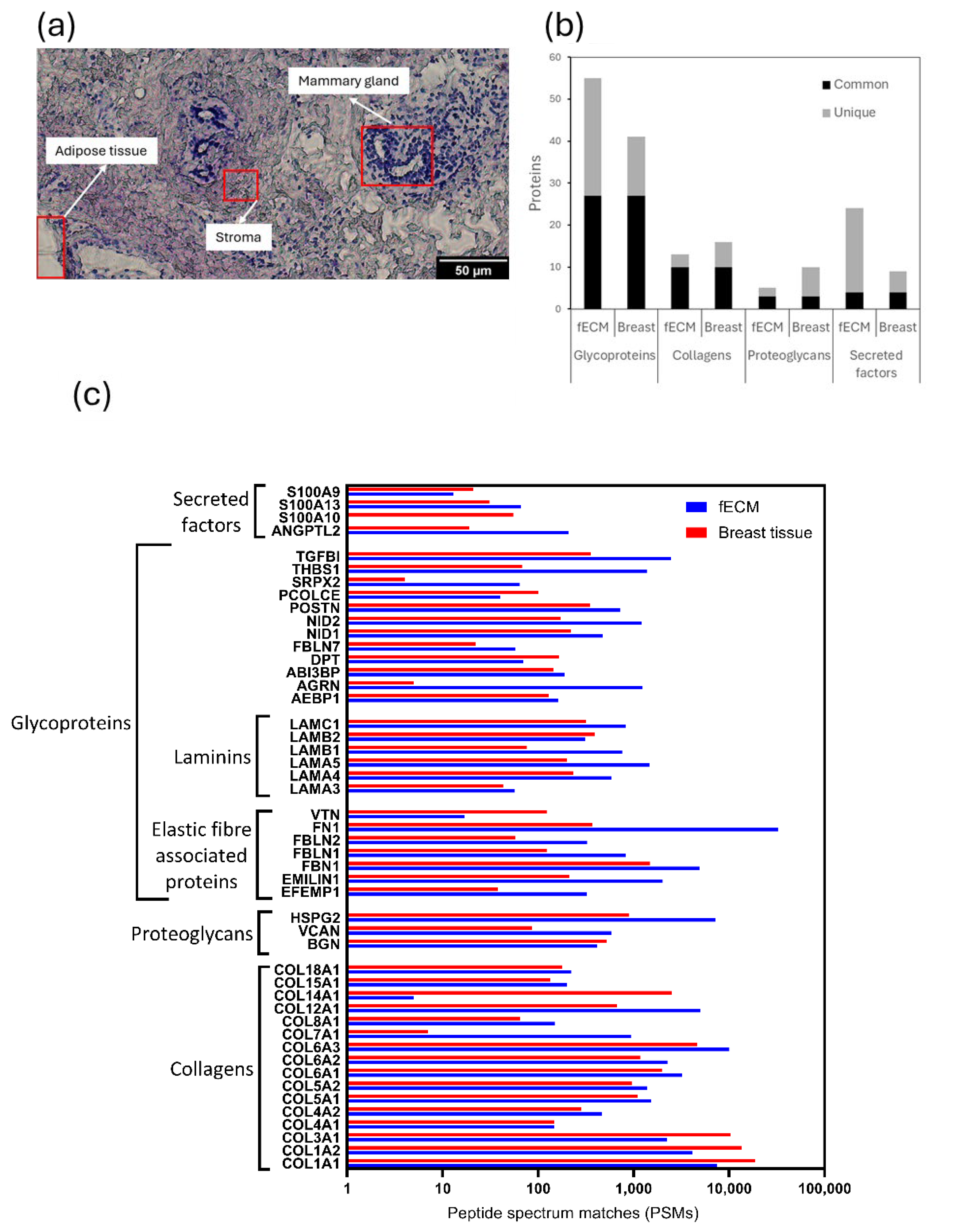
2.2.1. Fibroblast Cell Culture-Derived and Breast Tissue Samples Are Both Rich in ECM Proteins
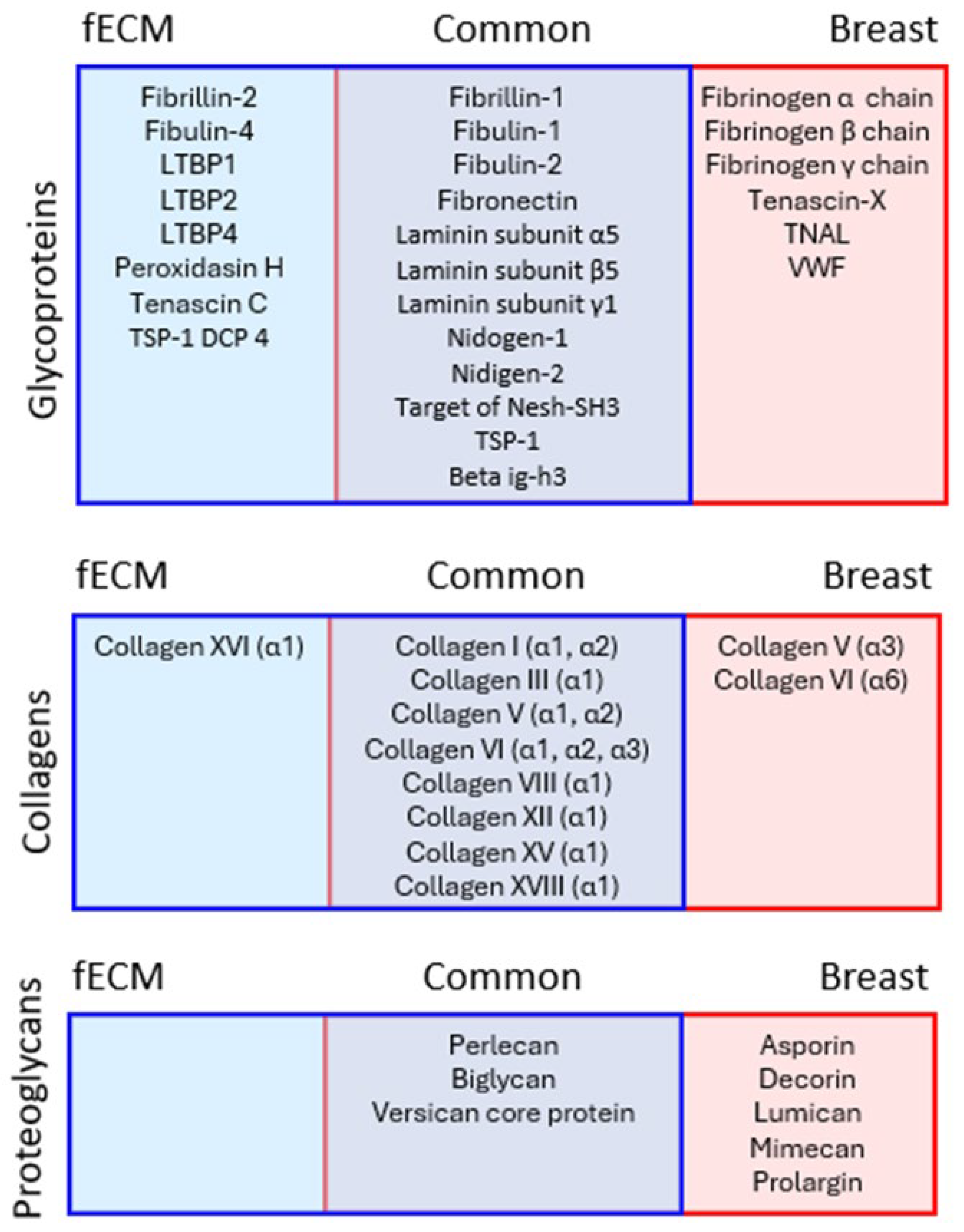
2.2.2. X-Ray Exposure Affects the Structure of Multiple ECM Proteins in Both Cell- and Tissue-Derived Proteomes
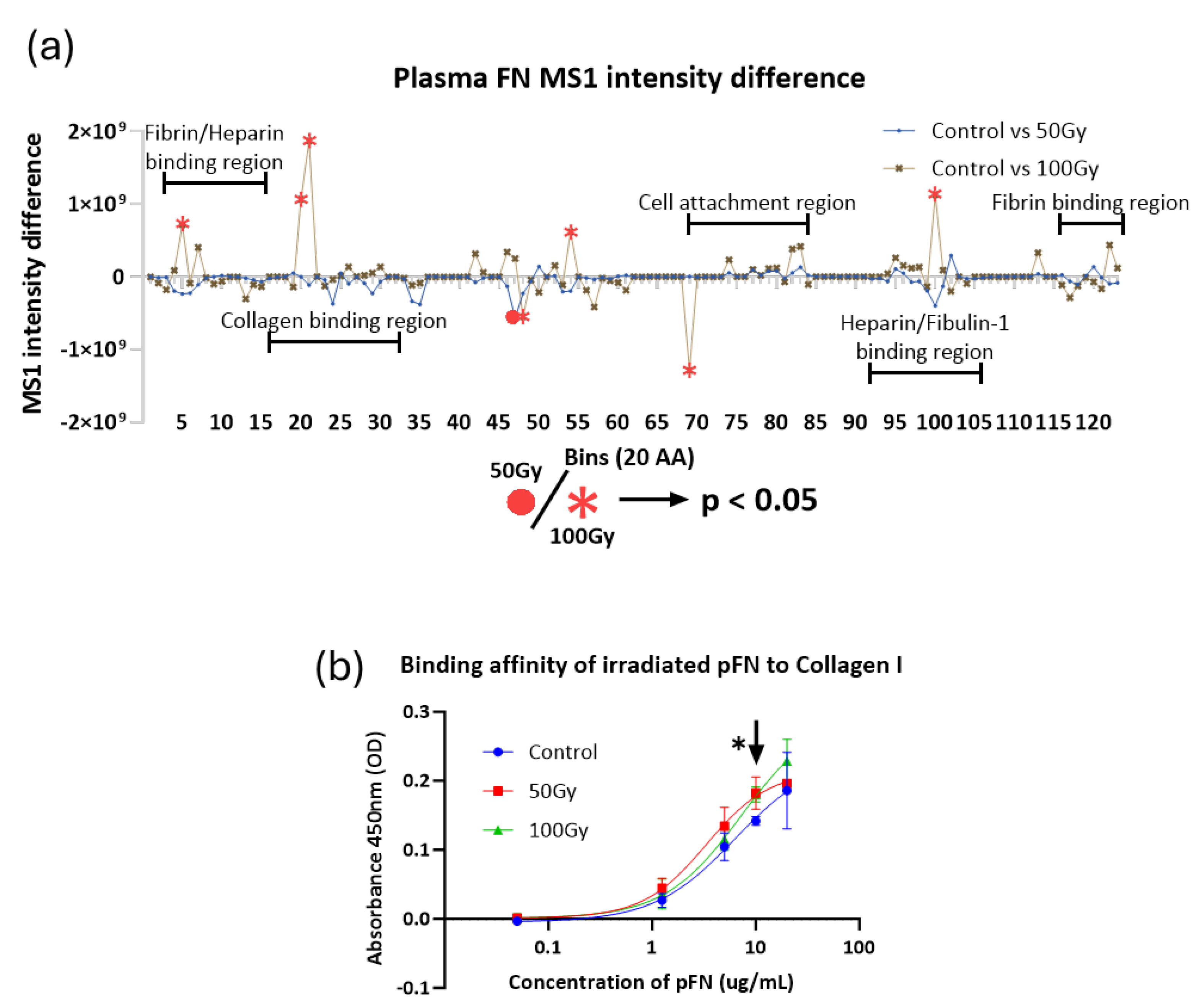
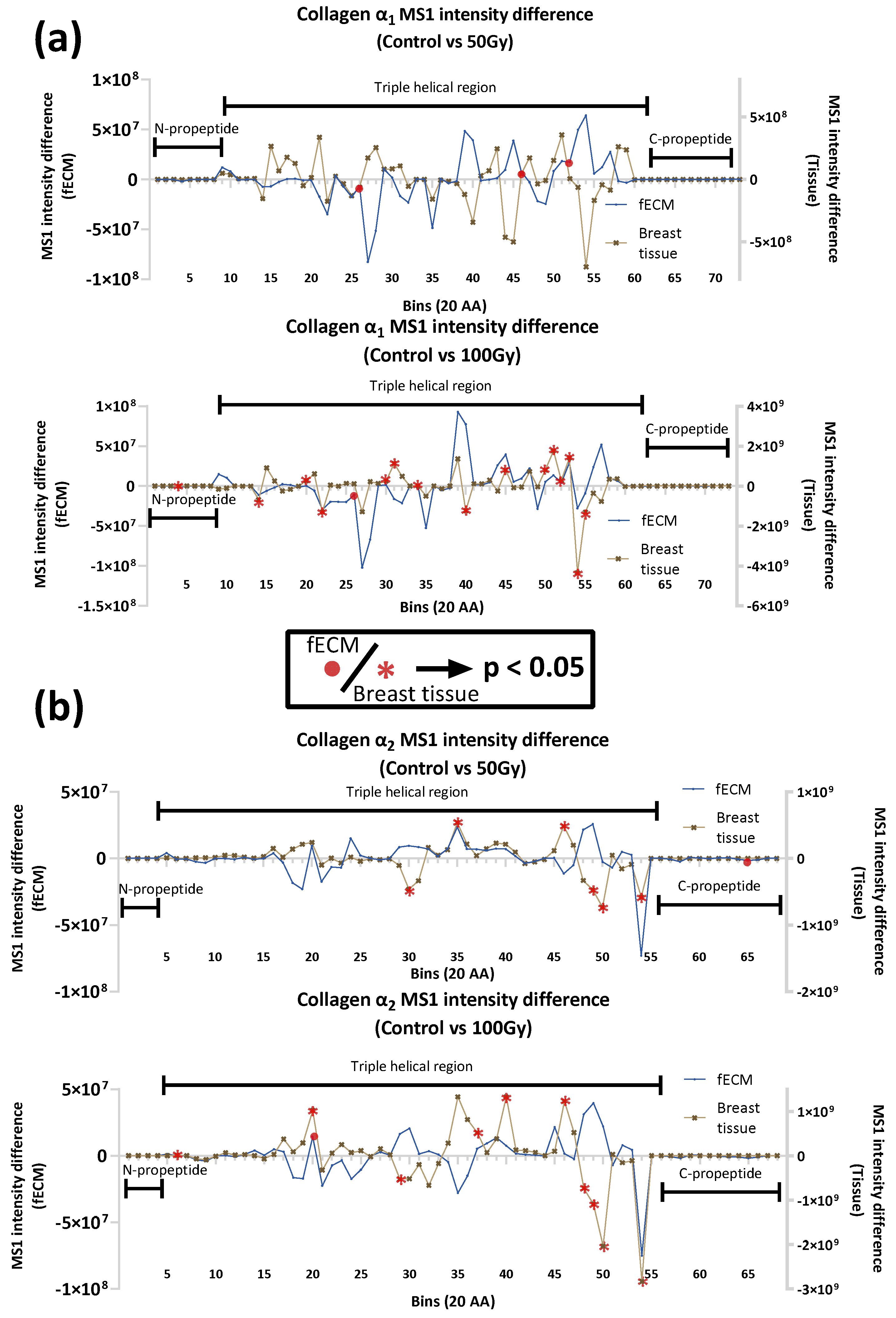
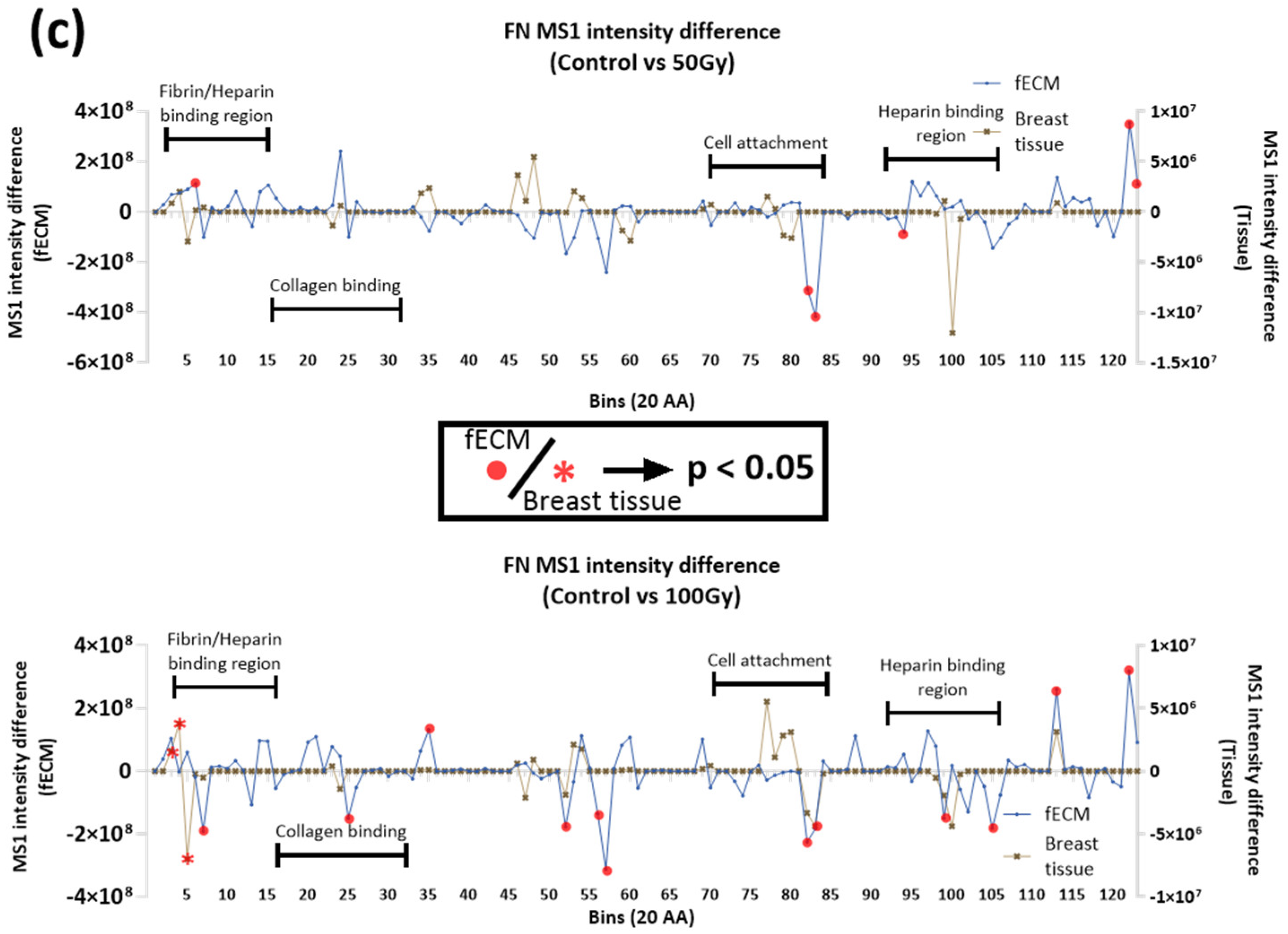
2.2.3. Impact of X-ray on Collagen I and Fibronectin in Complex Matrices
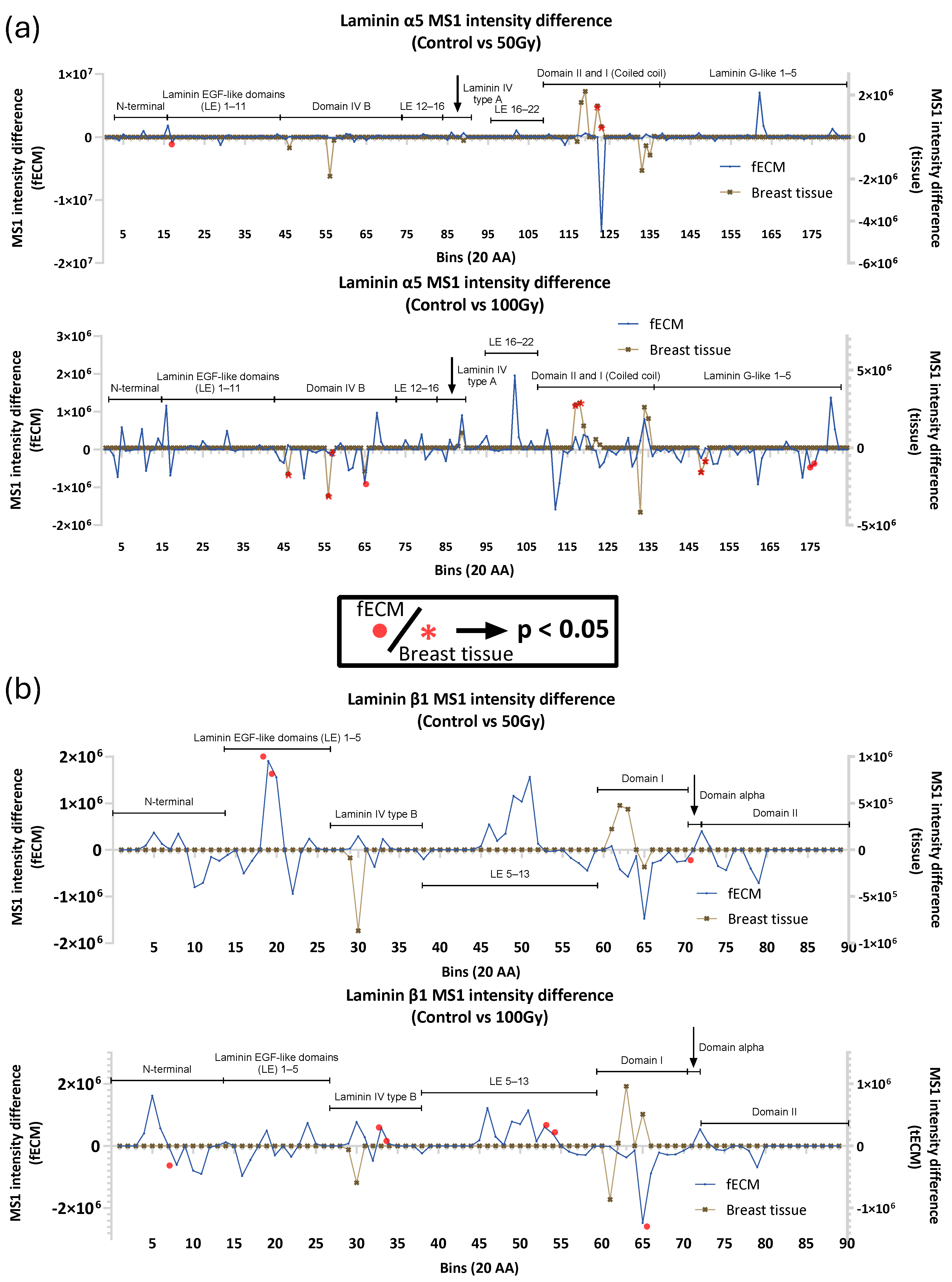
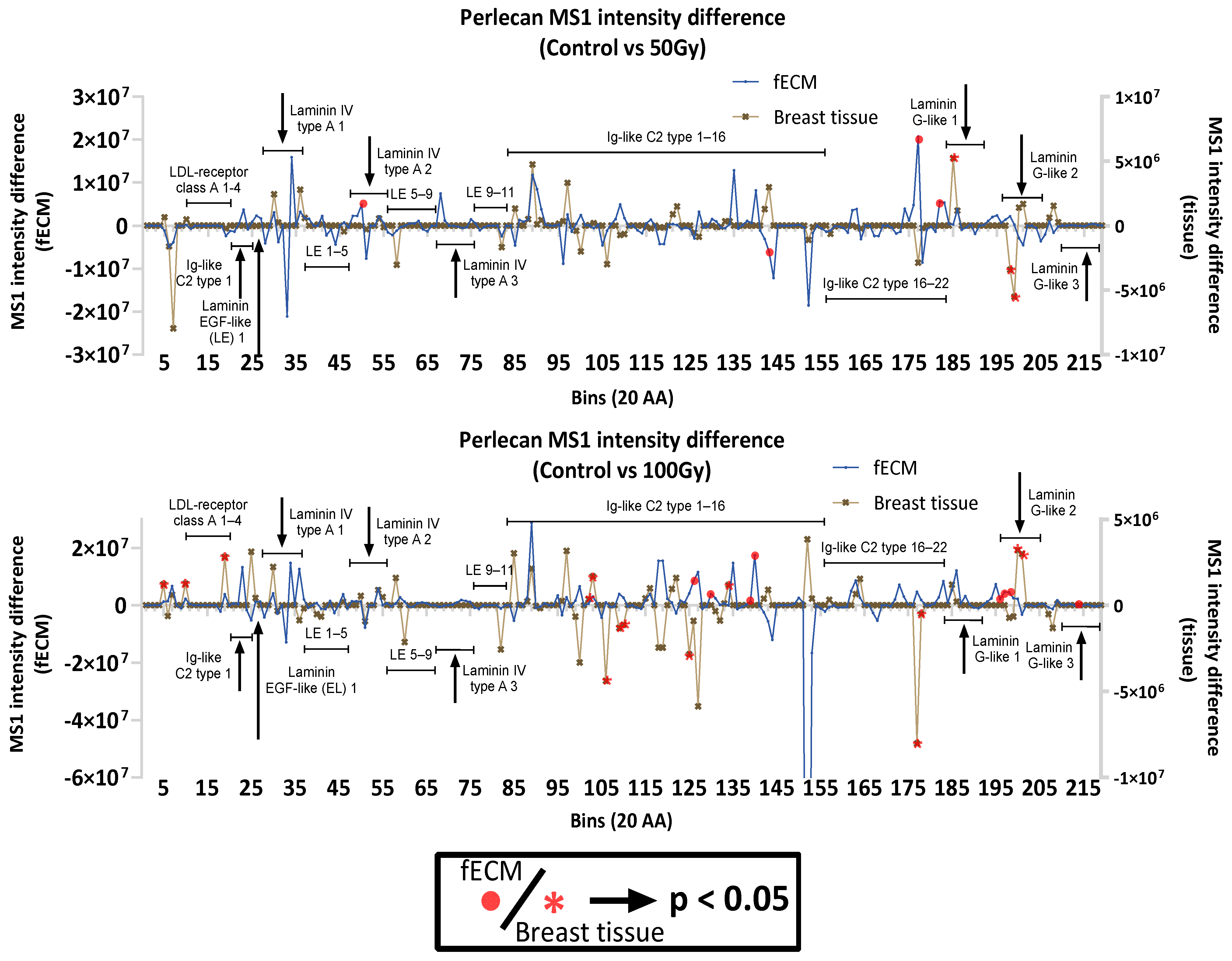
2.2.4. Impact of X-Ray on Basement Membrane Proteins in Complex Matrices
2.3. X-Ray Exposure Alters the Structure of Intracellular Proteins in Breast Tissue
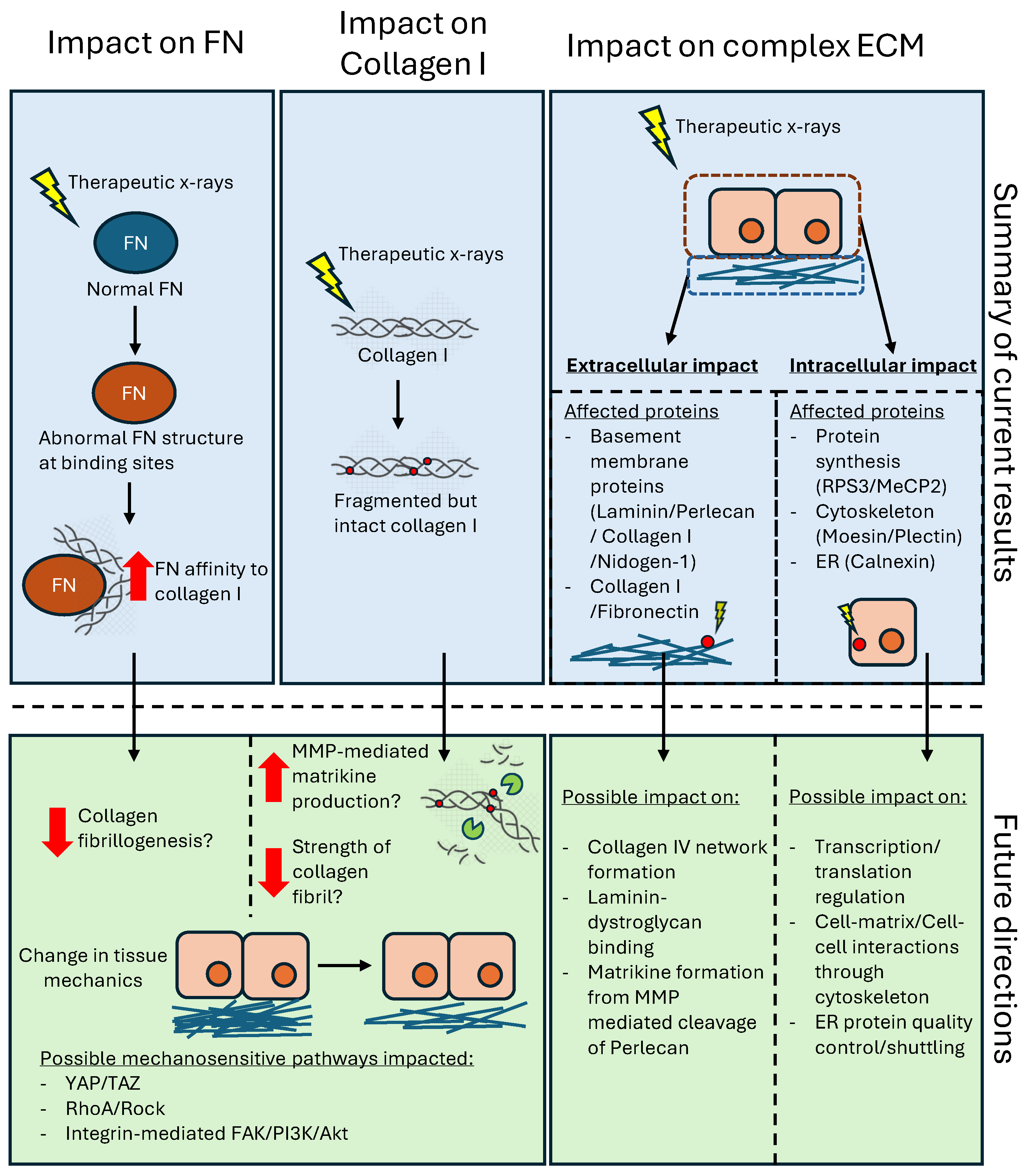
3. Discussion
3.1. Collagen I and Plasma Fibronectin Exhibit Differential Susceptibility to X-Rays
3.1.1. Impact of X-Ray Exposure on Collagen I
3.1.2. Impact of X-Ray Exposure on Fibronectin
3.1.3. Radiation Impact on ECM Proteins in Complex Environments
3.2. Radiation Impact to Intracellular Proteins in Complex Tissues
4. Materials and Methods
4.1. Biological Materials
4.2. X-Ray Irradiation
4.3. Protein Gel Electrophoresis of Purified Proteins
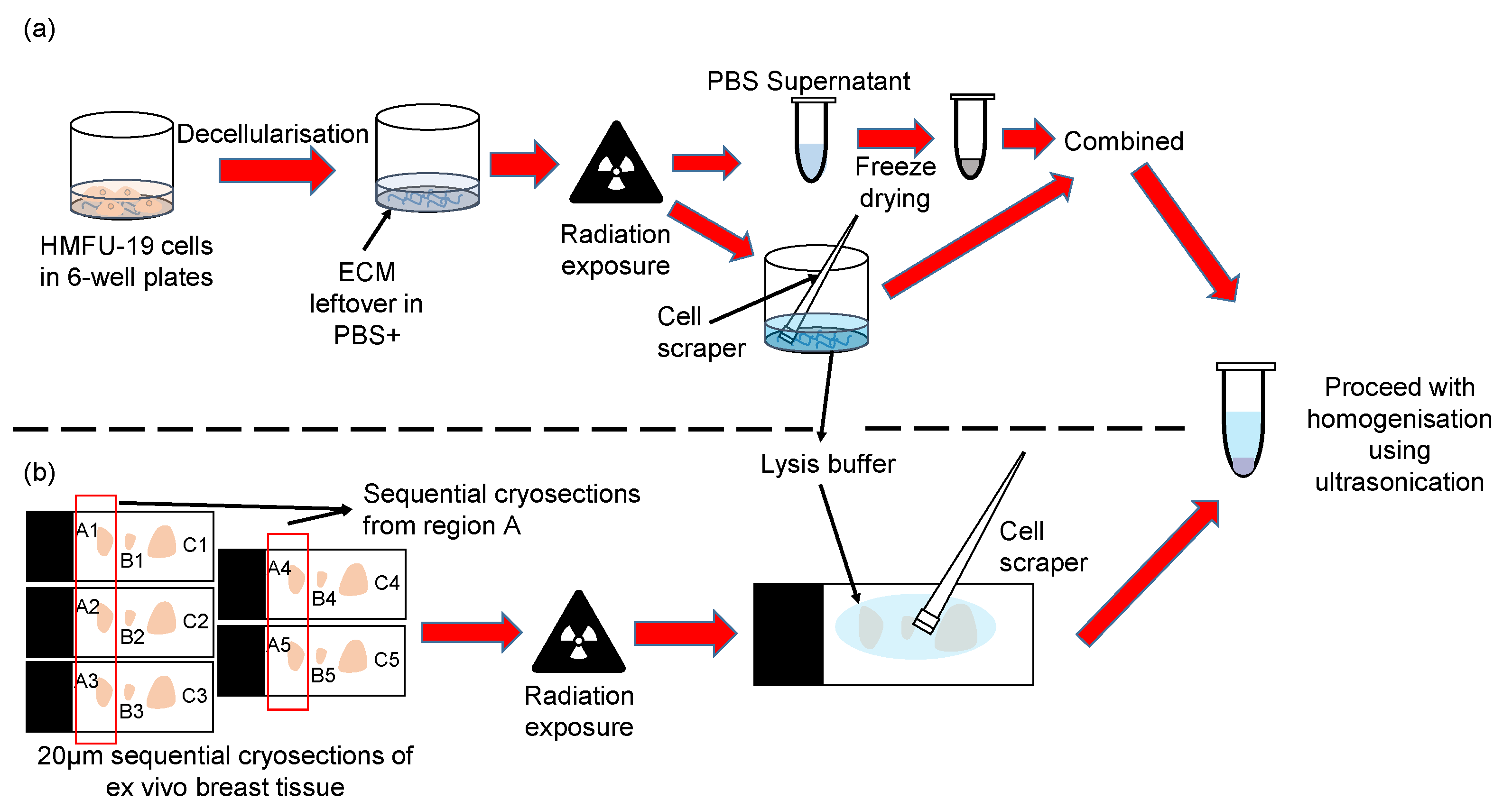
4.4. Fibroblast Extracellular Matrix Enrichment and Processing
4.5. Breast Tissue Preparation and Processing
4.6. Mass Spectrometry Sample Preparation
4.7. Mass Spectrometry Sample Runs
4.8. Mass Spectrometry Protein Identification and Quantification
4.9. Peptide Location Fingerprinting Analysis
4.10. Solid Phase Enzyme-Linked Immunosorbent Assay for Collagen–pFN Binding
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DSB | DNA double-strand break |
| DTT | Dithiothreitol |
| ECM | Extracellular matrix |
| ELISA | Enzyme-linked immunosorbent assay |
| ER | Endoplasmic reticulum |
| FAK | Focal adhesion kinase |
| fECM | ECM-enriched sample from cultured fibroblasts |
| FN | Fibronectin |
| H&E | Hematoxylin and eosin |
| HMFU-19 | Normal primary human mammary fibroblasts |
| hTERT | Human telomerase reverse transcriptase |
| IAM | Iodoacetamide |
| LCC | Laminin coiled coil domain |
| LC-MS/MS | Liquid chromatography with tandem mass spectrometry |
| LG | Laminin G-like (LG) domains |
| MOPS | 3-morpholinopropane-1-sulfonic acid |
| OCT | Optimal cutting temperature (OCT) compound |
| PGP | Proline–glycine–proline |
| PLF | Peptide location fingerprinting |
| pFN | Plasma fibronectin |
| ROCK | RhoA/rho-associated coiled-coil forming kinase |
| ROS | Reactive oxygen species |
| RT | Room temperature |
| SDS-PAGE | Sodium dodecyl sulphate–polyacrylamide gel electrophoresis |
| TAZ | Yes-associated protein-1 (YAP)/transcriptional coactivator with PDZ-binding motive |
| TEAB | Triethylammonium bicarbonate |
| TBST | Tris-buffered saline with Tween 20, |
| TMB | 3,3′,5,5′-tetramethylbenzidine |
References
- Tuieng, R.J.; Cartmell, S.H.; Kirwan, C.C.; Sherratt, M.J. The effects of ionising and non-ionising electromagnetic radiation on extracellular matrix proteins. Cells 2021, 10, 3041. [Google Scholar] [CrossRef]
- Patel, A.; Jackson, B. Low-dose radiation use in diagnostic imaging and cancer therapy settings. Radiol. Medica 2018, 123, 618–619. [Google Scholar] [CrossRef]
- Uselmann, A.J.; Thomadsen, B.R. On effective dose for radiotherapy based on doses to nontarget organs and tissues. Med. Phys. 2015, 42, 977–982. [Google Scholar] [CrossRef]
- Feldberg, R.S.; Carew, J.A. Water radiolysis products and nucleotide damage in γ-irradiated DNA. Int. J. Radiat. Biol. 1981, 40, 11–17. [Google Scholar] [CrossRef]
- Le Caër, S. Water radiolysis: Influence of oxide surfaces on H2 production under ionizing radiation. Water 2011, 3, 235–253. [Google Scholar] [CrossRef]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef]
- Pietraforte, D.; Paulicelli, E.; Patrono, C.; Gambardella, L.; Scorza, G.; Testa, A.; Fattibene, P. Protein oxidative damage and redox imbalance induced by ionising radiation in CHO cells. Free Radic. Res. 2018, 52, 465–479. [Google Scholar] [CrossRef]
- Wang, K.; Tepper, J.E. Radiation therapy-associated toxicity: Etiology, management, and prevention. CA Cancer J. Clin. 2021, 71, 437–454. [Google Scholar] [CrossRef]
- Hille-Betz, U.; Vaske, B.; Bremer, M.; Soergel, P.; Kundu, S.; Klapdor, R.; Hillemanns, P.; Henkenberens, C. Late radiation side effects, cosmetic outcomes and pain in breast cancer patients after breast-conserving surgery and three-dimensional conformal radiotherapy. Strahlenther. Onkol. 2016, 192, 8–16. [Google Scholar] [CrossRef]
- Henson, K.E.; McGale, P.; Taylor, C.; Darby, S.C. Radiation-related mortality from heart disease and lung cancer more than 20 years after radiotherapy for breast cancer. Br. J. Cancer 2013, 108, 179–182. [Google Scholar] [CrossRef]
- Pignol, J.-P.; Keller, B.M.; Ravi, A. Doses to internal organs for various breast radiation techniques—Implications on the risk of secondary cancers and cardiomyopathy. Radiat. Oncol. 2011, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Mettler, F.A. Medical effects and risks of exposure to ionising radiation. J. Radiol. Prot. 2012, 32, N9–N13. [Google Scholar] [CrossRef]
- Fernando, I.N.; Ford, H.T.; Powles, T.J.; Ashley, S.; Glees, J.P.; Torr, M.; Grafton, D.; Harmer, C.L. Factors affecting acute skin toxicity in patients having breast irradiation after conservative surgery: A prospective study of treatment practice at the Royal Marsden Hospital. Clin. Oncol. 1996, 8, 226–233. [Google Scholar] [CrossRef]
- Dörr, W.; Hendry, J.H. Consequential late effects in normal tissues. Radiother. Oncol. 2001, 61, 223–231. [Google Scholar] [CrossRef]
- Tsoutsou, P.G.; Koukourakis, M.I. Radiation pneumonitis and fibrosis: Mechanisms underlying its pathogenesis and implications for future research. Int. J. Radiat. Oncol. Biol. Phys. 2006, 66, 1281–1293. [Google Scholar] [CrossRef]
- Ejaz, A.; Greenberger, J.S.; Rubin, P.J. Understanding the mechanism of radiation induced fibrosis and therapy options. Pharmacol. Ther. 2019, 204, 107399. [Google Scholar] [CrossRef]
- Hu, M.; Ling, Z.; Ren, X. Extracellular matrix dynamics: Tracking in biological systems and their implications. J. Biol. Eng. 2022, 16, 13. [Google Scholar] [CrossRef]
- Jain, I.; Brougham-Cook, A.; Underhill, G.H. Effect of distinct ECM microenvironments on the genome-wide chromatin accessibility and gene expression responses of hepatic stellate cells. Acta Biomater. 2023, 167, 278–292. [Google Scholar] [CrossRef]
- Gjorevski, N.; Piotrowski, A.S.; Varner, V.D.; Nelson, C.M. Dynamic tensile forces drive collective cell migration through three-dimensional extracellular matrices. Sci. Rep. 2015, 5, 11458. [Google Scholar] [CrossRef]
- Hubbard, B.; Buczek-Thomas, J.A.; Nugent, M.A.; Smith, M.L. Fibronectin Fiber Extension Decreases Cell Spreading and Migration. J. Cell. Physiol. 2016, 231, 1728–1736. [Google Scholar] [CrossRef]
- Schultz, G.S.; Wysocki, A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 2009, 17, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S.; Salza, R. Matricryptins and matrikines: Biologically active fragments of the extracellular matrix. Exp. Dermatol. 2014, 23, 457–463. [Google Scholar] [CrossRef]
- Kisling, A.; Lust, R.M.; Katwa, L.C. What is the role of peptide fragments of collagen I and IV in health and disease? Life Sci. 2019, 228, 30–34. [Google Scholar] [CrossRef]
- Mohamed, F.; Bradley, D.A.; Winlove, C.P. Effects of ionizing radiation on extracellular matrix. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2007, 580, 566–569. [Google Scholar] [CrossRef]
- Kotova, S.L.; Timashev, P.S.; Belkova, G.V.; Kochueva, M.V.; Babak, K.V.; Timofeeva, V.A.; Kiseleva, E.B.; Vasilieva, O.O.; Maslennikova, A.V.; Solovieva, A.B. Early Effects of Ionizing Radiation on the Collagen Hierarchical Structure of Bladder and Rectum Visualized by Atomic Force Microscopy. Microsc. Microanal. 2018, 24, 38–48. [Google Scholar] [CrossRef]
- Paquette, B.; Baptiste, C.; Therriault, H.; Arguin, G.; Plouffe, B.; Lemay, R. In vitro irradiation of basement membrane enhances the invasiveness of breast cancer cells. Br. J. Cancer 2007, 97, 1505–1512. [Google Scholar] [CrossRef]
- Lalande, M.; Schwob, L.; Vizcaino, V.; Chirot, F.; Dugourd, P.; Schlathölter, T.; Poully, J.C. Direct Radiation Effects on the Structure and Stability of Collagen and Other Proteins. ChemBioChem 2019, 20, 2972–2980. [Google Scholar] [CrossRef]
- Pendleton, M.M.; Emerzian, S.R.; Liu, J.; Tang, S.Y.; O’Connell, G.D.; Alwood, J.S.; Keaveny, T.M. Effects of ex vivo ionizing radiation on collagen structure and whole-bone mechanical properties of mouse vertebrae. Bone 2019, 128, 115043. [Google Scholar] [CrossRef]
- Tuieng, R.J.; Disney, C.; Cartmell, S.H.; Kirwan, C.C.; Eckersley, A.; Newham, E.; Gupta, H.S.; Hoyland, J.A.; Lee, P.D.; Sherratt, M.J. Impact of therapeutic X-ray exposure on collagen I and associated proteins. Acta Biomater. 2025, 197, 294–311. [Google Scholar] [CrossRef]
- Ozols, M.; Eckersley, A.; Mellody, K.T.; Mallikarjun, V.; Warwood, S.; O’Cualain, R.; Knight, D.; Watson, R.E.B.; Griffiths, C.E.M.; Swift, J.; et al. Peptide location fingerprinting reveals modification-associated biomarker candidates of ageing in human tissue proteomes. Aging Cell 2021, 20, e13355. [Google Scholar] [CrossRef]
- Eckersley, A.; Ozols, M.; O’Cualain, R.; Keevill, E.J.; Foster, A.; Pilkington, S.; Knight, D.; Griffiths, C.E.M.; Watson, R.E.B.; Sherratt, M.J. Proteomic fingerprints of damage in extracellular matrix assemblies. Matrix Biol. Plus 2020, 5, 100027. [Google Scholar] [CrossRef] [PubMed]
- Eckersley, A.; Morais, M.R.; Ozols, M.; Lennon, R. Peptide location fingerprinting identifies structural alterations within basement membrane components in ageing kidney. Matrix Biol. 2023, 121, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Birk, D.E.; Bruckner, P. Collagen suprastructures. Top. Curr. Chem. 2005, 247, 185–205. [Google Scholar] [CrossRef]
- Danielson, K.G.; Baribault, H.; Holmes, D.F.; Graham, H.; Kadler, K.E.; Iozzo, R.V. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J. Cell Biol. 1997, 136, 729–743. [Google Scholar] [CrossRef]
- Robinson, K.A.; Sun, M.; Barnum, C.E.; Weiss, S.N.; Huegel, J.; Shetye, S.S.; Lin, L.; Saez, D.; Adams, S.M.; Iozzo, R.V.; et al. Decorin and biglycan are necessary for maintaining collagen fibril structure, fiber realignment, and mechanical properties of mature tendons. Matrix Biol. 2017, 64, 81–93. [Google Scholar] [CrossRef]
- Chia, J.; Kusuma, N.; Anderson, R.; Parker, B.; Bidwell, B.; Zamurs, L.; Nice, E.; Pouliot, N. Evidence for a role of tumor-derived laminin-511 in the metastatic progression of breast cancer. Am. J. Pathol. 2007, 170, 2135–2148. [Google Scholar] [CrossRef]
- Pouliot, N.; Kusuma, N. Laminin-511. Cell Adh. Migr. 2013, 7, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.Y.; Lee, J.Y.; Kim, J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004, 560, 81–85. [Google Scholar] [CrossRef]
- Wan, F.; Anderson, D.E.; Barnitz, R.A.; Snow, A.; Bidere, N.; Zheng, L.; Hegde, V.; Lam, L.T.; Staudt, L.M.; Levens, D.; et al. Ribosomal Protein S3: A KH Domain Subunit in NF-κB Complexes that Mediates Selective Gene Regulation. Cell 2007, 131, 927–939. [Google Scholar] [CrossRef]
- Adkins, N.L.; Georgel, P.T. MeCP2: Structure and function. Biochem. Cell Biol. 2011, 89, 1–11. [Google Scholar] [CrossRef]
- Paskevicius, T.; Farraj, R.A.; Michalak, M.; Agellon, L.B. Calnexin, More Than Just a Molecular Chaperone. Cells 2023, 12, 403. [Google Scholar] [CrossRef] [PubMed]
- Forrester, A.; De Leonibus, C.; Grumati, P.; Fasana, E.; Piemontese, M.; Staiano, L.; Settembre, C. A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM 134B complex. EMBO J. 2019, 38, e99847. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, G.; Muñoz-Escobar, J.; Castro, K.; Gehring, K. Mapping the ER Interactome: The P Domains of Calnexin and Calreticulin as Plurivalent Adapters for Foldases and Chaperones. Structure 2017, 25, 1415–1422.e3. [Google Scholar] [CrossRef]
- Neisch, A.L.; Fehon, R.G. Ezrin, Radixin and Moesin: Key regulators of membrane-cortex interactions and signaling. Curr. Opin. Cell Biol. 2011, 23, 377–382. [Google Scholar] [CrossRef]
- Fehon, R.G.; McClatchey, A.I.; Bretscher, A. Organizing the cell cortex: The role of ERM proteins. Nat. Rev. Mol. Cell Biol. 2010, 11, 276–287. [Google Scholar] [CrossRef]
- Wiche, G.; Osmanagic-Myers, S.; Castañón, M.J. Networking and anchoring through plectin: A key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol. 2015, 32, 21–29. [Google Scholar] [CrossRef]
- Gregor, M.; Osmanagic-Myers, S.; Burgstaller, G.; Wolfram, M.; Fischer, I.; Walko, G.; Resch, G.P.; Jörgl, A.; Herrmann, H.; Wiche, G. Mechanosensing through focal adhesion-anchored intermediate filaments. FASEB J. 2014, 28, 715–729. [Google Scholar] [CrossRef]
- Vartio, T. Regular fragmentation of hydrogen peroxide-treated fibronectin. J. Biol. Chem. 1989, 264, 4471–4475. [Google Scholar] [CrossRef]
- Wells, J.M.; Gaggar, A.; Blalock, J.E. MMP generated matrikines. Matrix Biol. 2015, 44–46, 122–129. [Google Scholar] [CrossRef]
- Van Doren, S.R. Matrix metalloproteinase interactions with collagen and elastin. Matrix Biol. 2015, 44–46, 224–231. [Google Scholar] [CrossRef]
- Manka, S.W.; Carafoli, F.; Visse, R.; Bihan, D.; Raynal, N.; Farndale, R.W.; Murphy, G.; Enghild, J.J.; Hohenester, E.; Nagase, H. Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1. Proc. Natl. Acad. Sci. USA 2012, 109, 12461–12466. [Google Scholar] [CrossRef] [PubMed]
- Gaggar, A.; Jackson, P.L.; Noerager, B.D.; O’Reilly, P.J.; McQuaid, D.B.; Rowe, S.M.; Clancy, J.P.; Blalock, J.E. A Novel Proteolytic Cascade Generates an Extracellular Matrix-Derived Chemoattractant in Chronic Neutrophilic Inflammation. J. Immunol. 2008, 180, 5662–5669. [Google Scholar] [CrossRef] [PubMed]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef]
- Park, J.S.; Chu, J.S.; Tsou, A.D.; Diop, R.; Tang, Z.; Wang, A.; Li, S. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials 2011, 32, 3921–3930. [Google Scholar] [CrossRef]
- Fioravanti, E.; Vellieux, F.M.D.; Amara, P.; Madern, D.; Weik, M. Specific radiation damage to acidic residues and its relation to their chemical and structural environment. J. Synchrotron Radiat. 2007, 14, 84–91. [Google Scholar] [CrossRef]
- Sharp, J.S.; Becker, J.M.; Hettich, R.L. Analysis of Protein Solvent Accessible Surfaces by Photochemical Oxidation and Mass Spectrometry. Anal. Chem. 2004, 76, 672–683. [Google Scholar] [CrossRef]
- Kennett, E.C.; Chuang, C.Y.; Degendorfer, G.; Whitelock, J.M.; Davies, M.J. Mechanisms and consequences of oxidative damage to extracellular matrix. Biochem. Soc. Trans 2012, 39, 1279–1287. [Google Scholar] [CrossRef]
- Bettinger, J.; Ghaemmaghami, S. Methionine oxidation within the prion protein. Prion 2020, 14, 193–205. [Google Scholar] [CrossRef]
- Nomura, T.; Kamada, R.; Ito, I.; Chuman, Y.; Shimohigashi, Y.; Sakaguchi, K. Oxidation of methionine residue at hydrophobic core destabilitizes p53 tetrameric structure. Biopolymers 2009, 91, 78–84. [Google Scholar] [CrossRef]
- Erat, M.C.; Schwarz-Linek, U.; Pickford, A.R.; Farndale, R.W.; Campbell, I.D.; Vakonakis, I. Implications for collagen binding from the crystallographic structure of fibronectin 6FnI1-2FnII7FnI. J. Biol. Chem. 2010, 285, 33764–33770. [Google Scholar] [CrossRef]
- Kubow, K.E.; Vukmirovic, R.; Zhe, L.; Klotzsch, E.; Smith, M.L.; Gourdon, D.; Luna, S.; Vogel, V. Mechanical forces regulate the interactions of fibronectin and collagen i in extracellular matrix. Nat. Commun. 2015, 6, 8026. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, H.K.; Wilkes, C.M.; Martin, G.R. Interaction of fibronectin with collagen fibrils. Biochemistry 1981, 20, 2325–2330. [Google Scholar] [CrossRef] [PubMed]
- Paten, J.A.; Martin, C.L.; Wanis, J.T.; Siadat, S.M.; Figueroa-Navedo, A.M.; Ruberti, J.W.; Deravi, L.F. Molecular Interactions Between Collagen and Fibronectin: A Reciprocal Relationship that Regulates De Novo Fibrillogenesis. Chem 2019, 5, 2126–2145. [Google Scholar] [CrossRef]
- Speranza, M.L.; Valentini, G.; Calligaro, A. Influence of Fibronectin on the Fibrillogenesis of Type I and Type III Collagen. Top. Catal. 1987, 7, 115–123. [Google Scholar] [CrossRef]
- Lessey, E.C.; Guilluy, C.; Burridge, K. From mechanical force to RhoA activation. Biochemistry 2012, 51, 7420–7432. [Google Scholar] [CrossRef]
- Pang, X.; He, X.; Qiu, Z.; Zhang, H.; Xie, R.; Liu, Z.; Gu, Y.; Zhao, N.; Xiang, Q.; Cui, Y. Targeting integrin pathways: Mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2023, 8, 1. [Google Scholar] [CrossRef]
- Amirrah, I.N.; Lokanathan, Y.; Zulkiflee, I.; Wee, M.F.M.R.; Motta, A.; Fauzi, M.B. A Comprehensive Review on Collagen Type I Development of Biomaterials for Tissue Engineering: From Biosynthesis to Bioscaffold. Biomedicines 2022, 10, 2307. [Google Scholar] [CrossRef]
- Harrison, D.; Hussain, S.A.; Combs, A.C.; Ervasti, J.M.; Yurchenco, P.D.; Hohenester, E. Crystal structure and cell surface anchorage sites of laminin α1LG4-5. J. Biol. Chem. 2007, 282, 11573–11581. [Google Scholar] [CrossRef]
- Wizemann, H.; Garbe, J.H.O.; Friedrich, M.V.K.; Timpl, R.; Sasaki, T.; Hohenester, E. Distinct requirements for heparin and α-dystroglycan binding revealed by structure-based mutagenesis of the laminin α2 LG4-LG5 domain pair. J. Mol. Biol. 2003, 332, 635–642. [Google Scholar] [CrossRef]
- Hohenester, E. Laminin G-like domains: Dystroglycan-specific lectins. Curr. Opin. Struct. Biol. 2019, 56, 56–63. [Google Scholar] [CrossRef]
- Moran, T.; Gat, Y.; Fass, D. Laminin L4 domain structure resembles adhesion modules in ephrin receptor and other transmembrane glycoproteins. FEBS J. 2015, 282, 2746–2757. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Pal, N.; Concannon, M.; Paul, M.; Doran, M.; Poluzzi, C.; Sekiguchi, K.; Whitelock, J.M.; Neill, T.; Iozzo, R.V. Endorepellin, the angiostatic module of perlecan, interacts with both the α2β1 integrin and vascular endothelial growth factor receptor 2 (VEGFR2): A dual receptor antagonism. J. Biol. Chem. 2011, 286, 25947–25962. [Google Scholar] [CrossRef]
- Jung, M.; Lord, M.S.; Cheng, B.; Lyons, J.G.; Alkhouri, H.; Hughes, J.M.; McCarthy, S.J.; Iozzo, R.V.; Whitelock, J.M. Mast cells produce novel shorter forms of perlecan that contain functional endorepellin a role in angiogenesis and wound healing. J. Biol. Chem. 2013, 288, 3289–3304. [Google Scholar] [CrossRef] [PubMed]
- Babitzke, P.; Baker, C.S.; Romeo, T. Regulation of translation initiation by RNA binding proteins. Annu. Rev. Microbiol. 2009, 63, 27–44. [Google Scholar] [CrossRef]
- Park, Y.J.; Kim, T.S.; Kim, E.H.; Kim, H.D.; Kim, J. Ribosomal protein S3 is a novel negative regulator of non-homologous end joining repair of DNA double-strand breaks. FASEB J. 2020, 34, 8102–8113. [Google Scholar] [CrossRef]
- Wahba, A.; Lehman, S.L.; Tofilon, P.J. Radiation-induced translational control of gene expression. Translation 2017, 5, e1265703. [Google Scholar] [CrossRef]
- Tamaddondoust, R.N.; Wang, Y.; Jafarnejad, S.M.; Graber, T.E.; Alain, T. The highs and lows of ionizing radiation and its effects on protein synthesis. Cell Signal. 2022, 89, 110169. [Google Scholar] [CrossRef]
- Mottareale, R.; Frascogna, C.; La Verde, G.; Arrichiello, C.; Muto, P.; Netti, P.A.; Fusco, S.; Panzetta, V.; Pugliese, M. Impact of ionizing radiation on cell-ECM mechanical crosstalk in breast cancer. Front. Bioeng. Biotechnol. 2024, 12, 1408789. [Google Scholar] [CrossRef]
- La Verde, G.; Artiola, V.; Panzetta, V.; Pugliese, M.; Netti, P.A.; Fusco, S. Cytoskeleton response to ionizing radiation: A brief review on adhesion and migration effects. Biomedicines 2021, 9, 1102. [Google Scholar] [CrossRef]
- Pearson, M.A.; Reczek, D.; Bretscher, A.; Karplus, P.A. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 2000, 101, 259–270. [Google Scholar] [CrossRef]
- Fontao, L.; Geerts, D.; Kulkman, I.; Koster, J.; Kramer, D.; Sonnenberg, A. The interaction of plectin with actin: Evidence for cross-linking of actin filaments by dimerization of the actin-binding domain of plectin. J. Cell Sci. 2001, 114, 2065–2076. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Röder, C.; Sendtner, M.; Lüningschrör, P. An Essential Role for Calnexin in ER-Phagy and the Unfolded Protein Response. Cells 2024, 13, 1498. [Google Scholar] [CrossRef] [PubMed]
- Nishikaze, T.; Takayama, M. Study of factors governing negative molecular ion yields of amino acid and peptide in FAB, MALDI and ESI mass spectrometry. Int. J. Mass Spectrom. 2007, 268, 47–59. [Google Scholar] [CrossRef]
- Kapp, E.A.; Schütz, F.; Reid, G.E.; Eddes, J.S.; Moritz, R.L.; O’Hair, R.A.J.; Speed, T.P.; Simpson, R.J. Mining a Tandem Mass Spectrometry Database to Determine the Trends and Global Factors Influencing Peptide Fragmentation. Anal. Chem. 2003, 75, 6251–6264. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Hassabis, D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- O’Hare, M.J.; Bond, J.; Clarke, C.; Takeuchi, Y.; Atherton, A.J.; Berry, C.; Moody, J.; Silver, A.R.J.; Davies, D.C.; Alsop, A.E.; et al. Conditional immortalization of freshly isolated human mammary fibroblasts and endothelial cells. Proc. Natl. Acad. Sci. USA 2001, 98, 646–651. [Google Scholar] [CrossRef]
- Franco-Barraza, J.; Beacham, D.A.; Amatangelo, M.D.; Cukierman, E. Preparation of extracellular matrices produced by cultured and primary fibroblasts. Curr. Protoc. Cell Biol. 2016, 71, 10.9.1–10.9.34. [Google Scholar] [CrossRef]
- Platt, C.I.; Stewart-McGuinness, C.; Eckersley, A.; Wilkins, L.; Sherratt, M.J. Acute exposure to ultraviolet radiation targets proteins involved in collagen fibrillogenesis. Front. Physiol. 2024, 15, 1352161. [Google Scholar] [CrossRef]
- Eckersley, A.; Mellody, K.T.; Pilkington, S.; Griffiths, C.E.M.; Watson, R.E.B.; O’Cualain, R.; Baldock, C.; Knight, D.; Sherratt, M.J. Structural and compositional diversity of fibrillin microfibrils in human tissues. J. Biol. Chem. 2018, 293, 5117–5133. [Google Scholar] [CrossRef]
- Biesiadecki, B.J.; Jin, J.P. A high-throughput solid-phase microplate protein-binding assay to investigate interactions between myofilament proteins. J. Biotechnol. Biomed. 2011, 2011, 421701. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuieng, R.J.; Cartmell, S.H.; Kirwan, C.C.; Eckersley, A.; Sherratt, M.J. X-Ray Exposure Induces Structural Changes in Human Breast Proteins. Int. J. Mol. Sci. 2025, 26, 5696. https://doi.org/10.3390/ijms26125696
Tuieng RJ, Cartmell SH, Kirwan CC, Eckersley A, Sherratt MJ. X-Ray Exposure Induces Structural Changes in Human Breast Proteins. International Journal of Molecular Sciences. 2025; 26(12):5696. https://doi.org/10.3390/ijms26125696
Chicago/Turabian StyleTuieng, Ren Jie, Sarah H. Cartmell, Cliona C. Kirwan, Alexander Eckersley, and Michael J. Sherratt. 2025. "X-Ray Exposure Induces Structural Changes in Human Breast Proteins" International Journal of Molecular Sciences 26, no. 12: 5696. https://doi.org/10.3390/ijms26125696
APA StyleTuieng, R. J., Cartmell, S. H., Kirwan, C. C., Eckersley, A., & Sherratt, M. J. (2025). X-Ray Exposure Induces Structural Changes in Human Breast Proteins. International Journal of Molecular Sciences, 26(12), 5696. https://doi.org/10.3390/ijms26125696




