Genome-Wide Characterization and Expression Profile of the Jumonji-C Family Genes in Populus alba × Populus glandulosa Reveal Their Potential Roles in Wood Formation
Abstract
1. Introduction
2. Results
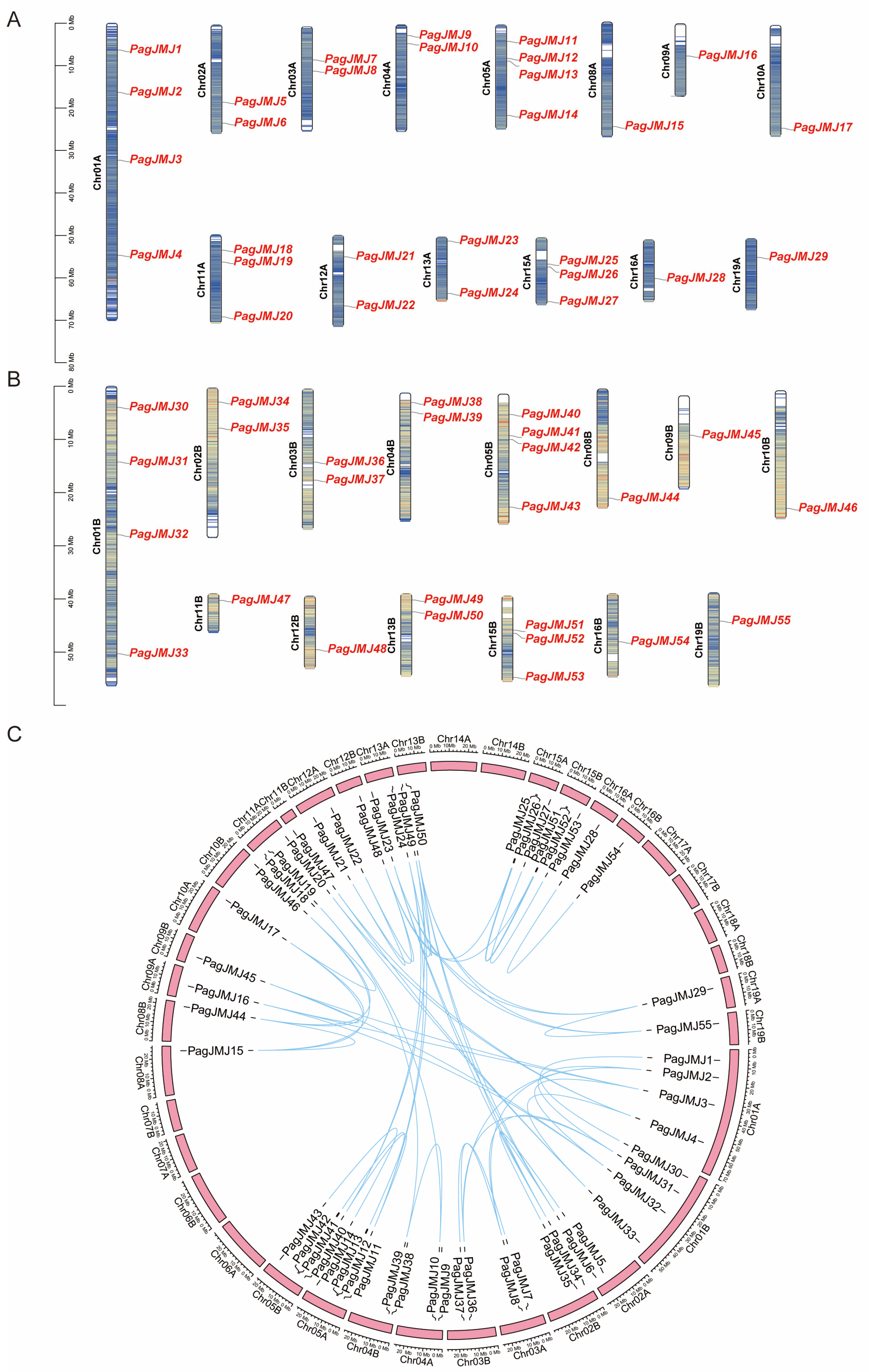
2.1. Chromosomal Location and Homologous Gene Analysis of JMJ-C Genes in P. alba × P. glandulosa
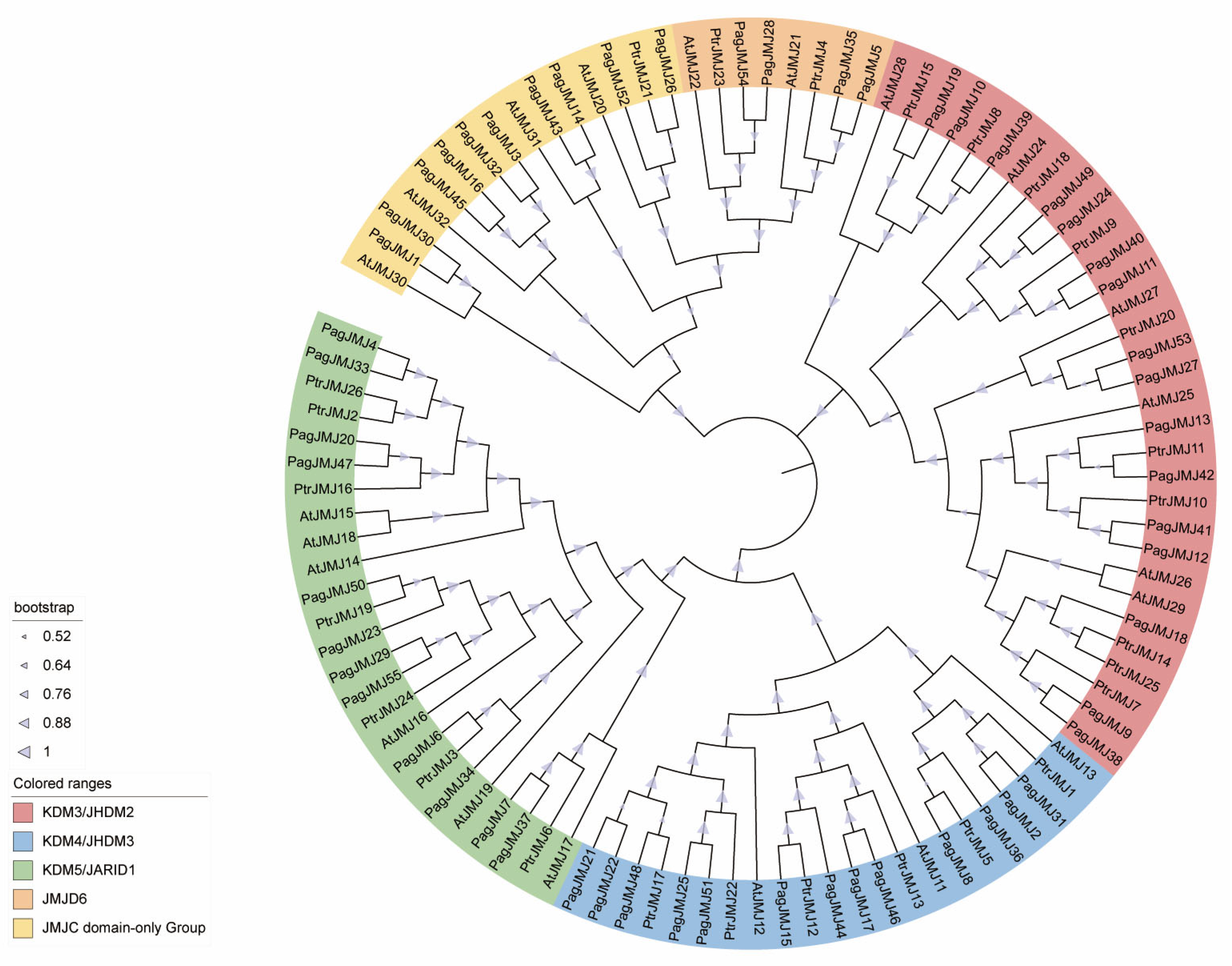
2.2. Phylogenetic Analysis of PagJMJ Genes
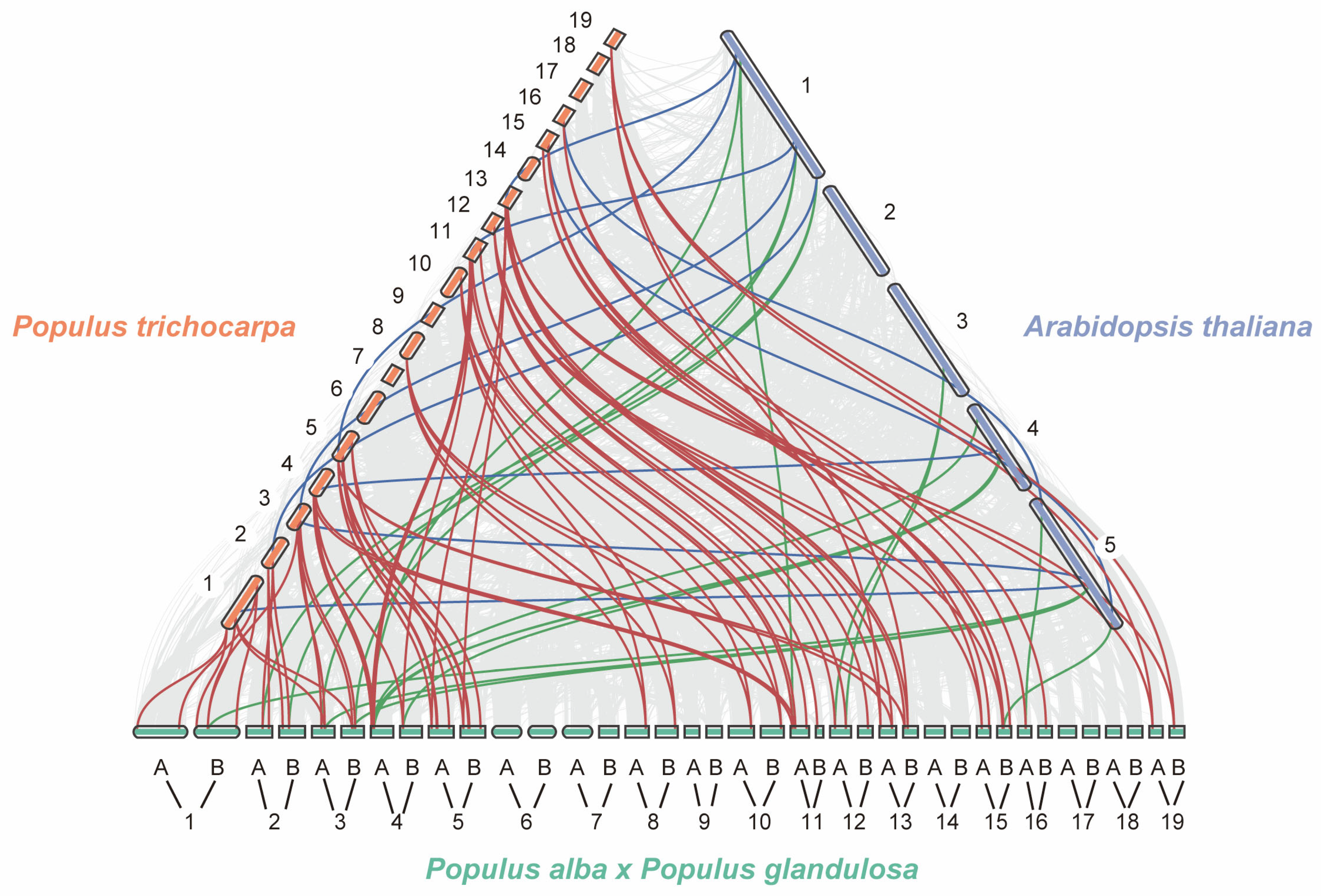
2.3. Collinearity Analysis of the PagJMJ Genes Family Across Species
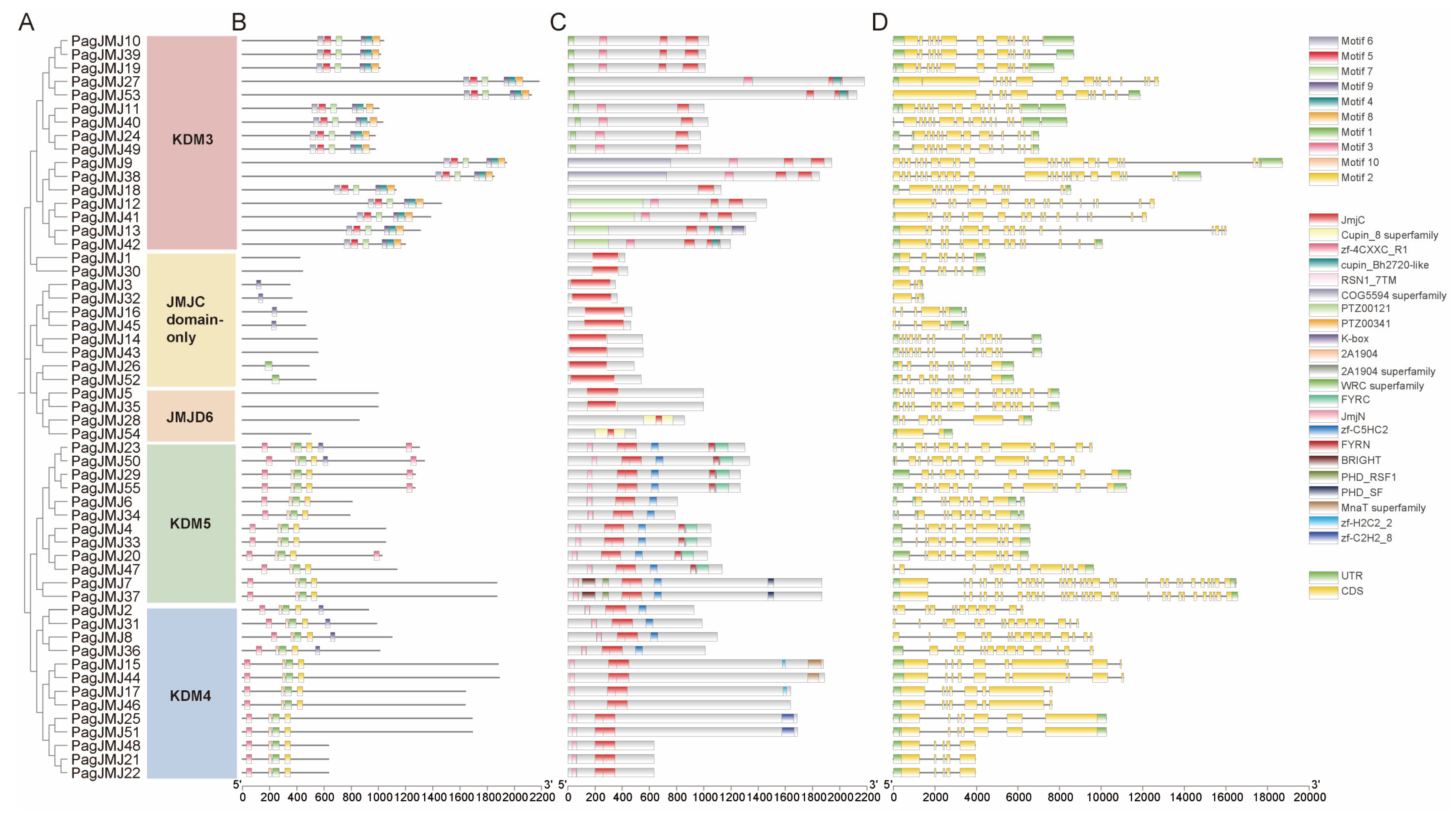
2.4. Gene Structure and Conserved Motif Analysis of PagJMJ Genes
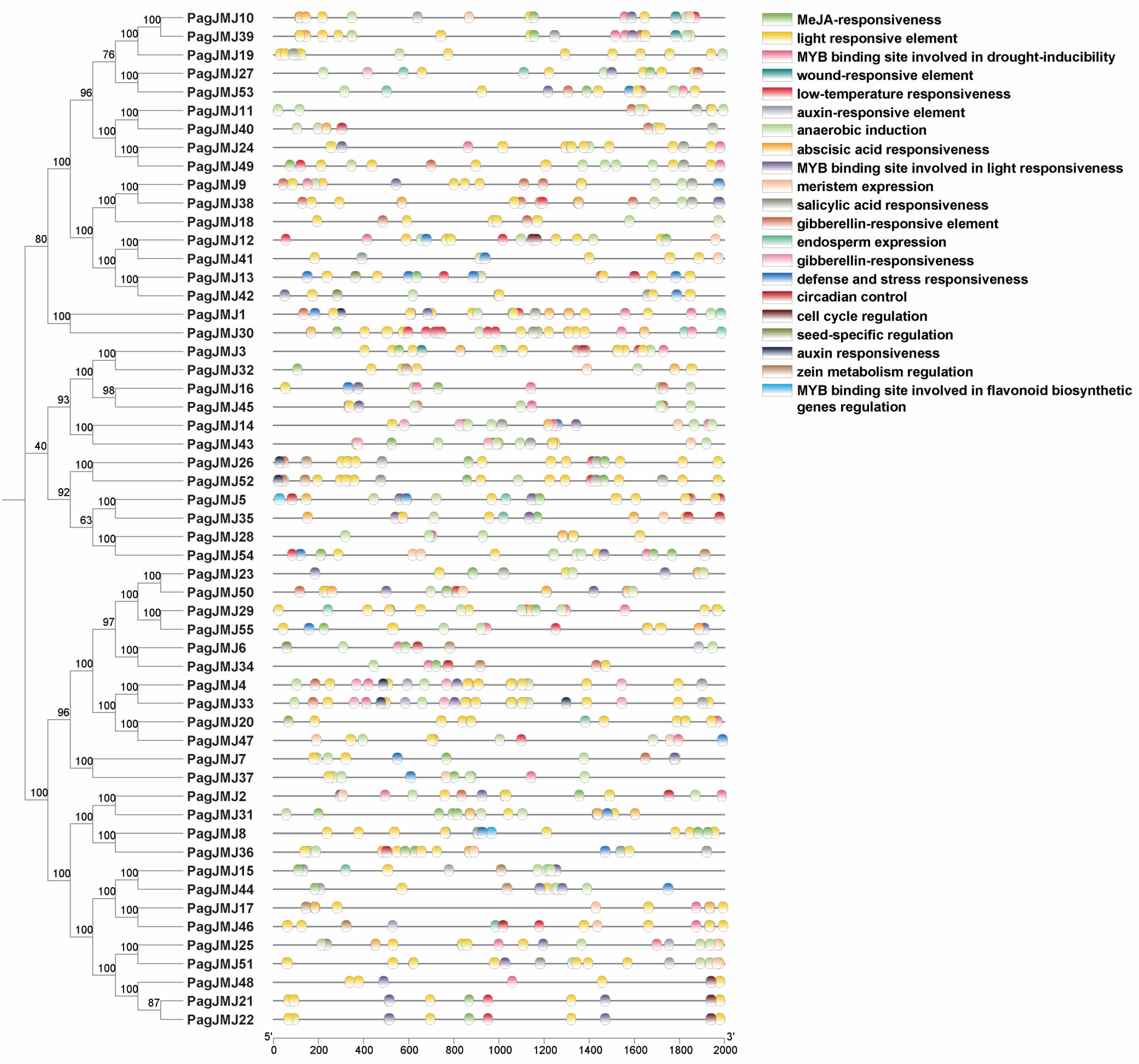
2.5. Cis-Element Analysis of PagJMJ Genes Promoter Regions
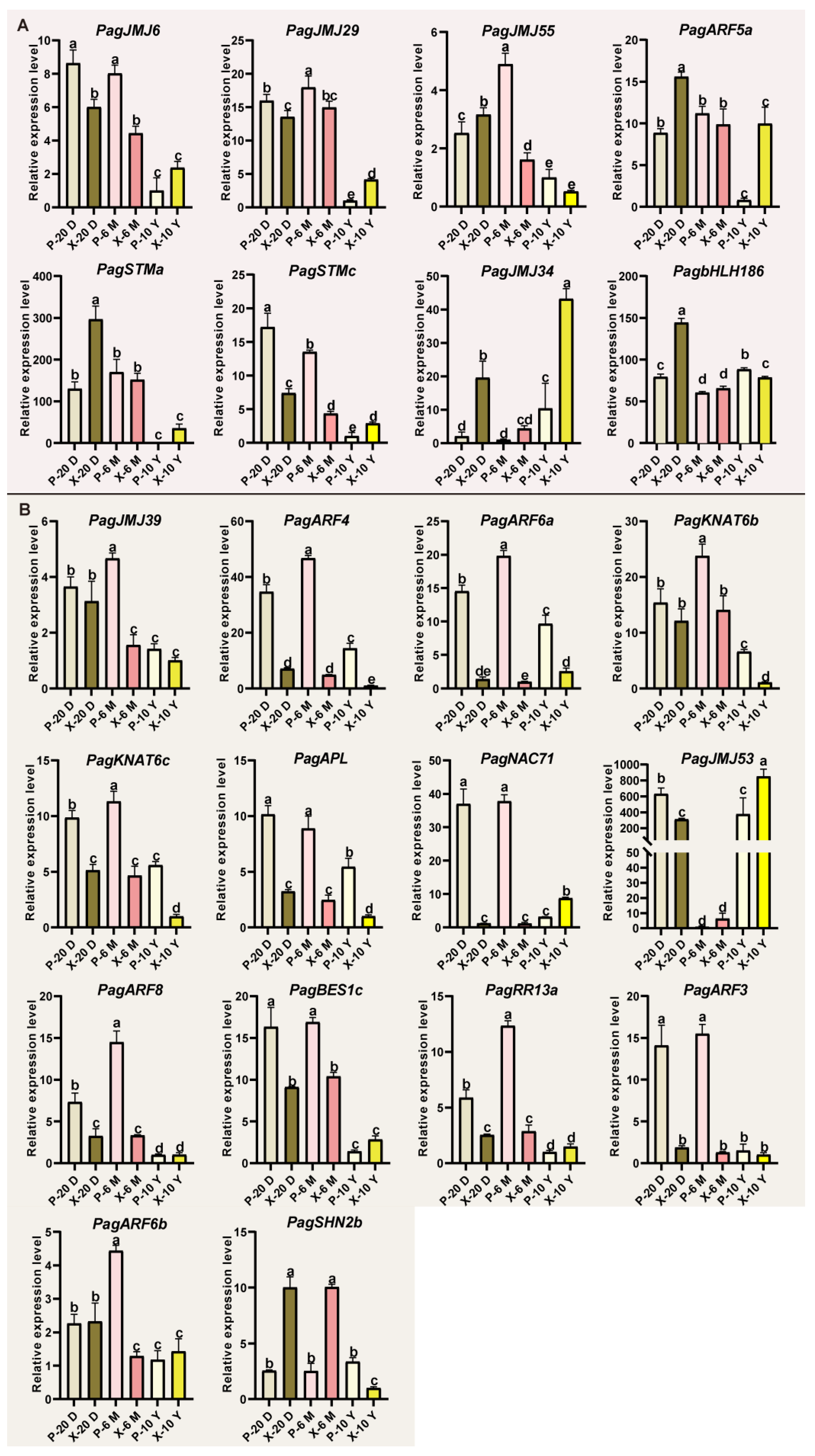
2.6. Expression Patterns of PagJMJ Genes in Different Tissues
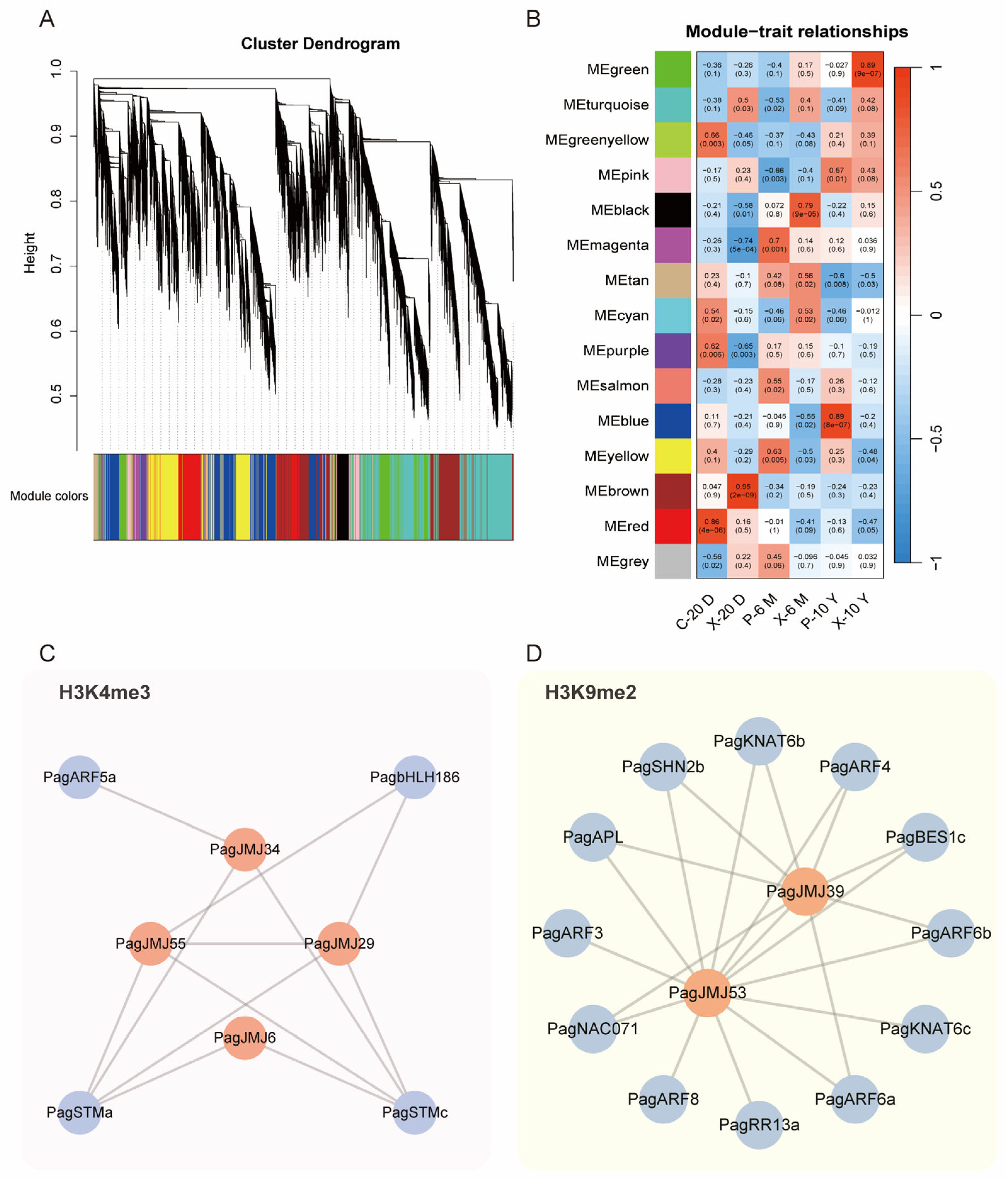
2.7. Co-Expression Analysis of PagJMJ Genes and Wood Formation-Related Genes
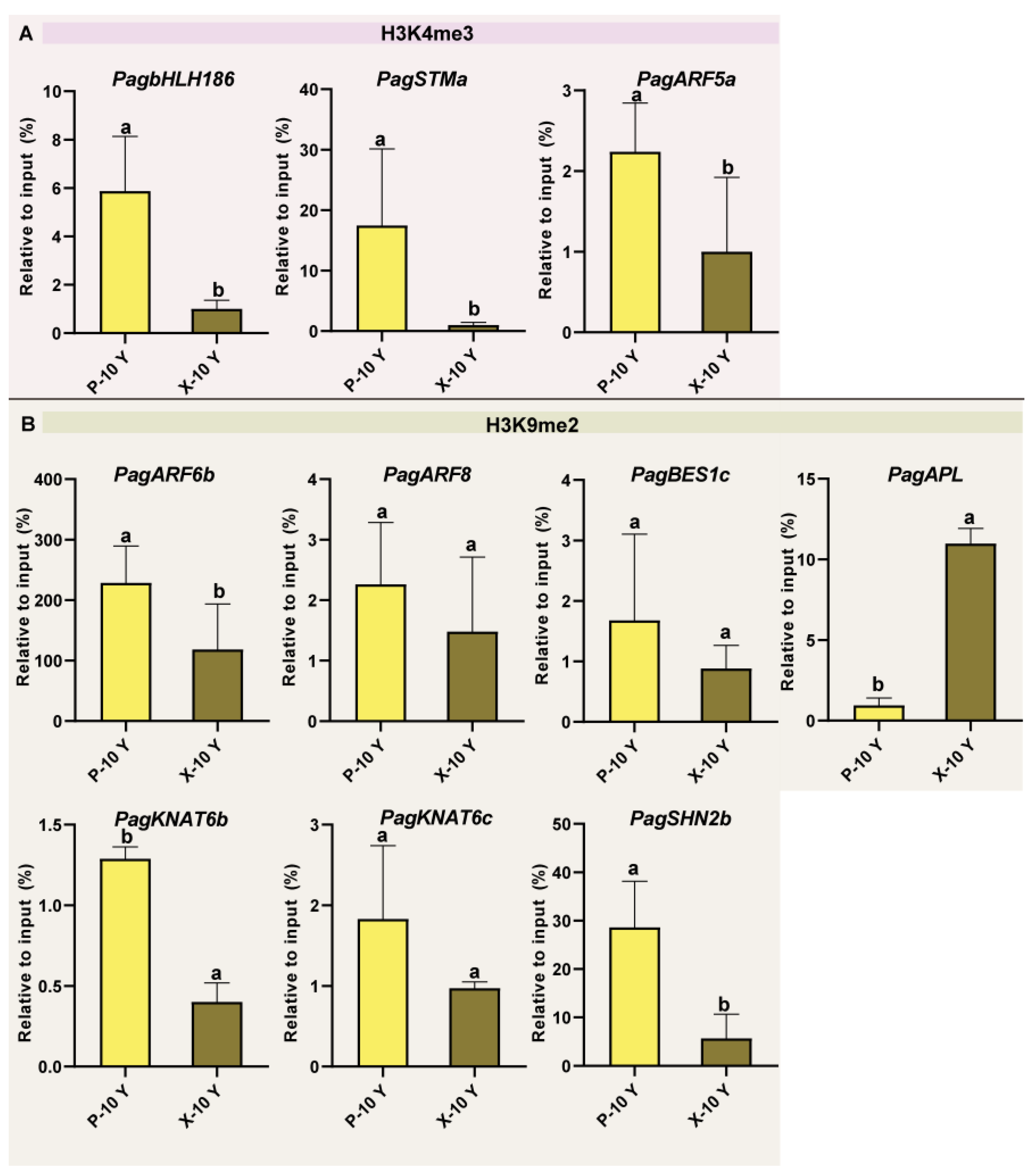
2.8. Analysis of Histone Modification Levels in Wood Formation-Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification and Physicochemical Analysis of PagJMJ Proteins
4.3. Chromosomal Localization and Collinearity Analysis of PagJMJ Genes
4.4. Analysis of Conserved Domains, Motifs, Gene Structures and Cis-Acting Elements
4.5. Collinearity Analyses of JMJ-C Genes Across Different Species
4.6. Transcriptomic Resources
4.7. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.8. RNA Extraction and Quantitative RT-PCR
4.9. Chromatin Immunoprecipitation and Quantitative PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Skvortsova, K.; Iovino, N.; Bogdanović, O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018, 19, 774–790. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.-K. Epigenetic gene regulation in plants and its potential applications in crop improvement. Nat. Rev. Mol. Cell Biol. 2024, 26, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Cao, X.; Chen, X.; Deng, X.; Deng, X.W.; Ding, Y.; Dong, A.; Duan, C.G.; Fang, X.; Gong, L.; et al. Epigenetics in the modern era of crop improvements. Sci. China Life Sci. 2025, 68, 1570–1609. [Google Scholar]
- Cheng, K.; Xu, Y.; Yang, C.; Ouellette, L.; Niu, L.; Zhou, X.; Chu, L.; Zhuang, F.; Liu, J.; Wu, H.; et al. Histone tales: Lysine methylation, a protagonist in Arabidopsis development. J. Exp. Bot. 2019, 71, 793–807. [Google Scholar] [CrossRef]
- Fuchs, J.; Demidov, D.; Houben, A.; Schubert, I. Chromosomal histone modification patterns--from conservation to diversity. Trends Plant Sci. 2006, 11, 199–208. [Google Scholar] [CrossRef]
- Klose, R.J.; Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 307–318. [Google Scholar] [CrossRef]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef]
- Qian, S.; Wang, Y.; Ma, H.; Zhang, L. Expansion and functional divergence of Jumonji C-containing histone demethylases: Significance of duplications in ancestral angiosperms and vertebrates. Plant Physiol. 2015, 168, 1321–1337. [Google Scholar] [CrossRef]
- Chen, Z.; Zang, J.; Whetstine, J.; Hong, X.; Davrazou, F.; Kutateladze, T.G.; Simpson, M.; Mao, Q.; Pan, C.-H.; Dai, S.; et al. Structural insights into histone demethylation by JMJD2 family members. Cell 2006, 125, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Li, G.; Cui, X.; Liu, C.; Wang, X.; Cao, X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 2008, 50, 886–896. [Google Scholar] [CrossRef]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef]
- Yang, C.; Bratzel, F.; Hohmann, N.; Koch, M.; Turck, F.; Calonje, M. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 2013, 23, 1324–1329. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Müller, K.; Kermode, A.R.; Finch-Savage, W.E. Seed dormancy cycling in Arabidopsis: Chromatin remodelling and regulation of DOG1 in response to seasonal environmental signals. Plant J. 2014, 81, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-N.; Ryu, J.-Y.; Jeong, Y.-M.; Park, J.; Song, J.-J.; Amasino, R.M.; Noh, B.; Noh, Y.-S. Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 2012, 22, 736–748. [Google Scholar] [CrossRef]
- Kumpf, R.; Thorstensen, T.; Rahman, M.A.; Heyman, J.; Nenseth, H.Z.; Lammens, T.; Herrmann, U.; Swarup, R.; Veiseth, S.V.; Emberland, G.; et al. The ASH1-RELATED3 SET-domain protein controls cell division competence of the meristem and the quiescent center of the Arabidopsis primary root. Plant Physiol. 2014, 166, 632–643. [Google Scholar] [CrossRef]
- Xu, Y.; Guo, C.; Zhou, B.; Li, C.; Wang, H.; Zheng, B.; Ding, H.; Zhu, Z.; Peragine, A.; Cui, Y.; et al. Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant. Physiol. 2016, 172, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Xu, T.; He, Y. A histone H3 lysine-27 methyltransferase complex represses lateral root formation in Arabidopsis thaliana. Mol. Plant 2014, 7, 977–988. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Cuttriss, A.J.; Cossetto, S.B.; Pye, W.; Crisp, P.; Whelan, J.; Finnegan, E.J.; Turnbull, C.; Pogson, B.J. Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 2009, 21, 39–53. [Google Scholar] [CrossRef]
- Dong, G.; Ma, D.-P.; Li, J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 2008, 373, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Smith, M.R.; Poethig, R.S. Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 2015, 28, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Deng, S.; Xu, H.; Mao, H.; Liu, J.; Niu, Q.; Wang, H.; Chua, N. A noncoding RNA transcribed from the AGAMOUS (AG) second intron binds to CURLY LEAF and represses AG expression in leaves. New Phytol. 2018, 219, 1480–1491. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, X.; Zhu, J.-K.; Dong, J. Demethylation of ERECTA receptor genes by IBM1 histone demethylase affects stomatal development. Development 2016, 143, 4452–4461. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Cui, X.; Zhang, S.; Liu, C.; Cao, X. JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 2010, 20, 387–390. [Google Scholar] [CrossRef]
- Wu, M.-F.; Sang, Y.; Bezhani, S.; Yamaguchi, N.; Han, S.-K.; Li, Z.; Su, Y.; Slewinski, T.L.; Wagner, D. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. USA 2012, 109, 3576–3581. [Google Scholar] [CrossRef]
- Xing, L.; Liu, Y.; Xu, S.; Xiao, J.; Wang, B.; Deng, H.; Lu, Z.; Xu, Y.; Chong, K. Arabidopsis O-GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. EMBO J. 2018, 37, e98115. [Google Scholar] [CrossRef]
- Berr, A.; Xu, L.; Gao, J.; Cognat, V.; Steinmetz, A.; Dong, A.; Shen, W.-H. SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009, 151, 1476–1485. [Google Scholar] [CrossRef]
- Chowrasia, S.; Panda, A.K.; Rawal, H.C.; Kaur, H.; Mondal, T.K. Identification of jumonjiC domain containing gene family among the Oryza species and their expression analysis in FL478, a salt tolerant rice genotype. Plant. Physiol. Biochem. 2018, 130, 43–53. [Google Scholar] [CrossRef]
- Su, S.; Ji, M.; Chen, J.; Zhang, M.; Xu, X.; Cheng, C.; Punnuri, S.; Raju, S.K.K. Genome-wide identfication and expression analysis of protein arginine methyltransferase and JmjC domain-containing family in apple. Front. Plant Sci. 2024, 15, 1381753. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, X.; Qiao, K.; Fan, S.; Ma, Q. Genome-wide analysis of JMJ-C histone demethylase family involved in salt-tolerance in Gossypium hirsutum L. Plant Physiol. Biochem. 2020, 158, 420–433. [Google Scholar]
- Qian, Y.; Chen, C.; Jiang, L.; Zhang, J.; Ren, Q. Genome-wide identification, classification and expression analysis of the JmjC domain-containing histone demethylase gene family in maize. BMC Genom. 2019, 20, 256. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, G.; Liu, X.; Ding, X.; Zhang, D.; Wang, X.; Zhou, Y.; Yan, H.; Li, T.; Wu, K.; et al. Histone demethylase SlJMJ6 promotes fruit ripening by removing H3K27 methylation of ripening-related genes in tomato. New Phytol. 2020, 227, 1138–1156. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, H.; Liu, Y.; Wang, X.; Xu, Q.; Deng, X. Genome-wide identification of sweet orange (Citrus sinensis) histone modification gene families and their expression analysis during the fruit development and fruit-blue mold infection process. Front. Plant Sci. 2015, 6, 607. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Yan, W.; Ma, M.; Hasi, A.; Che, G. Genome-wide identification and expression analysis of the JMJ-C gene family in melon (Cucumis melo L.) reveals their potential role in fruit development. BMC Genom. 2023, 24, 771. [Google Scholar] [CrossRef]
- Plomion, C.; Leprovost, G.; Stokes, A. Wood formation in trees. Plant Physiol. 2001, 127, 1513–1523. [Google Scholar] [CrossRef]
- Du, J.; Wang, Y.; Chen, W.; Xu, M.; Zhou, R.; Shou, H.; Chen, J. High-resolution anatomical and spatial transcriptome analyses reveal two types of meristematic cell pools within the secondary vascular tissue of poplar stem. Mol. Plant 2023, 16, 809–828. [Google Scholar] [CrossRef]
- Luo, L.; Li, L. Molecular understanding of wood formation in trees. For. Res. 2022, 2, 5. [Google Scholar] [CrossRef]
- Li, W.; Lin, Y.-C.J.; Chen, Y.-L.; Zhou, C.; Li, S.; De Ridder, N.; Oliveira, D.M.; Zhang, L.; Zhang, B.; Wang, J.P.; et al. Woody plant cell walls: Fundamentals and utilization. Mol. Plant 2023, 17, 112–140. [Google Scholar] [CrossRef]
- Demura, T.; Fukuda, H. Transcriptional regulation in wood formation. Trends Plant Sci. 2007, 12, 64–70. [Google Scholar] [CrossRef]
- Chen, H.; Wang, J.P.; Liu, H.; Li, H.; Lin, Y.-C.J.; Shi, R.; Yang, C.; Gao, J.; Zhou, C.; Li, Q.; et al. Hierarchical transcription factor and chromatin binding network for wood formation in black cottonwood (Populus trichocarpa). Plant Cell 2019, 31, 602–626. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Wei, H. Deciphering the intricate hierarchical gene regulatory network: Unraveling multi-level regulation and modifications driving secondary cell wall formation. Hortic. Res. 2023, 11, uhad281. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Su, L.; Zhang, W.; Sun, Y.; Li, D.; Li, S.; Lin, Y.J.; Zhou, C.; Li, W. Epigenetic regulation of lignin biosynthesis in wood formation. New Phytol. 2024, 245, 1589–1607. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhai, R.; Lin, J.; Wang, Z.; Meng, D.; Li, M.; Mao, Y.; Gao, B.; Ma, H.; Zhang, B.; et al. Cell-type-specific PtrWOX4a and PtrVCS2 form a regulatory nexus with a histone modification system for stem cambium development in Populus trichocarpa. Nat. Plants 2023, 9, 96–111. [Google Scholar] [CrossRef]
- Ma, S.; Zhang, Z.; Long, Y.; Huo, W.; Zhang, Y.; Yang, X.; Zhang, J.; Li, X.; Du, Q.; Liu, W.; et al. Evolutionary history and functional diversification of the JmjC domain-containing histone demethylase gene family in plants. Plants 2022, 11, 1041. [Google Scholar] [CrossRef]
- Zeng, D.; Dai, L.-J.; Li, X.; Li, W.; Qu, G.-Z.; Li, S. Genome-wide identification of the ERF transcription factor family for structure analysis, expression pattern, and response to drought stress in Populus alba × Populus glandulosa. Int. J. Mol. Sci. 2023, 24, 3697. [Google Scholar] [CrossRef]
- Li, H.; Dai, X.; Huang, X.; Xu, M.; Wang, Q.; Yan, X.; Sederoff, R.R.; Li, Q. Single-cell RNA sequencing reveals a high-resolution cell atlas of xylem in Populus. J. Integr. Plant Biol. 2021, 63, 1906–1921. [Google Scholar] [CrossRef]
- Li, H.; Chen, G.; Pang, H.; Wang, Q.; Dai, X. Investigation into different wood formation mechanisms between angiosperm and gymnosperm tree species at the transcriptional and post-transcriptional level. Front. Plant Sci. 2021, 12, 698602. [Google Scholar] [CrossRef]
- Nunez-Vazquez, R.; Desvoyes, B.; Gutierrez, C. Histone variants and modifications during abiotic stress response. Front. Plant Sci. 2022, 13, 984702. [Google Scholar] [CrossRef]
- Hu, H.; Du, J. Structure and mechanism of histone methylation dynamics in Arabidopsis. Curr. Opin. Plant Biol. 2022, 67, 102211. [Google Scholar] [CrossRef]
- Shi, W.; Li, Z.; Dong, K.; Ge, B.; Lu, C.; Chen, Y. Genome-wide identification, classification and expression analysis of the JmjC domain-containing histone demethylase gene family in Moso bamboo (Phyllostachys edulis). S. Afr. J. Bot. 2023, 157, 335–345. [Google Scholar] [CrossRef]
- Dunwell, J.M.; Purvis, A.; Khuri, S. Cupins: The most functionally diverse protein superfamily? Phytochemistry 2004, 65, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Yan, X.; Ma, L.; Pang, H.; Wang, P.; Liu, L.; Cheng, Y.; Cheng, J.; Guo, Y.; Li, Q. METHIONINE SYNTHASE1 Is Involved in Chromatin Silencing by Maintaining DNA and Histone Methylation. Plant Physiol. 2019, 181, 249–261. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, Z.; Liu, R.; Yan, X. Genome-Wide Characterization and Expression Profile of the Jumonji-C Family Genes in Populus alba × Populus glandulosa Reveal Their Potential Roles in Wood Formation. Int. J. Mol. Sci. 2025, 26, 5666. https://doi.org/10.3390/ijms26125666
Geng Z, Liu R, Yan X. Genome-Wide Characterization and Expression Profile of the Jumonji-C Family Genes in Populus alba × Populus glandulosa Reveal Their Potential Roles in Wood Formation. International Journal of Molecular Sciences. 2025; 26(12):5666. https://doi.org/10.3390/ijms26125666
Chicago/Turabian StyleGeng, Zhenghao, Rui Liu, and Xiaojing Yan. 2025. "Genome-Wide Characterization and Expression Profile of the Jumonji-C Family Genes in Populus alba × Populus glandulosa Reveal Their Potential Roles in Wood Formation" International Journal of Molecular Sciences 26, no. 12: 5666. https://doi.org/10.3390/ijms26125666
APA StyleGeng, Z., Liu, R., & Yan, X. (2025). Genome-Wide Characterization and Expression Profile of the Jumonji-C Family Genes in Populus alba × Populus glandulosa Reveal Their Potential Roles in Wood Formation. International Journal of Molecular Sciences, 26(12), 5666. https://doi.org/10.3390/ijms26125666




