Genome-Guided Metabolomic Profiling of Peptaibol-Producing Trichoderma
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Activity of Strain SK1-7 Is Associated with the Production of 11-Residue Peptaibols
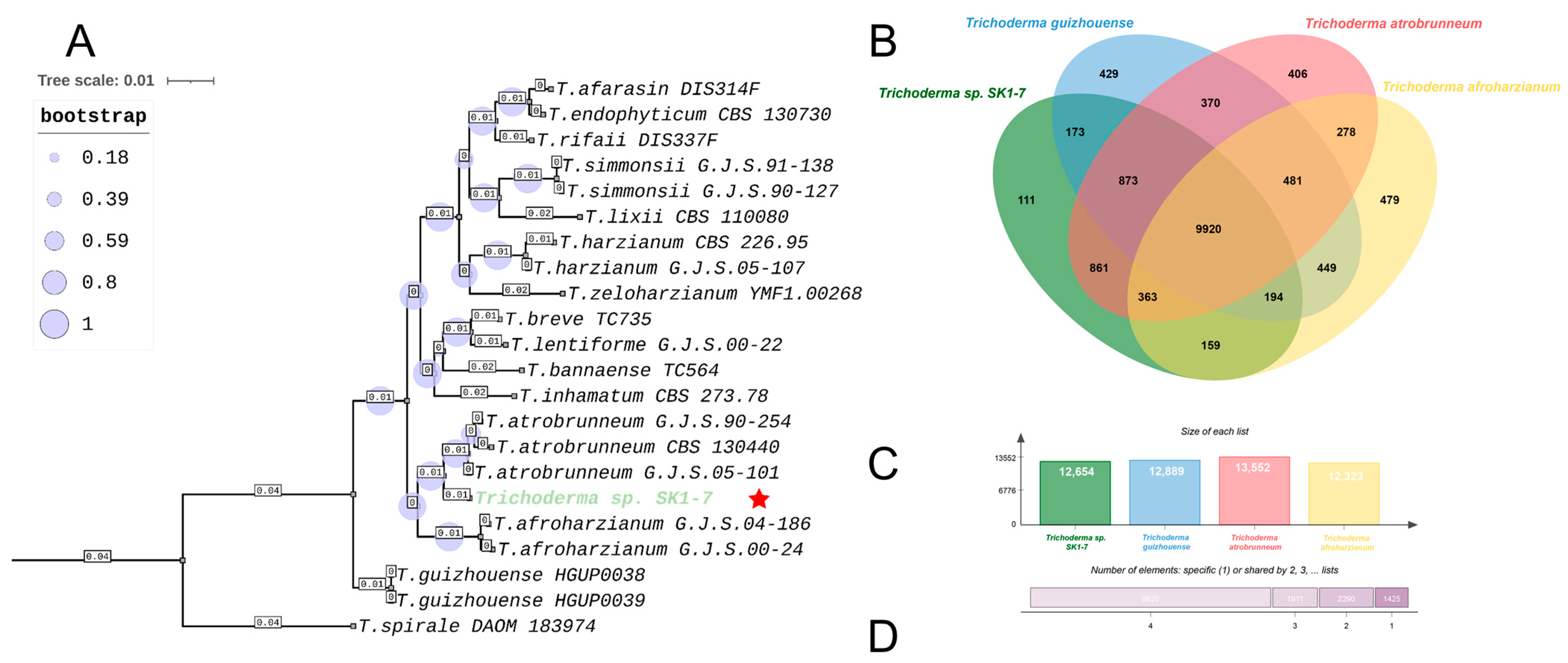
2.2. Complete Genome Sequencing Reveals Distinct Taxonomical Position of Trichoderma sp. SK1-7
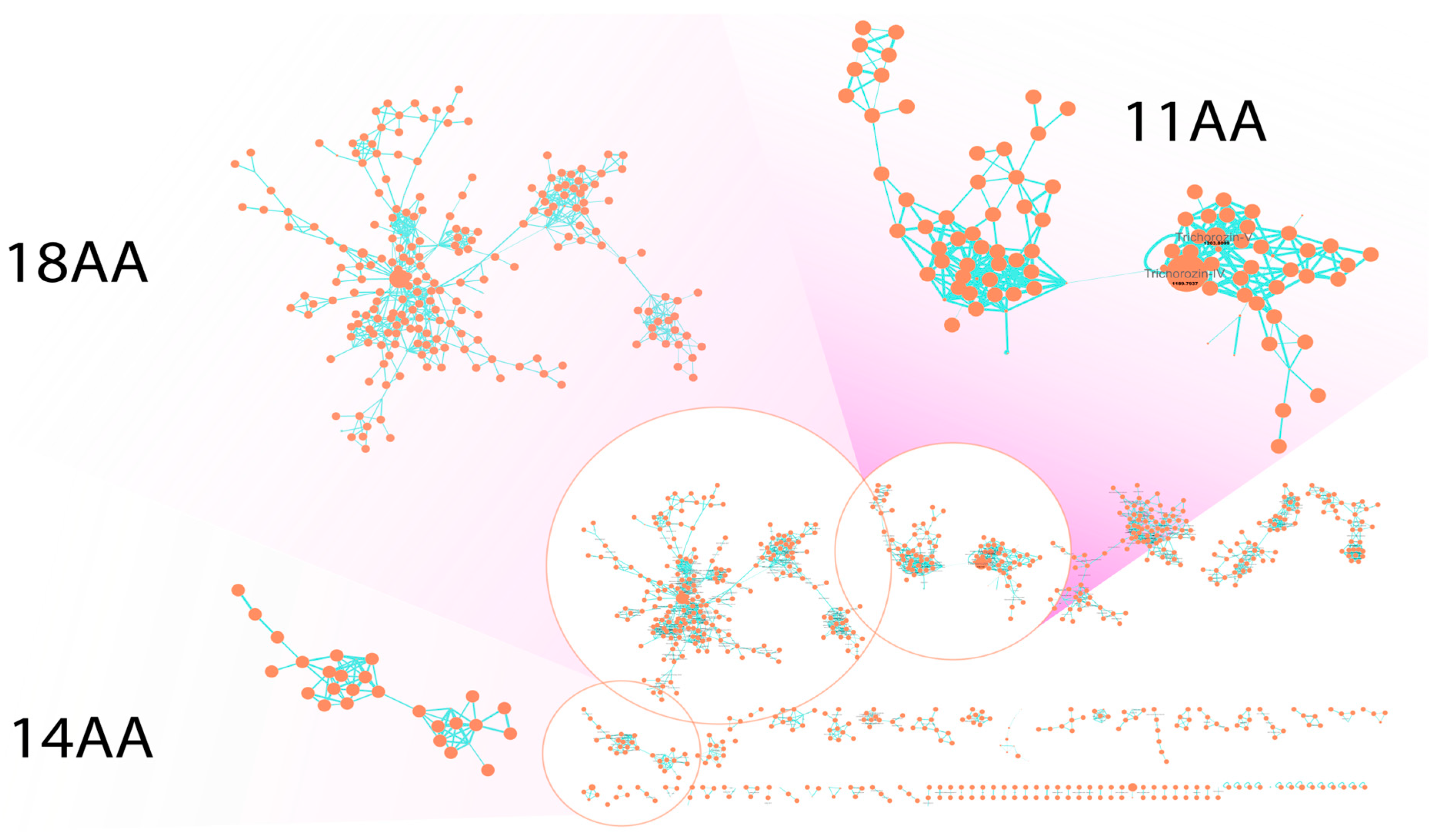
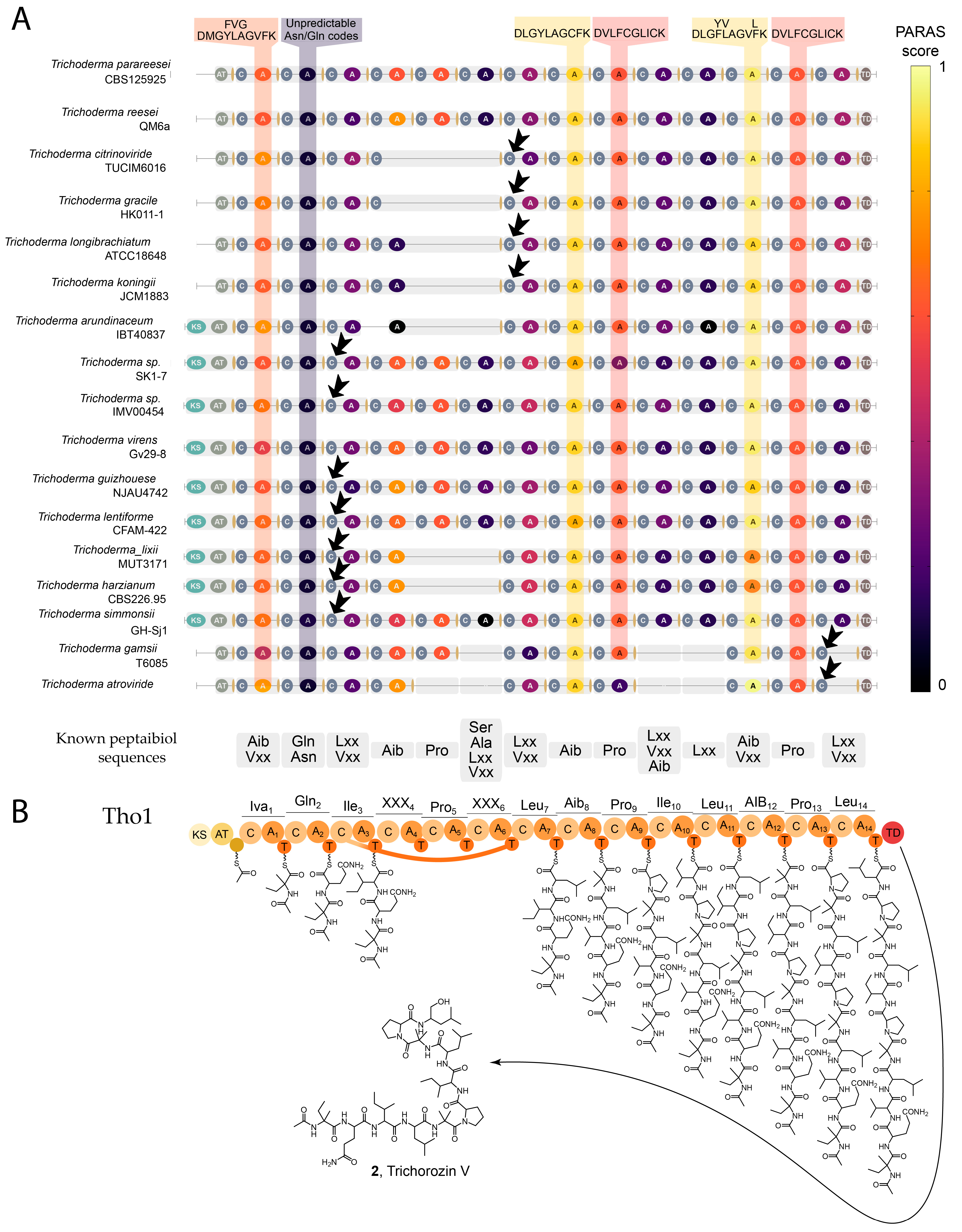
2.3. Bioinformatic Analysis Provides Insights into the Biosynthetic Origin of SF4-Type Peptaibols
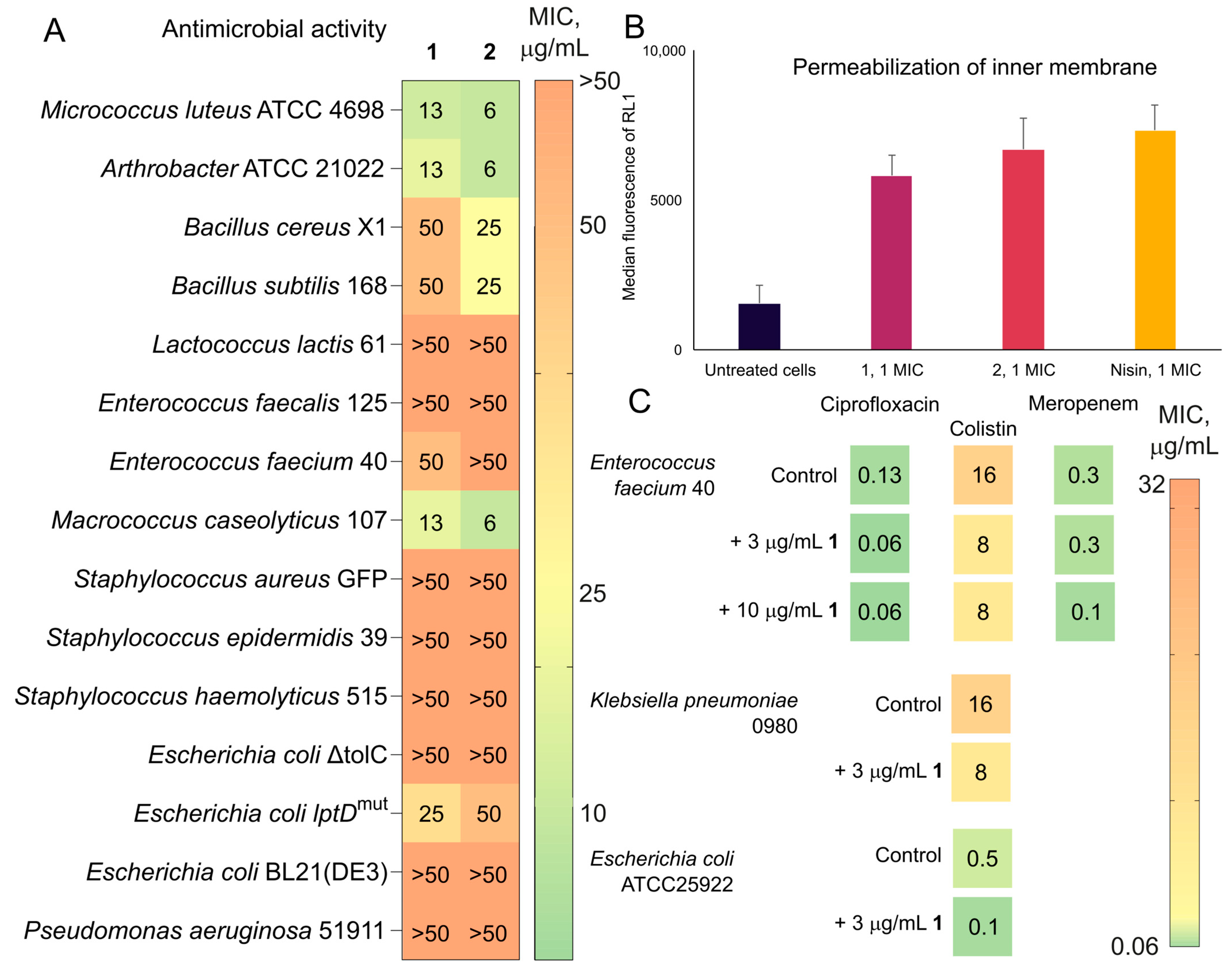
2.4. Trichorozins Exhibit Moderate Antibacterial and Cytotoxic Properties, Induce Membrane Permeabilization and Potentiate Membrane-Active Compounds
3. Discussion
4. Materials and Methods
4.1. Isolation and Fermentation of the Producing Strain Trichoderma sp. SK1-7
4.2. Bioactivity-Guided Fractionation, Isolation of Peptaibols
4.3. HRMS Analysis
4.4. Structure Elucidation
4.5. DNA Sequencing
4.6. Bioinformatic Analysis
4.7. Feature-Based Molecular Network (FBMN)
4.8. Amino Acid Supplementation Experiment
4.9. Bacterial Strains for Antimicrobial Activity Testing
4.10. Antibacterial Activity Assessment
4.11. Cell Viability Assay
4.12. Membrane Premeability Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hou, X.; Sun, R.; Feng, Y.; Zhang, R.; Zhu, T.; Che, Q.; Zhang, G.; Li, D. Peptaibols: Diversity, Bioactivity, and Biosynthesis. Eng. Microbiol. 2022, 2, 100026. [Google Scholar] [CrossRef]
- Pereira-Dias, L.; Oliveira-Pinto, P.R.; Fernandes, J.O.; Regalado, L.; Mendes, R.; Teixeira, C.; Mariz-Ponte, N.; Gomes, P.; Santos, C. Peptaibiotics: Harnessing the Potential of Microbial Secondary Metabolites for Mitigation of Plant Pathogens. Biotechnol. Adv. 2023, 68, 108223. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, D.; Wang, L.; Fan, A.; Wu, M.; Xu, N.; Zhu, K.; Lin, W. Marine-Derived New Peptaibols with Antibacterial Activities by Targeting Bacterial Membrane Phospholipids. Acta Pharm. Sin. B 2025, 15, 2764–2777. [Google Scholar] [CrossRef]
- Jiao, Y.; Ling, J.; Khan, R.A.A.; Luo, N.; Li, Z.; Li, Z.; Yang, Y.; Zhao, J.; Mao, Z.; Bills, G.F.; et al. Genome Mining Reveals Biosynthesis of the Antifungal Lipopeptaibols, Texenomycins, through a Hybrid PKS-NRPS System, in the Fungus Marian. Elegans. J. Agric. Food Chem. 2025, 73, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Kim, E.-S.; Cho, Y.B.; Kim, S.Y.; Lee, M.K.; Hwang, B.Y.; Lee, J.W. Cytotoxic Peptaibols from Trichoderma Guizhouense, a Fungus Isolated from an Urban Soil Sample. J. Nat. Prod. 2024, 87, 1994–2003. [Google Scholar] [CrossRef]
- Luo, N.; Jiao, Y.; Ling, J.; Li, Z.; Zhang, W.; Zhao, J.; Li, Y.; Mao, Z.; Li, H.; Xie, B. Synergistic Effect of Two Peptaibols from Biocontrol Fungus Trichoderma Longibrachiatum Strain 40418 on CO-Induced Plant Resistance. J. Agric. Food Chem. 2024, 72, 20763–20774. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Yang, J.-F.; Huang, Z.-H.; Ma, X.; Yan, Y.; Qi, S.-H. New Antibacterial Peptaibiotics against Plant and Fish Pathogens from the Deep-Sea-Derived Fungus Simplicillium Obclavatum EIODSF 020. J. Agric. Food Chem. 2024, 72, 6402–6413. [Google Scholar] [CrossRef]
- Song, K.; Ai, Y.; Zhou, J.; Dun, B.; Yue, Q.; Zhang, L.; Xu, Y.; Wang, C. Isolation, Characterization, and Bioherbicidal Potential of the 16-Residue Peptaibols from Emericellopsis Sp. XJ1056. J. Agric. Food Chem. 2024, 72, 6315–6326. [Google Scholar] [CrossRef]
- Collins, J.E.; Lee, J.W.; Rocamora, F.; Saggu, G.S.; Wendt, K.L.; Pasaje, C.F.A.; Smick, S.; Santos, N.M.; Paes, R.; Jiang, T.; et al. Antiplasmodial Peptaibols Act through Membrane Directed Mechanisms. Cell Chem. Biol. 2024, 31, 312–325.e9. [Google Scholar] [CrossRef]
- Lee, J.W.; Collins, J.E.; Wendt, K.L.; Chakrabarti, D.; Cichewicz, R.H. Leveraging Peptaibol Biosynthetic Promiscuity for Next-Generation Antiplasmodial Therapeutics. J. Nat. Prod. 2021, 84, 503–517. [Google Scholar] [CrossRef]
- Wada, S.; Tanaka, R. A Novel 11-Residual Peptaibol-Derived Carrier Peptide for in Vitro Oligodeoxynucleotide Delivery into Cell. Bioorg. Med. Chem. Lett. 2004, 14, 2563–2566. [Google Scholar] [CrossRef] [PubMed]
- Wada, S.; Tanaka, R. A Functionalized 20-Residue Peptaibol Derivative for Nucleic Acid Delivery. Chem. Biodivers. 2007, 4, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Wada, S.; Hitora, Y.; Yokoe, S.; Nakagawa, O.; Urata, H. Cellular Uptake of Covalent Conjugates of Oligonucleotide with Membrane-Modifying Peptide, Peptaibol. Bioorg. Med. Chem. 2012, 20, 3219–3222. [Google Scholar] [CrossRef]
- Wada, S.; Urase, T.; Hasegawa, Y.; Ban, K.; Sudani, A.; Kawai, Y.; Hayashi, J.; Urata, H. Aib-Containing Peptide Analogs: Cellular Uptake and Utilization in Oligonucleotide Delivery. Bioorg. Med. Chem. 2014, 22, 6776–6780. [Google Scholar] [CrossRef]
- Du, L.; Risinger, A.L.; Mitchell, C.A.; You, J.; Stamps, B.W.; Pan, N.; King, J.B.; Bopassa, J.C.; Judge, S.I.V.; Yang, Z.; et al. Unique Amalgamation of Primary and Secondary Structural Elements Transform Peptaibols into Potent Bioactive Cell-Penetrating Peptides. Proc. Natl. Acad. Sci. USA 2017, 114, E8957–E8966. [Google Scholar] [CrossRef]
- Chugh, J.K.; Wallace, B.A. Peptaibols: Models for Ion Channels. Biochem. Soc. Trans. 2001, 29, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm Portal: Gearing up for 1000 Fungal Genomes. Nucl. Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Wu, G.; Dentinger, B.T.M.; Nielson, J.R.; Peterson, R.T.; Winter, J.M. Emerimicins V–X, 15-Residue Peptaibols Discovered from an Acremonium Sp. through Integrated Genomic and Chemical Approaches. J. Nat. Prod. 2021, 84, 1113–1126. [Google Scholar] [CrossRef]
- Nakatani, T.; Koga, A.; Goto, S.; Inoue, M.; Shigedomi, K.; Seki, K.; Araki, K.; Taira, J.; Kodama, H.; Osada, S. Importance of Isoleucine Residue in Ion Channel Formation Ability of 11-Residue Peptaibols. Bioorg. Med. Chem. 2024, 110, 117839. [Google Scholar] [CrossRef]
- Dalla Torre, C.; Sannio, F.; Battistella, M.; Docquier, J.-D.; De Zotti, M. Peptaibol Analogs Show Potent Antibacterial Activity against Multidrug Resistant Opportunistic Pathogens. Int. J. Mol. Sci. 2023, 24, 7997. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Wiest, A.; Ruiz, N.; Keightley, A.; Moran-Diez, M.E.; McCluskey, K.; Pouchus, Y.F.; Kenerley, C.M. Two Classes of New Peptaibols Are Synthesized by a Single Non-Ribosomal Peptide Synthetase of Trichoderma Virens. J. Biol. Chem. 2011, 286, 4544–4554. [Google Scholar] [CrossRef] [PubMed]
- Berg, A.; Grigoriev, P.A.; Degenkolb, T.; Neuhof, T.; Härtl, A.; Schlegel, B.; Gräfe, U. Isolation, Structure Elucidation and Biological Activities of Trichofumins A, B, C and D, New 11 and 13mer Peptaibols from Trichoderma Sp. HKI 0276. J. Pept. Sci. 2003, 9, 810–816. [Google Scholar] [CrossRef]
- Degenkolb, T.; Karimi Aghcheh, R.; Dieckmann, R.; Neuhof, T.; Baker, S.E.; Druzhinina, I.S.; Kubicek, C.P.; Brückner, H.; von Döhren, H. The Production of Multiple Small Peptaibol Families by Single 14-Module Peptide Synthetases in Trichoderma/Hypocrea. Chem. Biodivers. 2012, 9, 499–535. [Google Scholar] [CrossRef]
- Neuhof, T.; Dieckmann, R.; Druzhinina, I.S.; Kubicek, C.P.; Von Döhren, H. Intact-Cell MALDI-TOF Mass Spectrometry Analysis of Peptaibol Formation by the Genus Trichoderma/Hypocrea: Can Molecular Phylogeny of Species Predict Peptaibol Structures? Microbiology 2007, 153, 3417–3437. [Google Scholar] [CrossRef] [PubMed]
- Degenkolb, T.; Fog Nielsen, K.; Dieckmann, R.; Branco-Rocha, F.; Chaverri, P.; Samuels, G.J.; Thrane, U.; von Döhren, H.; Vilcinskas, A.; Brückner, H. Peptaibol, Secondary-Metabolite, and Hydrophobin Pattern of Commercial Biocontrol Agents Formulated with Species of the Trichoderma Harzianum Complex. Chem. Biodivers. 2015, 12, 662–684. [Google Scholar] [CrossRef] [PubMed]
- Iida, A.; Sanekata, M.; Fujita, T.; Tanaka, H.; Enoki, A.; Fuse, G.; Kanai, M.; Rudewicz, P.J.; Tachikawa, E. Fungal Metabolites. XVI. Structures of New Peptaibols, Trichokindins I-VII, from the Fungus Trichoderma Harzianum. Chem. Pharm. Bull. 1994, 42, 1070–1075. [Google Scholar] [CrossRef]
- Tehan, R.M.; Blount, R.R.; Goold, R.L.; Mattos, D.R.; Spatafora, N.R.; Tabima, J.F.; Gazis, R.; Wang, C.; Ishmael, J.E.; Spatafora, J.W.; et al. Tolypocladamide H and the Proposed Tolypocladamide NRPS in Tolypocladium Species. J. Nat. Prod. 2022, 85, 1363–1373. [Google Scholar] [CrossRef]
- Fan, J.; Ren, J.; He, R.; Wei, P.-L.; Li, Y.; Li, W.; Chen, D.; Druzhinina, I.S.; Li, Z.; Yin, W.-B. Biosynthetic Diversification of Peptaibol Mediates Fungus-Mycohost Interactions. bioRxiv 2022. [Google Scholar] [CrossRef]
- Castro, G.S.; Sousa, T.F.; Da Silva, G.F.; Pedroso, R.C.N.; Menezes, K.S.; Soares, M.A.; Dias, G.M.; Santos, A.O.; Yamagishi, M.E.B.; Faria, J.V.; et al. Characterization of Peptaibols Produced by a Marine Strain of the Fungus Trichoderma Endophyticum via Mass Spectrometry, Genome Mining and Phylogeny-Based Prediction. Metabolites 2023, 13, 221. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & Clustermap.Js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Terlouw, B.R.; Huang, C.; Meijer, D.; Cediel-Becerra, J.D.D.; Rothe, M.L.; Jenner, M.; Zhou, S.; Zhang, Y.; Fage, C.D.; Tsunematsu, Y.; et al. PARAS: High-Accuracy Machine-Learning of Substrate Specificities in Nonribosomal Peptide Synthetases. bioRxiv 2025. [Google Scholar] [CrossRef]
- Wenzel, S.C.; Kunze, B.; Höfle, G.; Silakowski, B.; Scharfe, M.; Blöcker, H.; Müller, R. Structure and Biosynthesis of Myxochromides S1–3 in Stigmatella Aurantiaca : Evidence for an Iterative Bacterial Type I Polyketide Synthase and for Module Skipping in Nonribosomal Peptide Biosynthesis. ChemBioChem 2005, 6, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Bihlmaier, C.; Welle, E.; Hofmann, C.; Welzel, K.; Vente, A.; Breitling, E.; Müller, M.; Glaser, S.; Bechthold, A. Biosynthetic Gene Cluster for the Polyenoyltetramic Acid α-Lipomycin. Antimicrob. Agents Chemother. 2006, 50, 2113–2121. [Google Scholar] [CrossRef]
- Baars, O.; Zhang, X.; Morel, F.M.M.; Seyedsayamdost, M.R. The Siderophore Metabolome of Azotobacter Vinelandii. Appl. Environ. Microbiol. 2016, 82, 27–39. [Google Scholar] [CrossRef]
- Masschelein, J.; Mattheus, W.; Gao, L.-J.; Moons, P.; Van Houdt, R.; Uytterhoeven, B.; Lamberigts, C.; Lescrinier, E.; Rozenski, J.; Herdewijn, P.; et al. A PKS/NRPS/FAS Hybrid Gene Cluster from Serratia Plymuthica RVH1 Encoding the Biosynthesis of Three Broad Spectrum, Zeamine-Related Antibiotics. PLoS ONE 2013, 8, e54143. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.; Kiess, M.; Brückner, H. Structures of Peptaibol Antibiotics Hypomurocin A and B from the Ascomycetous Fungus Hypocrea Muroiana Hino et Katsumoto. Liebigs Ann. 1997, 1997, 767–772. [Google Scholar] [CrossRef]
- Raap, J.; Erkelens, K.; Ogrel, A.; Skladnev, D.A.; Brückner, H. Fungal Biosynthesis of Non-Ribosomal Peptide Antibiotics and α, α-Dialkylated Amino Acid Constituents. J. Pept. Sci. 2005, 11, 331–338. [Google Scholar] [CrossRef]
- Brückner, H.; Becker, D.; Gams, W.; Degenkolb, T. Aib and Iva in the Biosphere: Neither Rare nor Necessarily Extraterrestrial. Chem. Biodivers. 2009, 6, 38–56. [Google Scholar] [CrossRef]
- Bunno, R.; Awakawa, T.; Mori, T.; Abe, I. Aziridine Formation by a FeII /α-Ketoglutarate Dependent Oxygenase and 2-Aminoisobutyrate Biosynthesis in Fungi. Angew. Chem. Int. Ed. 2021, 60, 15827–15831. [Google Scholar] [CrossRef]
- Zhou, L.; Awakawa, T.; Ushimaru, R.; Kanaida, M.; Abe, I. Characterization of Aziridine-Forming α-Ketoglutarate-Dependent Oxygenase in L-Isovaline Biosynthesis. Org. Lett. 2024, 26, 724–727. [Google Scholar] [CrossRef]
- Cha, L.; Paris, J.C.; Zanella, B.; Spletzer, M.; Yao, A.; Guo, Y.; Chang, W. Mechanistic Studies of Aziridine Formation Catalyzed by Mononuclear Non-Heme Iron Enzymes. J. Am. Chem. Soc. 2023, 145, 6240–6246. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Analysis of Peptaibol Sequence Composition: Implications for in Vivo Synthesis and Channel Formation. Eur. Biophys. J. 2004, 33, 233–237. [Google Scholar] [CrossRef]
- Park, Y.S.; Kim, E.-S.; Deyrup, S.T.; Lee, J.W.; Shim, S.H. Cytotoxic Peptaibols from Trichoderma Strigosum. J. Nat. Prod. 2024, 87, 2081–2094. [Google Scholar] [CrossRef] [PubMed]
- Ancajas, C.M.F.; Oyedele, A.S.; Butt, C.M.; Walker, A.S. Advances, Opportunities, and Challenges in Methods for Interrogating the Structure Activity Relationships of Natural Products. Nat. Prod. Rep. 2024, 41, 1543–1578. [Google Scholar] [CrossRef]
- Yan, F.; Burgard, C.; Popoff, A.; Zaburannyi, N.; Zipf, G.; Maier, J.; Bernauer, H.S.; Wenzel, S.C.; Müller, R. Synthetic Biology Approaches and Combinatorial Biosynthesis towards Heterologous Lipopeptide Production. Chem. Sci. 2018, 9, 7510–7519. [Google Scholar] [CrossRef]
- Gao, L.; Guo, J.; Fan, Y.; Ma, Z.; Lu, Z.; Zhang, C.; Zhao, H.; Bie, X. Module and Individual Domain Deletions of NRPS to Produce Plipastatin Derivatives in Bacillus Subtilis. Microb. Cell Fact. 2018, 17, 84. [Google Scholar] [CrossRef]
- Rawa, M.S.A.; Nogawa, T.; Okano, A.; Futamura, Y.; Wahab, H.A.; Osada, H. Zealpeptaibolin, an 11-Mer Cytotoxic Peptaibol Group with 3 Aib-Pro Motifs Isolated from Trichoderma Sp. RK10-F026. J. Antibiot. 2021, 74, 485–495. [Google Scholar] [CrossRef]
- Mikkola, R.; Andersson, M.A.; Kredics, L.; Grigoriev, P.A.; Sundell, N.; Salkinoja-Salonen, M.S. 20-Residue and 11-residue Peptaibols from the Fungus T Richoderma Longibrachiatum Are Synergistic in Forming Na+/K+-permeable Channels and Adverse Action towards Mammalian Cells. FEBS J. 2012, 279, 4172–4190. [Google Scholar] [CrossRef]
- Fragiadaki, I.; Katogiritis, A.; Calogeropoulou, T.; Brückner, H.; Scoulica, E. Synergistic Combination of Alkylphosphocholines with Peptaibols in Targeting Leishmania Infantum in Vitro. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 194–202. [Google Scholar] [CrossRef]
- Setargie, A.; Wang, C.; Zhang, L.; Xu, Y. Chromatographic and Mass Spectroscopic Guided Discovery of Trichoderma Peptaibiotics and Their Bioactivity. Eng. Microbiol. 2024, 4, 100135. [Google Scholar] [CrossRef] [PubMed]
- Keller, R. The Computer Aided Resonance Assignment Tutorial; Cantina: Goldau, Switzerland, 2004; ISBN 978-3-85600-112-4. [Google Scholar]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; Wiley-Interscience: New York, NY, USA, 1986; ISBN 978-0-471-82893-8. [Google Scholar]
- Bhushan, R.; Brückner, H. Marfey’s Reagent for Chiral Amino Acid Analysis: A Review. Amino Acids 2004, 27, 231–247. [Google Scholar] [CrossRef]
- Pinaka, A.; Vougioukalakis, G.; Dimotikali, D.; Psyharis, V.; Papadopoulos, K. A Convenient One-Step Synthesis of Stable β-Amino Alcohol N-Boranes from α-Amino Acids. Synthesis 2012, 44, 1057–1062. [Google Scholar] [CrossRef]
- Kim, C.-K.; Krumpe, L.R.H.; Smith, E.; Henrich, C.J.; Brownell, I.; Wendt, K.L.; Cichewicz, R.H.; O’Keefe, B.R.; Gustafson, K.R. Roseabol A, a New Peptaibol from the Fungus Clonostachys Rosea. Molecules 2021, 26, 3594. [Google Scholar] [CrossRef]
- Vijayasarathy, S.; Prasad, P.; Fremlin, L.J.; Ratnayake, R.; Salim, A.A.; Khalil, Z.; Capon, R.J. C3 and 2D C3 Marfey’s Methods for Amino Acid Analysis in Natural Products. J. Nat. Prod. 2016, 79, 421–427. [Google Scholar] [CrossRef]
- Nanoporetech. GitHub—Nanoporetech/Dorado: Oxford Nanopore’s Basecaller. GitHub. Available online: https://Github.Com/Nanoporetech/Dorado (accessed on 21 March 2025).
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Keller, O.; Kollmar, M.; Stanke, M.; Waack, S. A Novel Hybrid Gene Prediction Method Employing Protein Multiple Sequence Alignments. Bioinformatics 2011, 27, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Slater, G.S.C.; Birney, E. Automated Generation of Heuristics for Biological Sequence Comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. 38—Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Carbone, I.; and Kohn, L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic Relationships among Ascomycetes: Evidence from an RNA Polymerse II Subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Dou, K.; Lu, Z.; Wu, Q.; Ni, M.; Yu, C.; Wang, M.; Li, Y.; Wang, X.; Xie, H.; Chen, J.; et al. MIST: A Multilocus Identification System for Trichoderma. Appl. Environ. Microbiol. 2020, 86, e01532-20. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A Web Server for Whole-Genome Comparison and Annotation of Orthologous Clusters across Multiple Species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evolut. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef]
- Oberg, N.; Zallot, R.; Gerlt, J.A. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) Web Resource for Genomic Enzymology Tools. J. Mol. Biol. 2023, 435, 168018. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Herrera-Estrella, A.; Seidl-Seiboth, V.; Martinez, D.A.; Druzhinina, I.S.; Thon, M.; Zeilinger, S.; Casas-Flores, S.; Horwitz, B.A.; Mukherjee, P.K.; et al. Comparative Genome Sequence Analysis Underscores Mycoparasitism as the Ancestral Life Style of Trichoderma. Genome Biol. 2011, 12, R40. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; McCormick, S.P.; Kim, H.-S.; Cardoza, R.E.; Stanley, A.M.; Lindo, L.; Kelly, A.; Brown, D.W.; Lee, T.; Vaughan, M.M.; et al. Evolution of Structural Diversity of Trichothecenes, a Family of Toxins Produced by Plant Pathogenic and Entomopathogenic Fungi. PLoS Pathog. 2018, 14, e1006946. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Chenthamara, K.; Zhang, J.; Atanasova, L.; Yang, D.; Miao, Y.; Rahimi, M.J.; Grujic, M.; Cai, F.; Pourmehdi, S.; et al. Massive Lateral Transfer of Genes Encoding Plant Cell Wall-Degrading Enzymes to the Mycoparasitic Fungus Trichoderma from Its Plant-Associated Hosts. PLoS Genet. 2018, 14, e1007322. [Google Scholar] [CrossRef]
- Maddau, L.; Cabras, A.; Franceschini, A.; Linaldeddu, B.T.; Crobu, S.; Roggio, T.; Pagnozzi, D. Occurrence and Characterization of Peptaibols from Trichoderma Citrinoviride, an Endophytic Fungus of Cork Oak, Using Electrospray Ionization Quadrupole Time-of-Flight Mass Spectrometry. Microbiology 2009, 155, 3371–3381. [Google Scholar] [CrossRef] [PubMed]
- Baroncelli, R.; Zapparata, A.; Piaggeschi, G.; Sarrocco, S.; Vannacci, G. Draft Whole-Genome Sequence of Trichoderma Gamsii T6085, a Promising Biocontrol Agent of Fusarium Head Blight on Wheat. Genome Announc. 2016, 4, e01747-15. [Google Scholar] [CrossRef] [PubMed]
- Marik, T.; Tyagi, C.; Racić, G.; Rakk, D.; Szekeres, A.; Vágvölgyi, C.; Kredics, L. New 19-Residue Peptaibols from Trichoderma Clade Viride. Microorganisms 2018, 6, 85. [Google Scholar] [CrossRef]
- Niu, X.; Thaochan, N.; Hu, Q. Diversity of Linear Non-Ribosomal Peptide in Biocontrol Fungi. J. Fungi 2020, 6, 61. [Google Scholar] [CrossRef]
- Peltola, J.; Ritieni, A.; Mikkola, R.; Grigoriev, P.A.; Pócsfalvi, G.; Andersson, M.A.; Salkinoja-Salonen, M.S. Biological Effects of Trichoderma Harzianum Peptaibols on Mammalian Cells. Appl. Environ. Microbiol. 2004, 70, 4996–5004. [Google Scholar] [CrossRef]
- Duval, D.; Rebuffat, S.; Goulard, C.; Prigent, Y.; Becchi, M.; Bodo, B. Isolation and Sequence Analysis of the Peptide Antibiotics Trichorzins PA from Trichoderma Harzianum. J. Chem. Soc. Perkin Trans. 1997, 1, 2147–2154. [Google Scholar] [CrossRef]
- Goulard, C.; Hlimi, S.; Rebuffat, S.; Bodo, B. Trichorzins HA and MA, Antibiotic Peptides from Trichoderma Harzianum. I. Fermentation, Isolation and Biological Properties. J. Antibiot. 1995, 48, 1248–1253. [Google Scholar] [CrossRef]
- Huang, Q.; Tezuka, Y.; Hatanaka, Y.; Kikuchi, T.; Nishi, A.; Tubaki, K. Studies on Metabolites of Mycoparasitic Fungi. IV. Minor Peptaibols of Trichoderma Koningii. Chem. Pharm. Bull. 1995, 43, 1663–1667. [Google Scholar] [CrossRef]
- Rahimi Tamandegani, P.; Marik, T.; Zafari, D.; Balázs, D.; Vágvölgyi, C.; Szekeres, A.; Kredics, L. Changes in Peptaibol Production of Trichoderma Species during In Vitro Antagonistic Interactions with Fungal Plant Pathogens. Biomolecules 2020, 10, 730. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, G.; Goulard, C.; Prigent, Y.; Bodo, B.; Wróblewski, H.; Rebuffat, S. Sequences and Antimycoplasmic Properties of Longibrachins LGB II and LGB III, Two Novel 20-Residue Peptaibols from Trichoderma l Ongibrachiatum. J. Nat. Prod. 2001, 64, 164–170. [Google Scholar] [CrossRef]
- Mohamed-Benkada, M.; François Pouchus, Y.; Vérité, P.; Pagniez, F.; Caroff, N.; Ruiz, N. Identification and Biological Activities of Long-Chain Peptaibols Produced by a Marine-Derived Strain of Trichoderma Longibrachiatum. Chem. Biodivers. 2016, 13, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Berka, R.M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S.E.; Chapman, J.; Chertkov, O.; Coutinho, P.M.; Cullen, D.; et al. Genome Sequencing and Analysis of the Biomass-Degrading Fungus Trichoderma Reesei (Syn. Hypocrea Jecorina). Nat. Biotechnol. 2008, 26, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-C.; Huang, C.-H.; Chen, C.-L.; Chuang, Y.-C.; Tung, S.-Y.; Wang, T.-F. Trichoderma Reesei Complete Genome Sequence, Repeat-Induced Point Mutation, and Partitioning of CAZyme Gene Clusters. Biotechnol. Biofuels 2017, 10, 170. [Google Scholar] [CrossRef]
- Pócsfalvi, G.; Ritieni, A.; Ferranti, P.; Randazzo, G.; Vékey, K.; Malorni, A. Microheterogeneity Characterization of a Paracelsin Mixture fromTrichoderma Reesei Using High-Energy Collision-Induced Dissociation Tandem Mass Spectrometry. Rapid Commun. Mass Spectrom. 1997, 11, 922–930. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinichich, A.A.; Krivonos, D.V.; Baranova, A.A.; Zhitlov, M.Y.; Belozerova, O.A.; Lushpa, V.A.; Vvedensky, A.V.; Serebryakova, M.V.; Kalganova, A.I.; Kudzhaev, A.M.; et al. Genome-Guided Metabolomic Profiling of Peptaibol-Producing Trichoderma. Int. J. Mol. Sci. 2025, 26, 5599. https://doi.org/10.3390/ijms26125599
Sinichich AA, Krivonos DV, Baranova AA, Zhitlov MY, Belozerova OA, Lushpa VA, Vvedensky AV, Serebryakova MV, Kalganova AI, Kudzhaev AM, et al. Genome-Guided Metabolomic Profiling of Peptaibol-Producing Trichoderma. International Journal of Molecular Sciences. 2025; 26(12):5599. https://doi.org/10.3390/ijms26125599
Chicago/Turabian StyleSinichich, Arseniy A., Danil V. Krivonos, Anna A. Baranova, Mikhail Y. Zhitlov, Olga A. Belozerova, Vladislav A. Lushpa, Andrey V. Vvedensky, Marina V. Serebryakova, Anastasia I. Kalganova, Arsen M. Kudzhaev, and et al. 2025. "Genome-Guided Metabolomic Profiling of Peptaibol-Producing Trichoderma" International Journal of Molecular Sciences 26, no. 12: 5599. https://doi.org/10.3390/ijms26125599
APA StyleSinichich, A. A., Krivonos, D. V., Baranova, A. A., Zhitlov, M. Y., Belozerova, O. A., Lushpa, V. A., Vvedensky, A. V., Serebryakova, M. V., Kalganova, A. I., Kudzhaev, A. M., Prokopenko, Y. A., Sinelnikova, S. S., Trusova, E. A., Kovalchuk, S. I., Ilina, E. N., Terekhov, S. S., & Alferova, V. A. (2025). Genome-Guided Metabolomic Profiling of Peptaibol-Producing Trichoderma. International Journal of Molecular Sciences, 26(12), 5599. https://doi.org/10.3390/ijms26125599




