Mitochondrial Genome Variations and Possible Adaptive Implications in Some Tephritid Flies (Diptera, Tephritidae)
Abstract
1. Introduction
2. Results and Discussion
2.1. Assembled Mitogenomes and Features
2.2. AT/GC Content and Skew Analysis
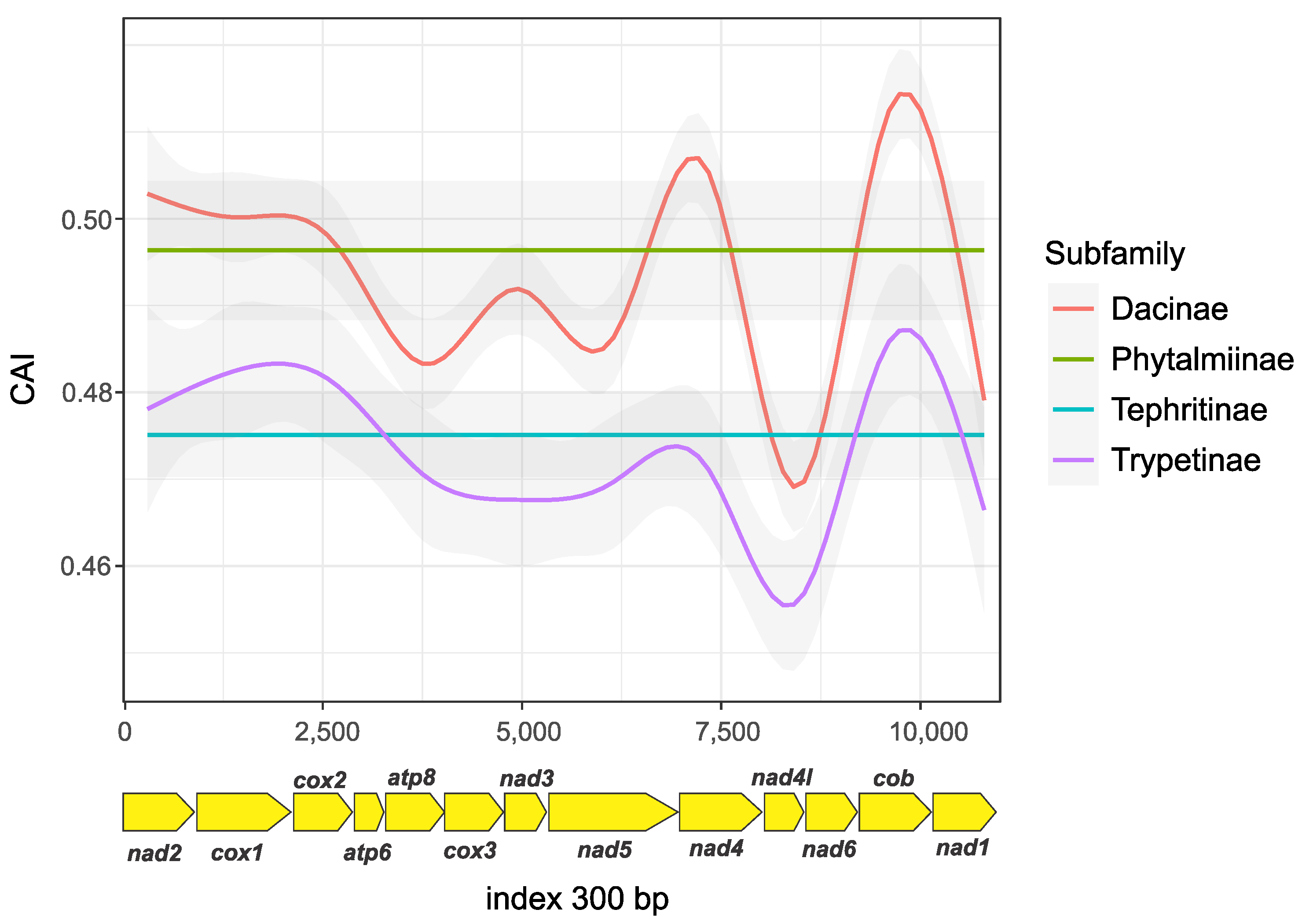
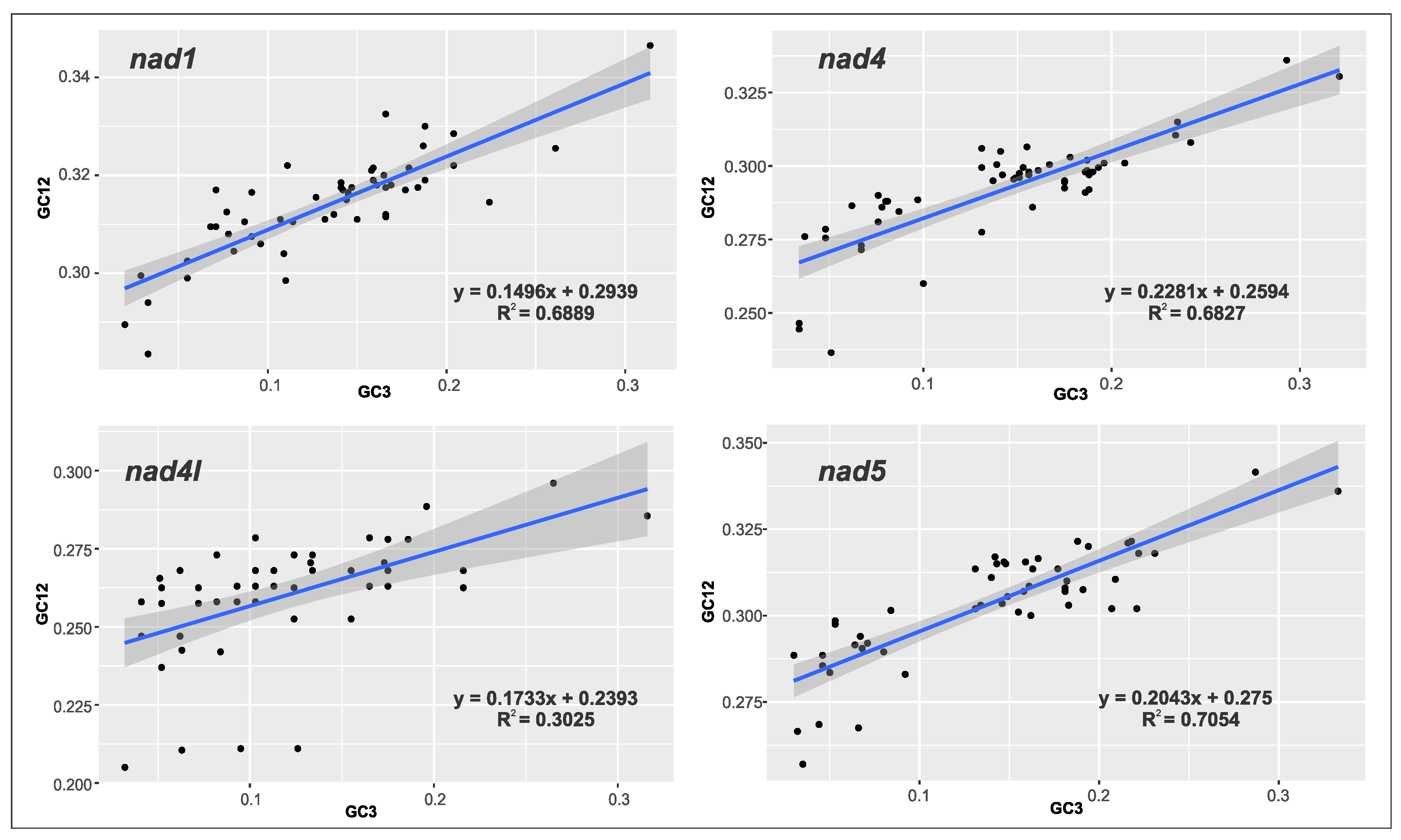
2.3. Relative Synonymous Codon Usage (RSCU) and Codon Adaptation Index (CAI) Analysis
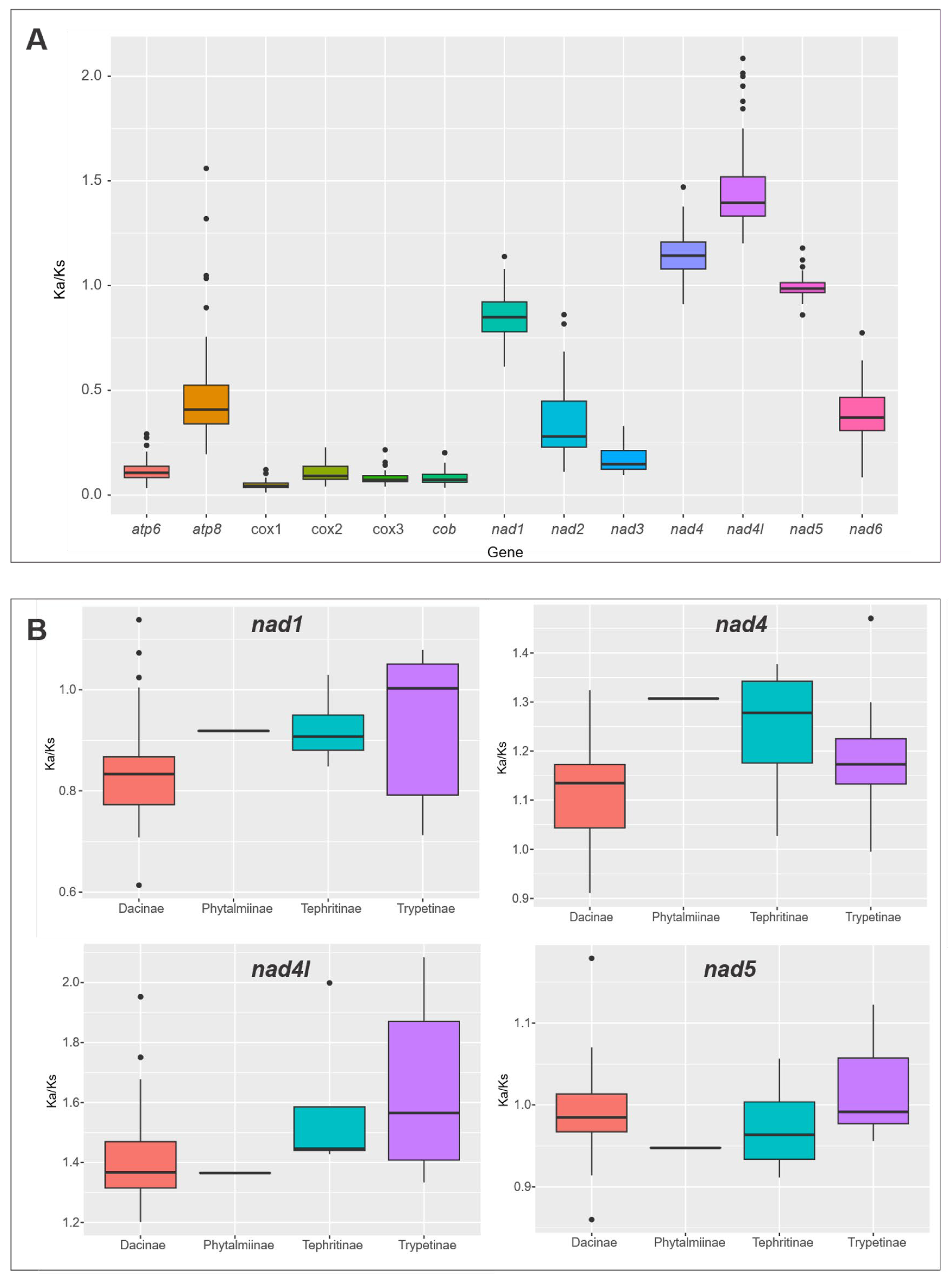
2.4. Ka/Ks Ratio and Neutrality Plot Analysis
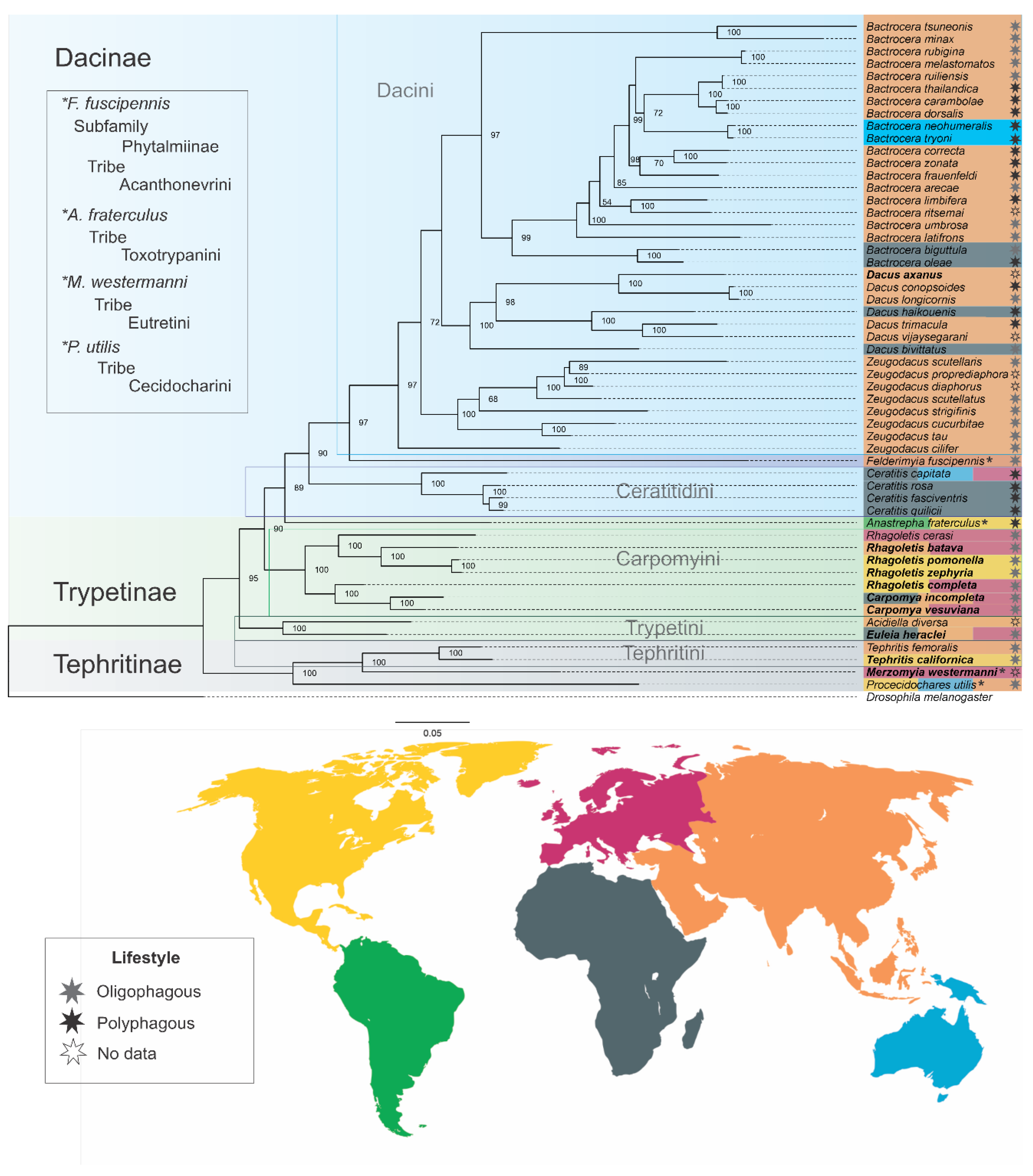
2.5. Phylogenetic Reconstruction of Tephritidae
3. Methods
3.1. Sampling of Genomic Data in Tephritidae
3.2. mtDNA Assembly and Extracted Read Assembly
3.3. mtDNA Sequence Manual Annotation
3.4. Sampling and Mitogenome Sequence Review
3.5. AT/GC Skew in Mitochondrial Genomes
3.6. Relative Synonymous Codon Usage (RSCU) and Codon Adaptation Index (CAI)
3.7. Ka/Ks Ratio and Neutrality Plot Analysis
3.8. Phylogenetic Inference, Lifestyle Adaptations, and Species Distribution in Tephritidae
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Norrbom, A. Tephritidae (fruit flies, moscas de frutas). Man. Cent. Am. Diptera 2010, 2, 909–954. [Google Scholar]
- Savaris, M.; Marinoni, L.; Norrbom, A.L. Family Tephritidae. Zootaxa 2016, 4122, 596. [Google Scholar] [CrossRef] [PubMed]
- White, I.M.; Elson-Harris, M.M. Fruit Flies of Economic Significance: Their Identification and Bionomics; Oxford University Press: New York, NY, USA, 1992. [Google Scholar]
- Aluja, M.; Norrbom, A. Fruit Flies (Tephritidae): Phylogeny and Evolution of Behavior; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]
- Headrick, D.H.; Goeden, R.D. The biology of nonfrugivorous tephritid fruit flies. Annu. Rev. Entomol. 1998, 43, 217–241. [Google Scholar] [CrossRef]
- Hancock, D.; Freidberg, A.; Friedman, A. Tephritidae. Chapter 71 (True fruit flies). Man. Afrotropical Diptera 2021, 3, 1669–1734. [Google Scholar]
- Aluja, M.; Mangan, R.L. Fruit fly (Diptera: Tephritidae) host status determination: Critical conceptual, methodological, and regulatory considerations. Annu. Rev. Entomol. 2008, 53, 473–502. [Google Scholar] [CrossRef]
- Korneyev, V.A. Phylogenetic relationships among higher groups of Tephritidae. In Fruit Flies (Tephritidae); CRC Press: Boca Raton, FL, USA, 1999; pp. 91–132. [Google Scholar]
- Han, H.-Y.; Ro, K.-E. Molecular phylogeny of the family Tephritidae (Insecta: Diptera): New insight from combined analysis of the mitochondrial 12S, 16S, and COII genes. Mol. Cells 2009, 27, 55–66. [Google Scholar] [CrossRef]
- Han, H.-Y.; McPheron, B.A. Molecular phylogenetic study of Tephritidae (Insecta: Diptera) using partial sequences of the mitochondrial 16S ribosomal DNA. Mol. Phylogenetics Evol. 1997, 7, 17–32. [Google Scholar] [CrossRef]
- Drew, R.A.; Yuval, B. The evolution of fruit fly feeding behavior. In Fruit Flies (Tephritidae); CRC Press: Boca Raton, FL, USA, 1999; pp. 749–768. [Google Scholar]
- Feder, J.L. The effects of parasitoids on sympatric host races of Rhagoletis pomonella (Diptera: Tephritidae). Ecology 1995, 76, 801–813. [Google Scholar] [CrossRef]
- Sivinski, J.; Aluja, M.; Dodson, G.N.; Freidberg, A.; Headrick, D.H.; Kaneshiro, K.Y.; Landolt, P.J. Topics in the Evolution of Sexual Behavior in the Tephritidae. In Fruit Flies (Tephritidae); CRC Press: Boca Raton, FL, USA, 1999; pp. 769–810. [Google Scholar]
- Sivinski, J.; Aluja, M.; Norrbom, A. Breeding habits and sex in families closely related to the Tephritidae: Opportunities for comparative studies of the evolution of fruit fly behavior. In Fruit Flies: Phylogeny the Evolution of Behavior; CRC Press: Boca Raton, FL, USA, 2000; pp. 23–37. [Google Scholar]
- Díaz-Fleischer, F.; Aluja, M. Behavior of tephritid flies: A historical perspective. In Fruit Flies (Tephritidae); CRC Press: Boca Raton, FL, USA, 1999; pp. 57–88. [Google Scholar]
- Eberhard, W.G. Sexual behavior and sexual selection in the Mediterranean fruit fly, Ceratitis capitata (Dacinae: Ceratitidini). In Fruit Flies (Tephritidae); CRC Press: Boca Raton, FL, USA, 1999; pp. 477–508. [Google Scholar]
- Shi, W.; Yang, T.-y.; Ye, H.; Cao, J. Impact of host plants on genetic variation in the Bactrocera tau (Diptera: Tephritidae) based on molecular markers. J. Entomol. Sci. 2017, 52, 411–426. [Google Scholar]
- Ghosh, A.; Sultana, N.; Hossain, M.F.; Khan, S.A.; Hossain, M.A.; Seheli, K. Molecular Identification of Ten Economically Important Fruit Flies (Diptera: Tephritidae) of Bangladesh by Using PCR-RFLP. Eur. J. Sci. Innov. Technol. 2022, 2, 41–48. [Google Scholar]
- Thanaphum, S.; Thaenkham, U. Relationships of forms within the Bactrocera tau (Walker) (Diptera: Tephritidae) taxon based on heat shock protein 70 cognate sequences. Ann. Entomol. Soc. Am. 2003, 96, 44–53. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, S.; Fekrat, L.; Jiang, F.; Khathutshelo, M.; Li, Z. The first two complete mitochondrial genome of Dacus bivittatus and Dacus ciliatus (Diptera: Tephritidae) by next-generation sequencing and implications for the higher phylogeny of Tephritidae. Int. J. Biol. Macromol. 2019, 140, 469–476. [Google Scholar] [CrossRef]
- Elson, J.L.; Lightowlers, R.N. Mitochondrial DNA clonality in the dock: Can surveillance swing the case? Trends Genet. 2006, 22, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- DeSalle, R.; Schierwater, B.; Hadrys, H. MtDNA: The small workhorse of evolutionary studies. Front. Biosci. 2017, 22, 873–887. [Google Scholar] [CrossRef]
- Dowling, D.K.; Abiega, K.C.; Arnqvist, G. Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution 2007, 61, 194–201. [Google Scholar] [CrossRef]
- James, J.E.; Piganeau, G.; Eyre-Walker, A. The rate of adaptive evolution in animal mitochondria. Mol. Ecol. 2016, 25, 67–78. [Google Scholar] [CrossRef] [PubMed]
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, R551–R560. [Google Scholar] [CrossRef]
- Herbeck, J.T.; Novembre, J. Codon usage patterns in cytochrome oxidase I across multiple insect orders. J. Mol. Evol. 2003, 56, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.-J.; Shi, M.; Chen, X.-X.; Sharkey, M.J.; van Achterberg, C.; Ye, G.-Y.; He, J.-H. New views on strand asymmetry in insect mitochondrial genomes. PLoS ONE 2010, 5, e12708. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; He, J.; Jia, X.; Qi, Q.; Liang, Z.; Zheng, H.; Ping, Y.; Liu, S.; Sun, J. Analysis of codon usage bias of mitochondrial genome in Bombyx mori and its relation to evolution. BMC Evol. Biol. 2014, 14, 262. [Google Scholar] [CrossRef]
- Athey, J.; Alexaki, A.; Osipova, E.; Rostovtsev, A.; Santana-Quintero, L.V.; Katneni, U.; Simonyan, V.; Kimchi-Sarfaty, C. A new and updated resource for codon usage tables. BMC Bioinform. 2017, 18, 391. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. A code within the genetic code: Codon usage regulates co-translational protein folding. Cell Commun. Signal. 2020, 18, 145. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.; Claassens, N.J.; Söll, D.; van der Oost, J. Codon bias as a means to fine-tune gene expression. Mol. Cell 2015, 59, 149–161. [Google Scholar] [CrossRef] [PubMed]
- da Costa, L.T.; Powell, C.; van Noort, S.; Costa, C.; Sinno, M.; Caleca, V.; Rhode, C.; Kennedy, R.J.; van Staden, M.; van Asch, B. The complete mitochondrial genome of Bactrocera biguttula (Bezzi) (Diptera: Tephritidae) and phylogenetic relationships with other Dacini. Int. J. Biol. Macromol. 2019, 126, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, J.S.; Naaz, N.; Prabhakar, C.S.; Rao, M.S.; Das, B. The mitochondrial genome of the peach fruit fly, Bactrocera zonata (Saunders) (Diptera: Tephritidae): Complete DNA sequence, genome organization, and phylogenetic analysis with other tephritids using next generation DNA sequencing. Gene 2015, 569, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zhang, Y.; Feng, S.; He, J.; Zhao, Z.; Bai, Z.; Liu, L.; Zhang, R.; Li, Z. The mitochondrial genome of the wolfberry fruit fly, Neoceratitis asiatica (Becker) (Diptera: Tephritidae) and the phylogeny of Neoceratitis Hendel genus. Sci. Rep. 2017, 7, 16612. [Google Scholar] [CrossRef]
- Yong, H.-S.; Chua, K.-O.; Song, S.-L.; Liew, Y.J.-M.; Eamsobhana, P.; Chan, K.-G. Complete mitochondrial genome of Dacus vijaysegarani and phylogenetic relationships with congeners and other tephritid fruit flies (Insecta: Diptera). Mol. Biol. reports 2021, 48, 6047–6056. [Google Scholar] [CrossRef]
- Congrains, C.; Zucchi, R.A.; de Brito, R.A. Phylogenomic approach reveals strong signatures of introgression in the rapid diversification of neotropical true fruit flies (Anastrepha: Tephritidae). Mol. Phylogenetics Evol. 2021, 162, 107200. [Google Scholar] [CrossRef] [PubMed]
- Rand, D.M. The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Syst. 2001, 32, 415–448. [Google Scholar] [CrossRef]
- Garvin, M.R.; Bielawski, J.P.; Sazanov, L.A.; Gharrett, A.J. Review and Meta-Analysis of Natural Selection in Mitochondrial Complex I in Metazoans; Wiley Online Library: Hoboken, NJ, USA, 2015; Volume 53, pp. 1–17. [Google Scholar]
- Slimen, H.B.; Awadi, A.; Makni, M. Ambient temperature and host specialization drive mitochondrial genome evolution in fruit flies of the genus Bactrocera (Diptera: Tephritidae). Evol. Ecol. Res. 2017, 18, 443–457. [Google Scholar]
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—A baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41, e129. [Google Scholar] [CrossRef]
- Smith, D.R. The past, present and future of mitochondrial genomics: Have we sequenced enough mtDNAs? Brief. Funct. Genom. 2016, 15, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Forni, G.; Puccio, G.; Bourguignon, T.; Evans, T.; Mantovani, B.; Rota-Stabelli, O.; Luchetti, A. Complete mitochondrial genomes from transcriptomes: Assessing pros and cons of data mining for assembling new mitogenomes. Sci. Rep. 2019, 9, 14806. [Google Scholar] [CrossRef] [PubMed]
- Sanitá Lima, M.; Smith, D.R. Pervasive transcription of mitochondrial, plastid, and nucleomorph genomes across diverse plastid-bearing species. Genome Biol. Evol. 2017, 9, 2650–2657. [Google Scholar] [CrossRef]
- Moreno-Carmona, M.; Cameron, S.L.; Quiroga, C.F.P. How are the mitochondrial genomes reorganized in Hexapoda? Differential evolution and the first report of convergences within Hexapoda. Gene 2021, 791, 145719. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, J.S.; Naaz, N.; Lemtur, M.; Das, B.; Singh, A.K.; Bhatt, B.P.; Prabhakar, C.S. Genetic analysis of Bactrocera zonata (Diptera: Tephritidae) populations from India based on cox1 and nad1 gene sequences. Mitochondrial DNA Part A 2018, 29, 727–736. [Google Scholar] [CrossRef]
- Jiang, F.; Pan, X.; Li, X.; Yu, Y.; Zhang, J.; Jiang, H.; Dou, L.; Zhu, S. The first complete mitochondrial genome of Dacus longicornis (Diptera: Tephritidae) using next-generation sequencing and mitochondrial genome phylogeny of Dacini tribe. Sci. Rep. 2016, 6, 36426. [Google Scholar] [CrossRef]
- Liu, J.-H.; Xu, J.; Li, Y.-H.; Dan, W.; Pan, Y. Complete mitochondrial genome of the guava fruit fly, Bactrocera correcta (Diptera: Tephritidae). Mitochondrial DNA Part A 2016, 27, 4553–4554. [Google Scholar] [CrossRef] [PubMed]
- Song, S.-L.; Yong, H.-S.; Suana, I.W.; Lim, P.-E. Complete mitochondrial genome of Bactrocera ritsemai (Insecta: Tephritidae) and phylogenetic relationship with its congeners and related tephritid taxa. J. Asia-Pac. Entomol. 2018, 21, 252–257. [Google Scholar] [CrossRef]
- Yong, H.-S.; Song, S.-L.; Lim, P.-E.; Eamsobhana, P.; Suana, I.W. Complete mitochondrial genome of three Bactrocera fruit flies of subgenus Bactrocera (Diptera: Tephritidae) and their phylogenetic implications. PLoS ONE 2016, 11, e0148201. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Xu, L.; Nardi, F.; Li, J.; Zhang, R. The complete nucleotide sequence of the mitochondrial genome of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Gene 2007, 396, 66–74. [Google Scholar] [CrossRef]
- Zhang, B.; Nardi, F.; Hull-Sanders, H.; Wan, X.; Liu, Y. The complete nucleotide sequence of the mitochondrial genome of Bactrocera minax (Diptera: Tephritidae). PLoS ONE 2014, 9, e100558. [Google Scholar] [CrossRef] [PubMed]
- Goussarov, G.; Mysara, M.; Vandamme, P.; Van Houdt, R. Introduction to the principles and methods underlying the recovery of metagenome-assembled genomes from metagenomic data. MicrobiologyOpen 2022, 11, e1298. [Google Scholar] [CrossRef] [PubMed]
- Schatz, M.C.; Delcher, A.L.; Salzberg, S.L. Assembly of large genomes using second-generation sequencing. Genome Res. 2010, 20, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.-J.; Liu, J.-H.; Wan, X.-S.; Zhang, Q.-L.; Fu, D.-Y.; Wang, X.-B.; Dan, W.-L.; Zhou, X.-H. Complete mitochondrial genome of the black-winged fly, Felderimyia fuscipennis (Diptera: Tephritidae) and its phylogenetic relationship within family Tephritidae. Mitochondrial DNA Part B 2020, 5, 3620–3621. [Google Scholar] [CrossRef]
- Yong, H.-S.; Song, S.-L.; Lim, P.-E.; Eamsobhana, P. Complete mitochondrial genome of Zeugodacus tau (Insecta: Tephritidae) and differentiation of Z. tau species complex by mitochondrial cytochrome c oxidase subunit I gene. PLoS ONE 2017, 12, e0189325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; De Meyer, M.; Virgilio, M.; Feng, S.; Badji, K.; Li, Z. Phylogenomic resolution of the Ceratitis FARQ complex (Diptera: Tephritidae). Mol. Phylogenetics Evol. 2021, 161, 107160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.-X.; Yu, G.; Li, T.-J.; He, Q.-Y.; Zhou, Y.; Si, F.-L.; Ren, S.; Chen, B. The complete mitochondrial genome of Delia antiqua and its implications in dipteran phylogenetics. PLoS ONE 2015, 10, e0139736. [Google Scholar] [CrossRef] [PubMed]
- Negrisolo, E.; Babbucci, M.; Patarnello, T. The mitochondrial genome of the ascalaphid owlfly Libelloides macaronius and comparative evolutionary mitochondriomics of neuropterid insects. BMC genomics 2011, 12, 221. [Google Scholar] [CrossRef] [PubMed]
- Morton, R.A.; Morton, B.R. Separating the effects of mutation and selection in producing DNA skew in bacterial chromosomes. BMC Genom. 2007, 8, 369. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L.; Johnson, K.P.; Whiting, M.F. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): Effects of extensive gene rearrangements on the evolution of the genome. J. Mol. Evol. 2007, 65, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, A.; Leger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Guo, S.; Chen, J.; Song, N. The First Complete Mitochondrial Genome of Sphaeniscus atilius (Walker, 1849) (Diptera: Tephritidae) and Implications for the Phylogenetic Relationships of Tephritidae; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2023. [Google Scholar]
- Kabiraj, D.; Chetia, H.; Nath, A.; Sharma, P.; Mosahari, P.V.; Singh, D.; Dutta, P.; Neog, K.; Bora, U. Mitogenome-wise codon usage pattern from comparative analysis of the first mitogenome of Blepharipa sp.(Muga uzifly) with other Oestroid flies. Sci. Rep. 2022, 12, 7028. [Google Scholar] [CrossRef] [PubMed]
- da Silva, F.S.; Cruz, A.C.R.; de Almeida Medeiros, D.B.; da Silva, S.P.; Nunes, M.R.T.; Martins, L.C.; Chiang, J.O.; da Silva Lemos, P.; Cunha, G.M.; de Araujo, R.F. Mitochondrial genome sequencing and phylogeny of Haemagogus albomaculatus, Haemagogus leucocelaenus, Haemagogus spegazzinii, and Haemagogus tropicalis (Diptera: Culicidae). Sci. Rep. 2020, 10, 16948. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, S.; Cameron, S.L.; Kang, Z.; Wang, Y.; Yang, D. The first mitochondrial genome of the sepsid fly Nemopoda mamaevi Ozerov, 1997 (Diptera: Sciomyzoidea: Sepsidae), with mitochondrial genome phylogeny of Cyclorrhapha. PLoS ONE 2015, 10, e0123594. [Google Scholar] [CrossRef]
- Sahyoun, A.H.; Bernt, M.; Stadler, P.F.; Tout, K. GC skew and mitochondrial origins of replication. Mitochondrion 2014, 17, 56–66. [Google Scholar] [CrossRef]
- Lessinger, A.; Martins Junqueira, A.; Lemos, T.; Kemper, E.; Da Silva, F.; Vettore, A.; Arruda, P.; Azeredo-Espin, A.L. The mitochondrial genome of the primary screwworm fly Cochliomyia hominivorax (Diptera: Calliphoridae). Insect Mol. Biol. 2000, 9, 521–529. [Google Scholar] [CrossRef]
- Xing, K.; Kang, C.; Zhao, F. The first complete mitochondrial genome sequences for Ulidiidae and phylogenetic analysis of Diptera. Mol. Biol. Rep. 2023, 50, 2501–2510. [Google Scholar] [CrossRef]
- Zhu, J.-c.; Tang, P.; Zheng, B.-Y.; Wu, Q.; Wei, S.-j.; Chen, X.-x. The first two mitochondrial genomes of the family Aphelinidae with novel gene orders and phylogenetic implications. Int. J. Biol. Macromol. 2018, 118, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L.; Brown, W.M. Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: Sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol. Biol. Evol. 2000, 17, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Castellana, S.; Vicario, S.; Saccone, C. Evolutionary patterns of the mitochondrial genome in Metazoa: Exploring the role of mutation and selection in mitochondrial protein–coding genes. Genome Biol. Evol. 2011, 3, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Montana-Lozano, P.; Balaguera-Reina, S.A.; Prada-Quiroga, C.F. Comparative analysis of codon usage of mitochondrial genomes provides evolutionary insights into reptiles. Gene 2023, 851, 146999. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Luo, Y.; Sha, A.; Xiao, W.; Xiong, Z.; Chen, X.; He, J.; Peng, L.; Zou, L. Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species. Front. Microbiol. 2023, 14, 1134228. [Google Scholar] [CrossRef]
- Behura, S.K.; Severson, D.W. Codon usage bias: Causative factors, quantification methods and genome-wide patterns: With emphasis on insect genomes. Biol. Rev. 2013, 88, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.R.; Sezzi, E.; Moriyama, E.N.; Gleason, J.M.; Caccone, A. Analysis of a shift in codon usage in Drosophila. J. Mol. Evol. 2003, 57, S214–S225. [Google Scholar] [CrossRef]
- Pandey, A.; Suman, S.; Chandna, S. Predictive role of mitochondrial genome in the stress resistance of insects and nematodes. Bioinformation 2010, 5, 21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gil Borlado, M.C.; Moreno Lastres, D.; Gonzalez Hoyuela, M.; Moran, M.; Blazquez, A.; Pello, R.; Marin Buera, L.; Gabaldon, T.; Garcia Penas, J.J.; Martin, M.A. Impact of the mitochondrial genetic background in complex III deficiency. PLoS ONE 2010, 5, e12801. [Google Scholar]
- Mitchell, A.L.; Elson, J.L.; Howell, N.; Taylor, R.W.; Turnbull, D.M. Sequence variation in mitochondrial complex I genes: Mutation or polymorphism? J. Med. Genet. 2006, 43, 175–179. [Google Scholar]
- Stein, L.R.; Imai, S.-i. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol. Metab. 2012, 23, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.M.; Bradley, T.J. Metabolic rate in female Culex tarsalis (Diptera: Culicidae): Age, size, activity, and feeding effects. J. Med. Entomol. 2003, 40, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Suarez, R.; Staples, J.; Lighton, J.; Mathieu-Costello, O. Mitochondrial function in flying honeybees (Apis mellifera): Respiratory chain enzymes and electron flow from complex III to oxygen. J. Exp. Biol. 2000, 203, 905–911. [Google Scholar] [CrossRef]
- Terblanche, J.; Klok, C.; Chown, S. Metabolic rate variation in Glossina pallidipes (Diptera: Glossinidae): Gender, ageing and repeatability. J. Insect Physiol. 2004, 50, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.R. Why so many polyphagous fruit flies (Diptera: Tephritidae)? A further contribution to the ‘generalism’ debate. Biol. J. Linn. Soc. 2017, 120, 245–257. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Fang, G.; Xu, P.; Gao, B.; Liu, X.; Qi, X.; Zhang, G.; Cao, S.; Li, Z.; Ren, X. Behavioral and genomic divergence between a generalist and a specialist fly. Cell Reports 2022, 41, 111654. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeister, T.S.; Vidal, S. The diversity of fruit fly (Diptera: Tephritidae) parasitoids. In Parasitoid Community Ecology; Oxford University Press: New York, NY, USA, 1994; pp. 47–76. [Google Scholar]
- Charlery De La Masseliere, M.; Ravigné, V.; Facon, B.; Lefeuvre, P.; Massol, F.; Quilici, S.; Duyck, P.F. Changes in phytophagous insect host ranges following the invasion of their community: Long-term data for fruit flies. Ecol. Evol. 2017, 7, 5181–5190. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P. Codon usage bias from tRNA’s point of view: Redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004, 14, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.M.; Erill, I. Relative codon adaptation: A generic codon bias index for prediction of gene expression. DNA Res. 2010, 17, 185–196. [Google Scholar] [CrossRef]
- Li, H.; Yan, Y.; Li, J. Eighteen mitochondrial genomes of Syrphidae (Insecta: Diptera: Brachycera) with a phylogenetic analysis of Muscomorpha. PLoS ONE 2023, 18, e0278032. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, D. Mitochondrial Genomes Provide New Phylogenetic and Evolutionary Insights into Psilidae (Diptera: Brachycera). Insects 2022, 13, 518. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Li, M.; Dong, P.; Cui, Y.; Xie, Q.; Bu, W. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genom. 2008, 9, 610. [Google Scholar] [CrossRef]
- Choudhary, J.S.; Naaz, N.; Prabhakar, C.S.; Lemtur, M. Genetic analysis of oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae) populations based on mitochondrial cox1 and nad1 gene sequences from India and other Asian countries. Genetica 2016, 144, 611–623. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends. Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Cvijović, I.; Good, B.H.; Desai, M.M. The effect of strong purifying selection on genetic diversity. Genetics 2018, 209, 1235–1278. [Google Scholar] [CrossRef]
- Jaworski, T.; Hilszczański, J. The Effect of Temperature and Humidity Changes on Insects Development Their Impact on Forest Ecosystems in the Context of Expected Climate Change. For. Res. Pap. 2013, 74, 345–355. [Google Scholar]
- Krosch, M.N.; Schutze, M.K.; Armstrong, K.F.; Graham, G.C.; Yeates, D.K.; Clarke, A.R. A molecular phylogeny for the Tribe Dacini (Diptera: Tephritidae): Systematic and biogeographic implications. Mol. Phylogenetics Evol. 2012, 64, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; Callejas, C.; Fernandez, M.; Ochando, M. New contributions towards the understanding of the phylogenetic relationships among economically important fruit flies (Diptera: Tephritidae). Bull. Entomol. Res. 2006, 96, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, H.; Feng, S.; Qin, Y.; De Meyer, M.; Virgilio, M.; Singh, S.; Jiang, F.; Kawi, A.P.; Susanto, A. Mitochondrial phylogenomics reveals the evolutionary and biogeographical history of fruit flies (Diptera: Tephritidae). Entomol. Gen. 2022, 43, 359–368. [Google Scholar] [CrossRef]
- Fernández, P.; Segura, D.; Callejas, C.; Ochando, M. A phylogenetic study of the family Tephritidae (Insecta: Diptera) using a mitochondrial DNA sequence. In Proceedings of 6th International Symposium on Fruit Flies of Economic Importance, Stellenbosch, South Africa, 6–10 May 2002; pp. 6–10. [Google Scholar]
- Nash, W.J.; Chapman, T. Effect of dietary components on larval life history characteristics in the Medfly (Ceratitis capitata: Diptera, Tephritidae). PLoS ONE 2014, 9, e86029. [Google Scholar] [CrossRef]
- Blacket, M.J.; Agarwal, A.; Zheng, L.; Cunningham, J.P.; Britton, D.; Schneider, I.; Rodoni, B.C. A LAMP assay for the detection of Bactrocera tryoni Queensland fruit fly (Diptera: Tephritidae). Sci. Rep. 2020, 10, 9554. [Google Scholar] [CrossRef]
- Jiang, F.; Liang, L.; Wang, J.; Zhu, S. Chromosome-level genome assembly of Bactrocera dorsalis reveals its adaptation and invasion mechanisms. Commun. Biol. 2022, 5, 25. [Google Scholar]
- Nisar, M.J.; Gogi, M.D.; Atta, B.; Tufail, M.; Ali, R.A.; Naveed, W.A.; Iqbal, M. Pathogenicity of fungal and bacterial bioinsecticides against adult peach fruit fly, Bactrocera zonata (Saunders) (Diptera: Tephritidae) admixed with adult diet under controlled conditions. Egypt. J. Biol. Pest Control 2021, 31, 146. [Google Scholar] [CrossRef]
- Sutton, B.D.; Steck, G.J.; Norrbom, A.L.; Rodriguez, E.J.; Srivastava, P.; Alvarado, N.N.; Colque, F.; Landa, E.Y.; Sánchez, J.J.L.; Quisberth, E. Nuclear ribosomal internal transcribed spacer 1 (ITS1) variation in the Anastrepha fraterculus cryptic species complex (Diptera, Tephritidae) of the Andean region. ZooKeys 2015, 540, 175–191. [Google Scholar] [CrossRef][Green Version]
- Harvey, J.A.; Tougeron, K.; Gols, R.; Heinen, R.; Abarca, M.; Abram, P.K.; Basset, Y.; Berg, M.; Boggs, C.; Brodeur, J. Scientists’ warning on climate change and insects. Ecol. Monogr. 2023, 93, e1553. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. FResearch 2018, 7, 1338. [Google Scholar]
- Nachtigall, P.G.; Grazziotin, F.G.; Junqueira-de-Azevedo, I.L. MITGARD: An automated pipeline for mitochondrial genome assembly in eukaryotic species using RNA-seq data. Brief. Bioinform. 2021, 22, bbaa429. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis; Taylor & Francis: New York, NY, USA, 2019. [Google Scholar]
- Allaire, J. RStudio: Integrated development environment for R. Boston MA 2012, 770, 165–171. [Google Scholar]
- Puigbò, P.; Bravo, I.G.; Garcia-Vallve, S. CAIcal: A combined set of tools to assess codon usage adaptation. Biol. Direct 2008, 3, 1–8. [Google Scholar] [CrossRef]
- Kolde, R.; Kolde, M.R. Package ‘Pheatmap’; R package: Madison, WI, USA, 2018; Volume 1. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Peden, J.F. Analysis of Codon Usage. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 1999. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Neher, R.A. Genetic draft, selective interference, and population genetics of rapid adaptation. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 195–215. [Google Scholar] [CrossRef]
- He, Y.; Xu, Y.; Chen, X. Biology, ecology and management of tephritid fruit flies in China: A review. Insects 2023, 14, 196. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina, N.S.; Moreno-Carmona, M.; Canal, N.A.; Prada-Quiroga, C.F. Mitochondrial Genome Variations and Possible Adaptive Implications in Some Tephritid Flies (Diptera, Tephritidae). Int. J. Mol. Sci. 2025, 26, 5560. https://doi.org/10.3390/ijms26125560
Medina NS, Moreno-Carmona M, Canal NA, Prada-Quiroga CF. Mitochondrial Genome Variations and Possible Adaptive Implications in Some Tephritid Flies (Diptera, Tephritidae). International Journal of Molecular Sciences. 2025; 26(12):5560. https://doi.org/10.3390/ijms26125560
Chicago/Turabian StyleMedina, Natalia S., Manuela Moreno-Carmona, Nelson A. Canal, and Carlos F. Prada-Quiroga. 2025. "Mitochondrial Genome Variations and Possible Adaptive Implications in Some Tephritid Flies (Diptera, Tephritidae)" International Journal of Molecular Sciences 26, no. 12: 5560. https://doi.org/10.3390/ijms26125560
APA StyleMedina, N. S., Moreno-Carmona, M., Canal, N. A., & Prada-Quiroga, C. F. (2025). Mitochondrial Genome Variations and Possible Adaptive Implications in Some Tephritid Flies (Diptera, Tephritidae). International Journal of Molecular Sciences, 26(12), 5560. https://doi.org/10.3390/ijms26125560





