Genome-Wide Characterization and Expression Analysis of the Cysteine-Rich Polycomb-like Protein Gene Family in Response to Hormone Signaling in Apple (Malus domestica)
Abstract
1. Introduction
2. Results
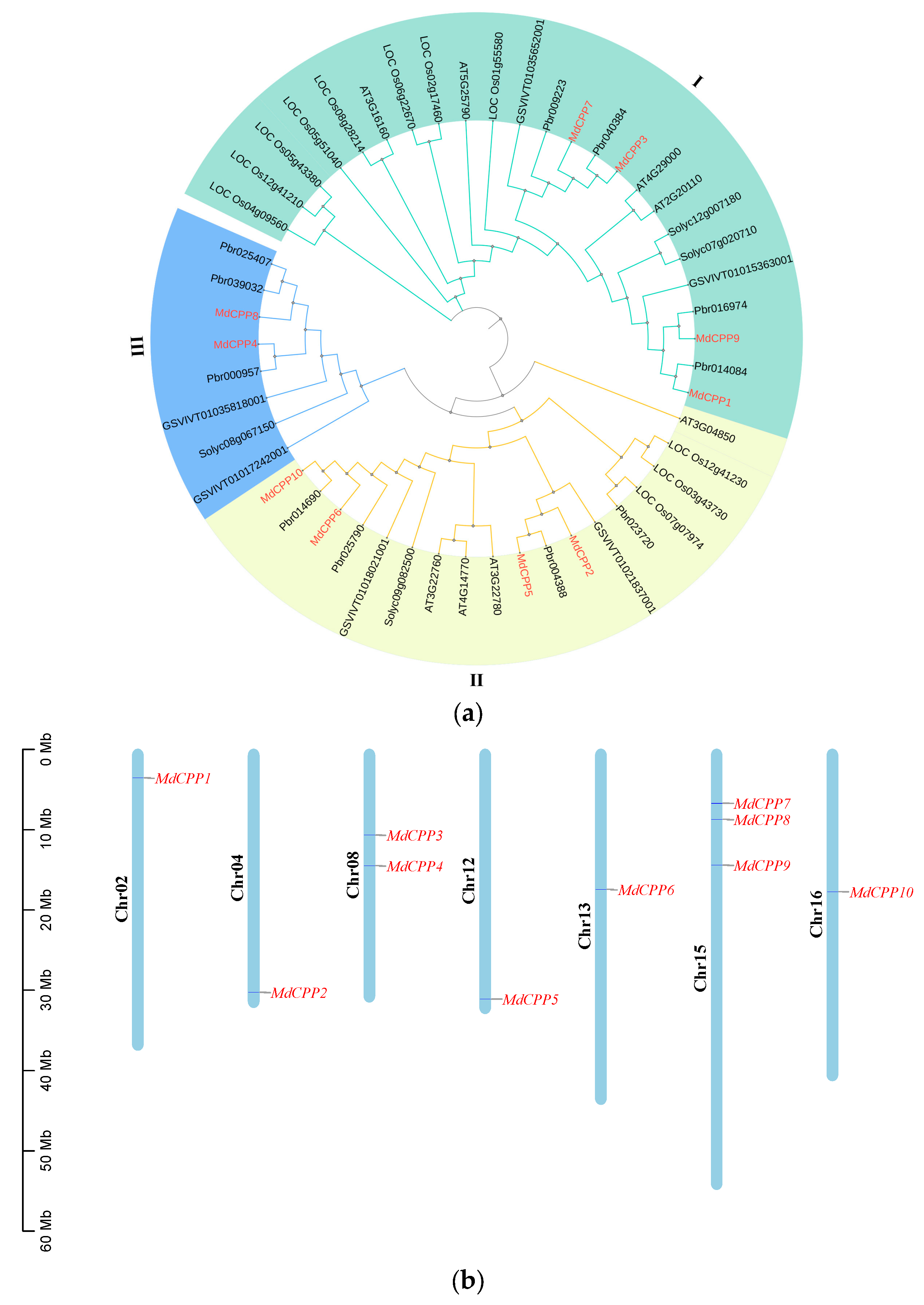
2.1. Genome-Wide Identification and Phylogenetic Analysis of the CPP in Apple
2.2. Multiple Sequence Alignment and Gene Structure Analysis of MdCPPs
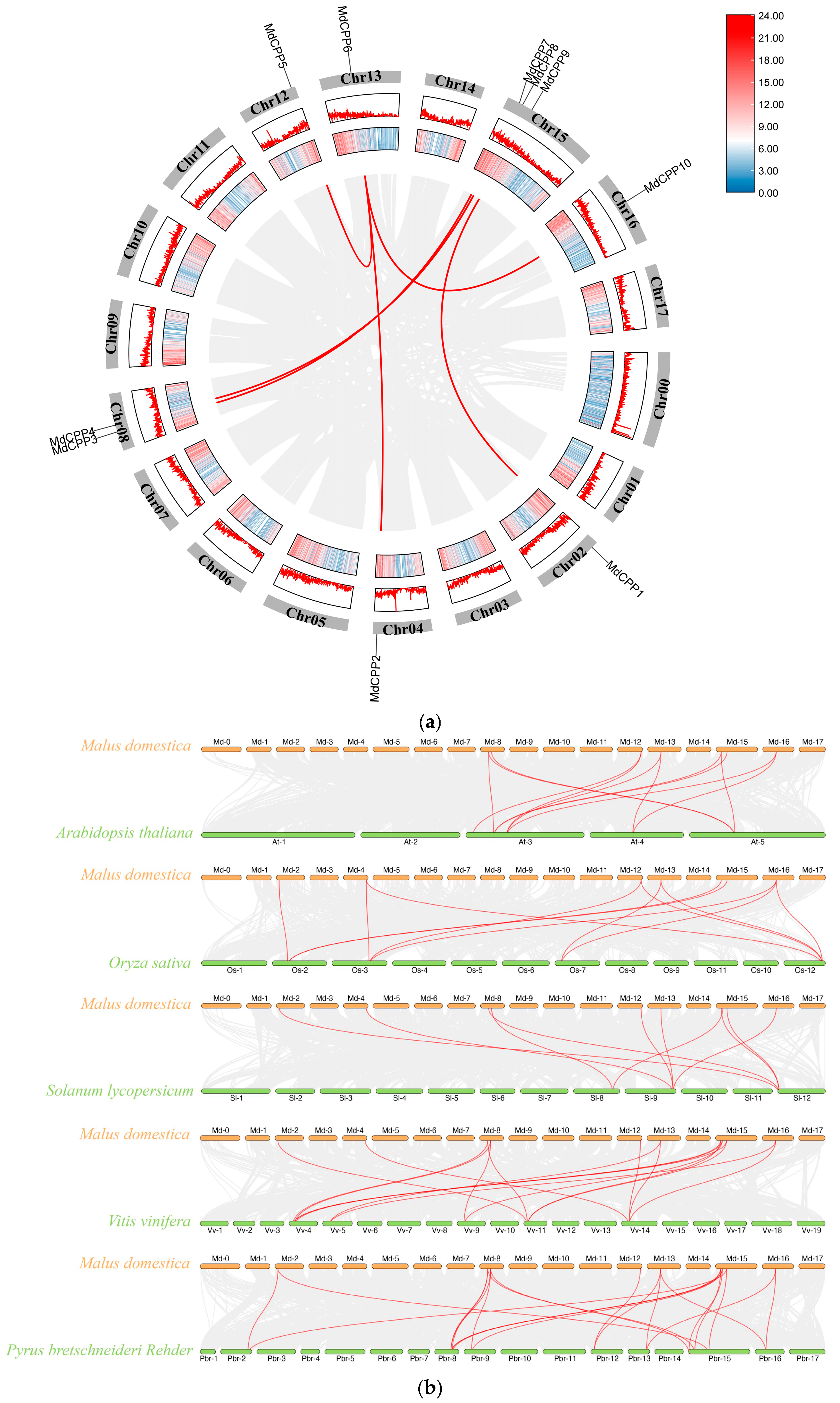
2.3. Synteny Analysis of MdCPPs
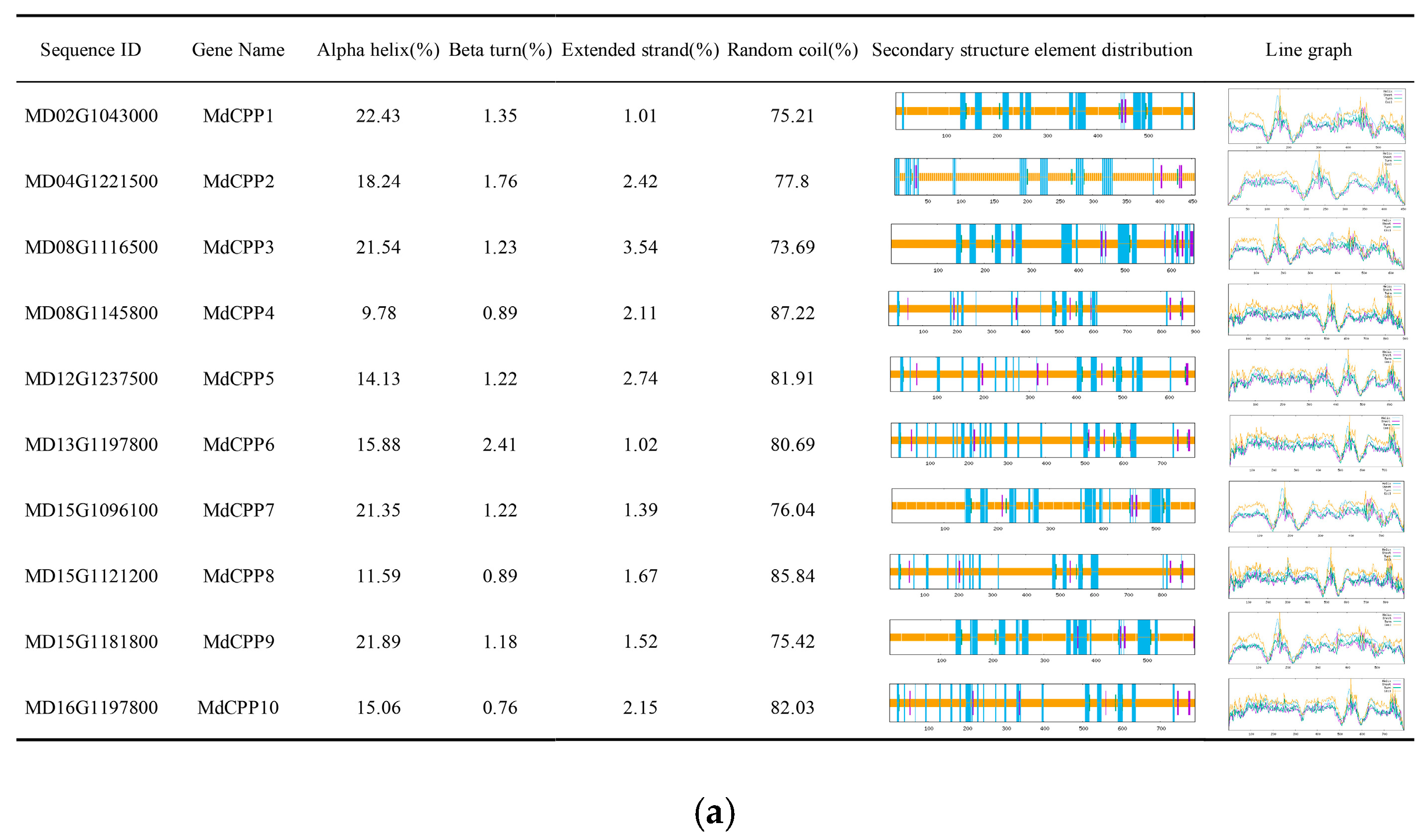
2.4. Secondary and Tertiary Structure Prediction of Apple CPP Proteins
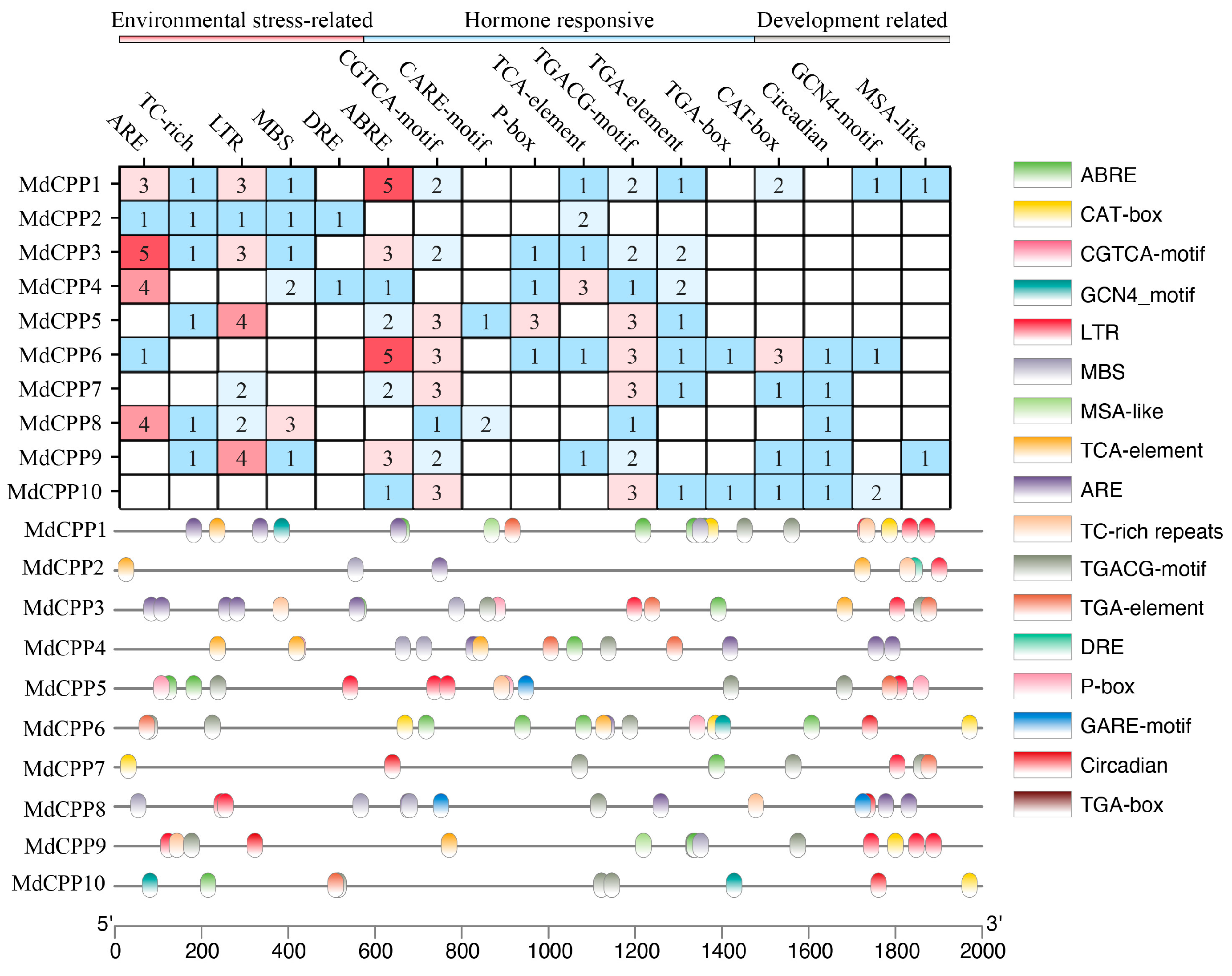
2.5. Cis-Acting Element Analysis of MdCPPs
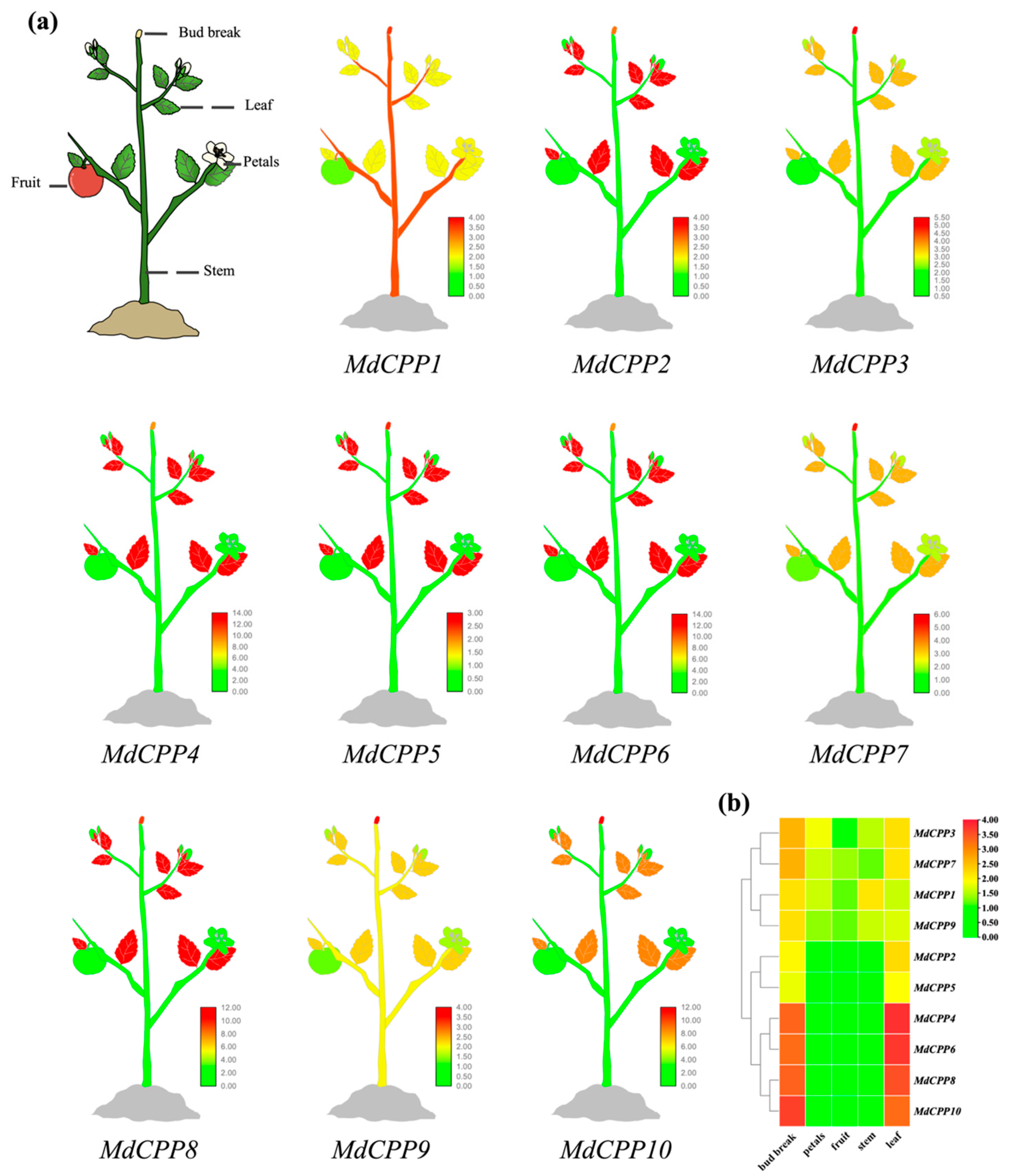
2.6. Expression Pattern of MdCPPs
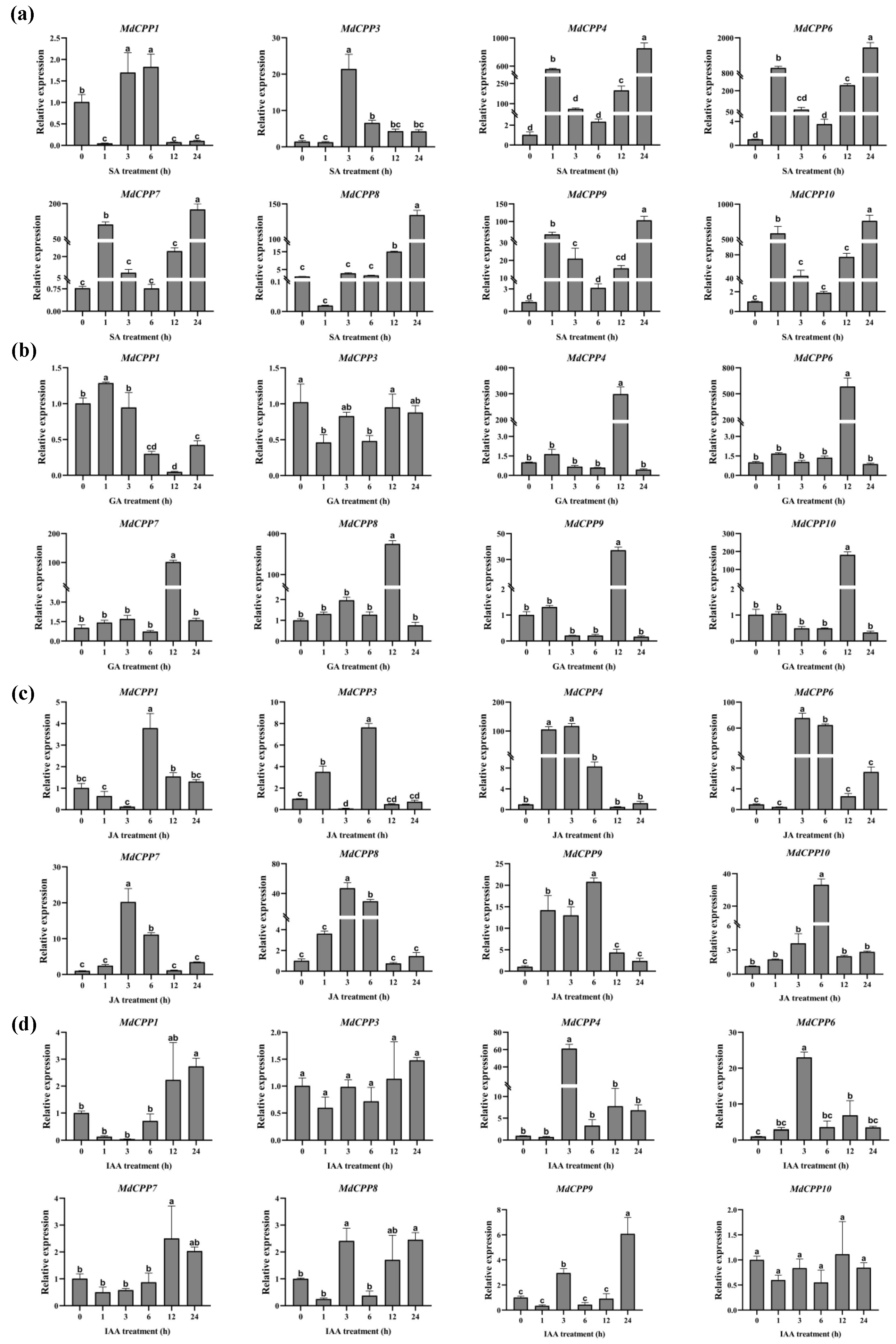
2.7. Expression Profiling of MdCPPs Under Phytohormone Treatments Using qRT-PCR
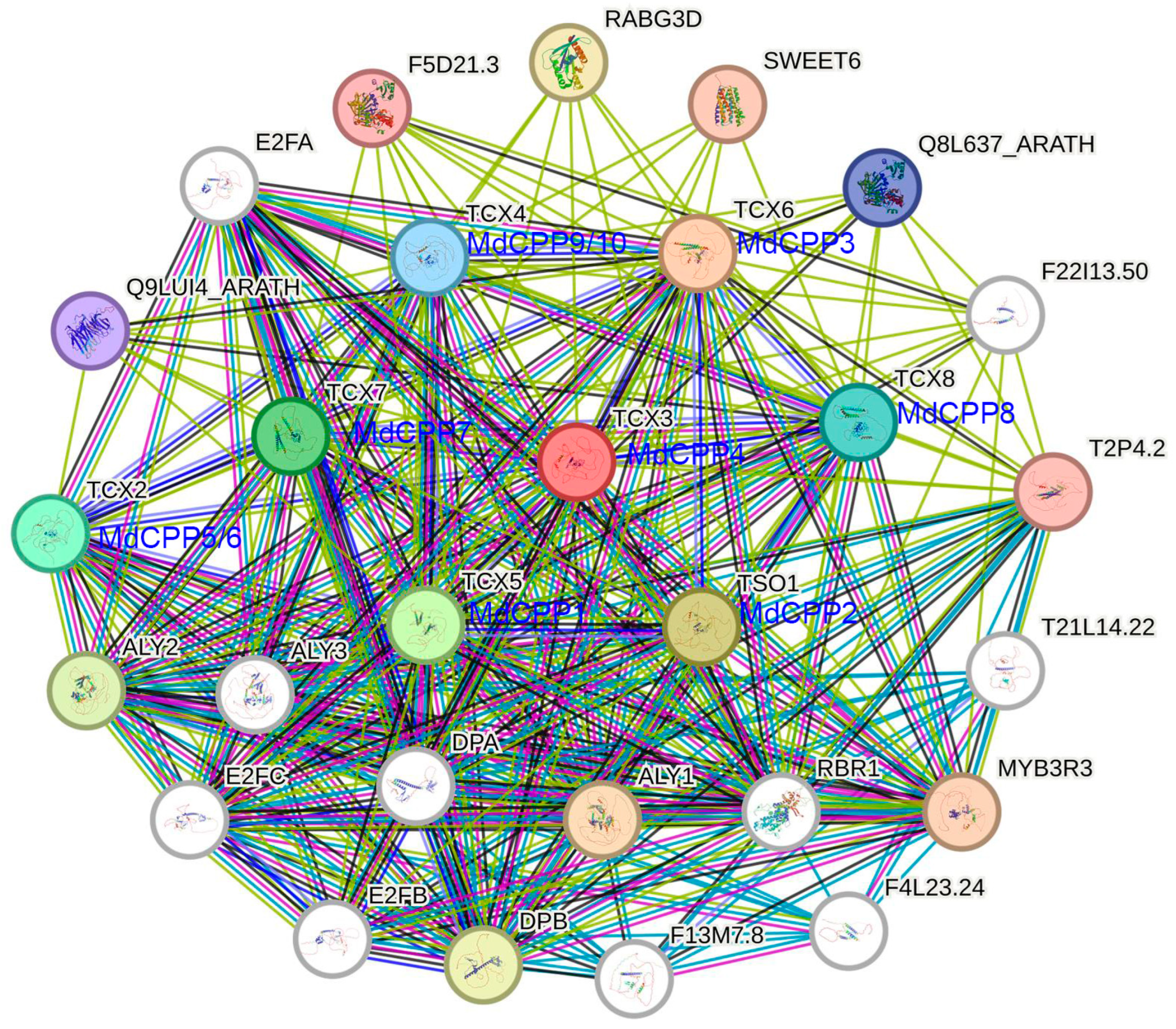
2.8. Protein–Protein Interaction Analysis of MdCPPs
3. Discussion
3.1. Identification and Characterization of CPPs in Apple
3.2. Roles of CPPs in Apple Development
3.3. Response to Hormone Signaling of CPPs in Apple
4. Materials and Methods
4.1. Identification of CPP Transcription Factor Family Members in Apple
4.2. Phylogenetic Tree Construction, Chromosomal Localization, and Protein Physicochemical Characterization
4.3. Multiple Sequence Alignment, Conserved Motifs, and Gene Structure Analysis
4.4. Collinearity Analysis of CPP Genes
4.5. Secondary and Tertiary Structure Prediction
4.6. Cis-Acting Element and Protein Interaction Network Analysis
4.7. Analysis of Expression Patterns in Different Organizations
4.8. Plant Material Treatment and qRT-PCR Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hoshikawa, K.; Pham, D.; Ezura, H.; Schafleitner, R.; Nakashima, K. Genetic and Molecular Mechanisms Conferring Heat Stress Tolerance in Tomato Plants. Front. Plant Sci. 2021, 12, 786688. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Wang, P.; Gou, H.; Ma, Z.; Chen, B.; Mao, J. Identification and Expression Analysis of the SKP1-Like Gene Family under Phytohormone and Abiotic Stresses in Apple (Malus domestica). Int. J. Mol. Sci. 2023, 24, 16414. [Google Scholar] [CrossRef]
- Kumar, A.; Mushtaq, M.; Kumar, P.; Sharma, D.P.; Gahlaut, V. Insights into flowering mechanisms in apple (Malus × domestica Borkh.) amidst climate change: An exploration of genetic and epigenetic factors. Biochim. Biophys. Acta Gen. Subj. 2024, 1868, 130593. [Google Scholar] [CrossRef]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Munns, R.; Millar, A.H. Seven plant capacities to adapt to abiotic stress. J. Exp. Bot. 2023, 74, 4308–4323. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription Factors and Plants Response to Drought Stress: Current Understanding and Future Directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K.; et al. Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 2021, 257, 153351. [Google Scholar] [CrossRef]
- Ma, Z.; Jin, Y.M.; Wu, T.; Hu, L.; Zhang, Y.; Jiang, W.; Du, X. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice. Front. Plant Sci. 2022, 13, 1007811. [Google Scholar] [CrossRef]
- Vodiasova, E.; Sinchenko, A.; Khvatkov, P.; Dolgov, S. Genome-Wide Identification, Characterisation, and Evolution of the Transcription Factor WRKY in Grapevine (Vitis vinifera): New View and Update. Int. J. Mol. Sci. 2024, 25, 6241. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Ohta, M.; Sato, A.; Renhu, N.; Yamamoto, T.; Oka, N.; Zhu, J.K.; Tada, Y.; Suzaki, T.; Miura, K. MYC-type transcription factors, MYC67 and MYC70, interact with ICE1 and negatively regulate cold tolerance in Arabidopsis. Sci. Rep. 2018, 8, 11622. [Google Scholar] [CrossRef]
- Andersen, S.U.; Algreen-Petersen, R.G.; Hoedl, M.; Jurkiewicz, A.; Cvitanich, C.; Braunschweig, U.; Schauser, L.; Oh, S.A.; Twell, D.; Jensen, E. The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 3657–3670. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Ye, S.; Jiang, L.; Liu, S. Genome-wide identification and characterization of cysteine-rich polycomb-like protein (CPP) family genes in cucumber (Cucumis sativus) and their roles in stress responses. Biologia 2018, 73, 425–435. [Google Scholar] [CrossRef]
- Yang, Z.; Gu, S.; Wang, X.; Li, W.; Tang, Z.; Xu, C. Molecular evolution of the CPP-like gene family in plants: Insights from comparative genomics of Arabidopsis and rice. J. Mol. Evol. 2008, 67, 266–277. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Li, A.; Guo, J.; Wang, H.; Qi, H.; Li, M.; Yang, P.; Song, S. An AP2/ERF transcription factor confers chilling tolerance in rice. Sci. Adv. 2024, 10, eado4788. [Google Scholar] [CrossRef]
- Lu, T.; Dou, Y.; Zhang, C. Fuzzy clustering of CPP family in plants with evolution and interaction analyses. BMC Bioinform. 2013, 14 (Suppl. 13), S10. [Google Scholar] [CrossRef]
- Sun, Y.; Jia, X.; Chen, D.; Fu, Q.; Chen, J.; Yang, W.; Yang, H.; Xu, X. Genome-Wide Identification and Expression Analysis of Cysteine-Rich Polycomb-like Protein (CPP) Gene Family in Tomato. Int. J. Mol. Sci. 2023, 24, 5762. [Google Scholar] [CrossRef]
- Li, M.; Wang, F.; Ma, J.; Liu, H.; Ye, H.; Zhao, P.; Wang, J. Comprehensive Evolutionary Analysis of CPP Genes in Brassica napus L. and Its Two Diploid Progenitors Revealing the Potential Molecular Basis of Allopolyploid Adaptive Advantage Under Salt Stress. Front. Plant Sci. 2022, 13, 873071. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cheng, L.; Guan, X.; Liang, Y.; Xue, Y.; Zhao, W.; Zhang, Z.; Chang, X.; Liang, L.; Gao, G. StCPP3 interacts with type III secretion protein HrpB7 and negatively regulates plant resistance against Ralstonia solanacearum. Biochem. Biophys. Res. Commun. 2025, 742, 151105. [Google Scholar] [CrossRef]
- Ullah, U.; Buttar, Z.A.; Shalmani, A.; Muhammad, I.; Ud-Din, A.; Ali, H. Genome-wide identification and expression analysis of CPP-like gene family in Triticum aestivum L. under different hormone and stress conditions. Open Life Sci. 2022, 17, 544–562. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Liu, S.; Guan, Y.; Weng, Y.; Liao, B.; Tong, L.; Hao, Z.; Chen, J.; Shi, J.; Cheng, T. Genome-wide identification of the NAC gene family and its functional analysis in Liriodendron. BMC Plant Biol. 2023, 23, 415. [Google Scholar] [CrossRef]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- Kwon, S.I.; Cho, H.J.; Kim, S.R.; Park, O.K. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 2013, 161, 1722–1736. [Google Scholar] [CrossRef]
- Liu, T.; Chen, T.; Kan, J.; Yao, Y.; Guo, D.; Yang, Y.; Ling, X.; Wang, J.; Zhang, B. The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants. Plant Biotechnol. J. 2022, 20, 722–735. [Google Scholar] [CrossRef]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 Contributes to Stress Resistance in Maize by Regulating ABA Synthesis and Root Development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, H.K.; Wang, Y.M.; Yuan, C.P.; Zhang, Y.Y.; Li, H.Y.; Yan, X.F.; Li, Q.Y.; Dong, Y.S. Genome-wide identification and expression analysis of the CPP-like gene family in soybean. Genet. Mol. Res. 2015, 14, 1260–1268. [Google Scholar] [CrossRef] [PubMed]
- Song, X.Y.; Zhang, Y.Y.; Wu, F.C.; Zhang, L. Genome-wide analysis of the maize (Zea may L.) CPP-like gene family and expression profiling under abiotic stress. Genet. Mol. Res. 2016, 15, 8023. [Google Scholar] [CrossRef]
- Tan, J.; Xuan, X.; Su, S.; Jiao, Y.; Guo, H.; Zhang, Z. Comprehensive analysis of the CPP gene family in Moso bamboo: Insights into their role in rapid shoot growth. BMC Genom. 2024, 25, 1173. [Google Scholar] [CrossRef]
- Wang, H.; Yan, H.; Liu, H.; Liu, R.; Chen, J.; Xiang, Y. GFDP: The gene family database in poplar. Database 2018, 2018, bay107. [Google Scholar] [CrossRef]
- Guo, R.; Xu, X.; Carole, B.; Li, X.; Gao, M.; Zheng, Y.; Wang, X. Genome-wide identification, evolutionary and expression analysis of the aspartic protease gene superfamily in grape. BMC Genom. 2013, 14, 554. [Google Scholar] [CrossRef]
- Wang, W.; Ma, J.; Liu, H.; Wang, Z.; Nan, R.; Zhong, T.; Sun, M.; Wang, S.; Yao, Y.; Sun, F.; et al. Genome-wide analysis of the switchgrass YABBY family and functional characterization of PvYABBY14 in response to ABA and GA stress in Arabidopsis. BMC Plant Biol. 2024, 24, 114. [Google Scholar] [CrossRef]
- Kent, W.J.; Baertsch, R.; Hinrichs, A.; Miller, W.; Haussler, D. Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. USA 2003, 100, 11484–11489. [Google Scholar] [CrossRef]
- Lan, T.; Xiong, W.; Chen, X.; Mo, B.; Tang, G. Plant cytoplasmic ribosomal proteins: An update on classification, nomenclature, evolution and resources. Plant J. 2022, 110, 292–318. [Google Scholar] [CrossRef]
- Bai, J.; Pennill, L.A.; Ning, J.; Lee, S.W.; Ramalingam, J.; Webb, C.A.; Zhao, B.; Sun, Q.; Nelson, J.C.; Leach, J.E.; et al. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002, 12, 1871–1884. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Li, L.; Meng, L.; Singh, J.; Jiang, N.; Deng, X.W.; He, Z.H.; Lemaux, P.G. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005, 139, 1107–1124. [Google Scholar] [CrossRef]
- Song, J.Y.; Leung, T.; Ehler, L.K.; Wang, C.; Liu, Z. Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development 2000, 127, 2207–2217. [Google Scholar] [CrossRef]
- Hauser, B.A.; He, J.Q.; Park, S.O.; Gasser, C.S. TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development 2000, 127, 2219–2226. [Google Scholar] [CrossRef]
- Tavares, G.G.; Santana, L.R.; da Silva, L.N.; Teixeira, M.B.; da Silva, A.A.; Cabral, J.S.R.; Souchie, E.L. Morpho-physiological traits of soybean plants in symbiosis with Gigaspora sp. and submitted to water restriction. Sci. Rep. 2025, 15, 7133. [Google Scholar] [CrossRef]
- Cheng, S.; Zou, Y.N.; Kuča, K.; Hashem, A.; Abd Allah, E.F.; Wu, Q.S. Elucidating the Mechanisms Underlying Enhanced Drought Tolerance in Plants Mediated by Arbuscular Mycorrhizal Fungi. Front. Microbiol. 2021, 12, 809473. [Google Scholar] [CrossRef]
- Beauvieux, R.; Wenden, B.; Dirlewanger, E. Bud Dormancy in Perennial Fruit Tree Species: A Pivotal Role for Oxidative Cues. Front. Plant Sci. 2018, 9, 657. [Google Scholar] [CrossRef]
- Pan, W.; Liang, J.; Sui, J.; Li, J.; Liu, C.; Xin, Y.; Zhang, Y.; Wang, S.; Zhao, Y.; Zhang, J.; et al. ABA and Bud Dormancy in Perennials: Current Knowledge and Future Perspective. Genes 2021, 12, 1635. [Google Scholar] [CrossRef]
- Liu, K.; Deng, F.; Zeng, F.; Chen, Z.-H.; Qin, Y.; Chen, G. Plant growth-promoting rhizobacteria improve drought tolerance of crops: A review. Plant Growth Regul. 2025, 105, 567–581. [Google Scholar] [CrossRef]
- Ghosh, A.; Agrawal, M.; Agrawal, S.B. Effect of water deficit stress on an Indian wheat cultivar (Triticum aestivum L. HD 2967) under ambient and elevated level of ozone. Sci. Total Environ. 2020, 714, 136837. [Google Scholar] [CrossRef]
- Tan, S.; Sha, Y.; Sun, L.; Li, Z. Abiotic Stress-Induced Leaf Senescence: Regulatory Mechanisms and Application. Int. J. Mol. Sci. 2023, 24, 11996. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, J.W. The Crosstalk between MicroRNAs and Gibberellin Signaling in Plants. Plant Cell Physiol. 2020, 61, 1880–1890. [Google Scholar] [CrossRef]
- Das, S.; Shil, S.; Rime, J.; Alice, A.K.; Yumkhaibam, T.; Mounika, V.; Singh, A.P.; Kundu, M.; Lalhmangaihzuali, H.P.; Hazarika, T.K.; et al. Phytohormonal signaling in plant resilience: Advances and strategies for enhancing abiotic stress tolerance. Plant Growth Regul. 2025, 105, 329–360. [Google Scholar] [CrossRef]
- Spoel, S.H.; Dong, X. Salicylic acid in plant immunity and beyond. Plant Cell 2024, 36, 1451–1464. [Google Scholar] [CrossRef]
- Mishra, A.K.; Baek, K.H. Salicylic Acid Biosynthesis and Metabolism: A Divergent Pathway for Plants and Bacteria. Biomolecules 2021, 11, 705. [Google Scholar] [CrossRef]
- Dong, Q.; Duan, D.; Wang, F.; Yang, K.; Song, Y.; Wang, Y.; Wang, D.; Ji, Z.; Xu, C.; Jia, P.; et al. The MdVQ37-MdWRKY100 complex regulates salicylic acid content and MdRPM1 expression to modulate resistance to Glomerella leaf spot in apples. Plant Biotechnol. J. 2024, 22, 2364–2376. [Google Scholar] [CrossRef]
- Rao, M.V.; Lee, H.; Creelman, R.A.; Mullet, J.E.; Davis, K.R. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 2000, 12, 1633–1646. [Google Scholar] [CrossRef]
- Noh, M.; Shin, J.S.; Hong, J.C.; Kim, S.Y.; Shin, J.S. Arabidopsis TCX8 functions as a senescence modulator by regulating LOX2 expression. Plant Cell Rep. 2021, 40, 677–689. [Google Scholar] [CrossRef]
- Wang, L.; Liu, S.; Gao, M.; Wang, L.; Wang, L.; Wang, Y.; Dai, L.; Zhao, J.; Liu, M.; Liu, Z. The Crosstalk of the Salicylic Acid and Jasmonic Acid Signaling Pathways Contributed to Different Resistance to Phytoplasma Infection Between the Two Genotypes in Chinese Jujube. Front. Microbiol. 2022, 13, 800762. [Google Scholar] [CrossRef]
- Tian, H.; Xu, L.; Li, X.; Zhang, Y. Salicylic acid: The roles in plant immunity and crosstalk with other hormones. J. Integr. Plant Biol. 2024, 67, 773–785. [Google Scholar] [CrossRef]
- Mu, T.; Luo, S.; Li, L.; Zhang, R.; Wang, P.; Zhang, G. A review of the interaction mechanisms between jasmonic acid (JA) and various plant hormones, as well as the core regulatory role of MYC2. Plant Sci. 2025, 353, 112407. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Tsuda, K. Salicylic acid and jasmonic acid crosstalk in plant immunity. Essays Biochem. 2022, 66, 647–656. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Maurya, A.; Gupta, R.; Joshi, P.; Rajkumar, S.; Singh, A.K.; Bhardwaj, R.; Singh, G.P.; Singh, R. Genome-wide identification and expression profiling of WRKY gene family in grain Amaranth (Amaranthus hypochondriacus L.) under salinity and drought stresses. BMC Plant Biol. 2025, 25, 265. [Google Scholar] [CrossRef]
- Gao, W.; Zhang, L.; Zhang, Y.; Zhang, P.; Shahinnia, F.; Chen, T.; Yang, D. Genome-wide identification and expression analysis of the UBC gene family in wheat (Triticum aestivum L.). BMC Plant Biol. 2024, 24, 341. [Google Scholar] [CrossRef] [PubMed]
- Da, L.; Liu, Y.; Yang, J.; Tian, T.; She, J.; Ma, X.; Xu, W.; Su, Z. AppleMDO: A Multi-Dimensional Omics Database for Apple Co-Expression Networks and Chromatin States. Front. Plant Sci. 2019, 10, 1333. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Gu, K.D.; Cheng, L.; Wang, J.H.; Yu, J.Q.; Wang, X.F.; You, C.X.; Hu, D.G.; Hao, Y.J. BTB-TAZ Domain Protein MdBT2 Modulates Malate Accumulation and Vacuolar Acidification in Response to Nitrate. Plant Physiol. 2020, 183, 750–764. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Ma, C.N.; Gu, K.D.; Wang, J.H.; Yu, J.Q.; Liu, B.; Wang, Y.; He, J.X.; Hu, D.G.; Sun, Q. The BTB-BACK-TAZ domain protein MdBT2 reduces drought resistance by weakening the positive regulatory effect of MdHDZ27 on apple drought tolerance via ubiquitination. Plant J. 2024, 119, 283–299. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Ren, Y.R.; Yang, Y.Y.; Zhang, R.; You, C.X.; Zhao, Q.; Hao, Y.J. MdGRF11, an apple 14-3-3 protein, acts as a positive regulator of drought and salt tolerance. Plant Sci. 2019, 288, 110219. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Zhu, M.; Huang, Y.; Zhang, Q. Genome-Wide Characterization and Expression Analysis of the Cysteine-Rich Polycomb-like Protein Gene Family in Response to Hormone Signaling in Apple (Malus domestica). Int. J. Mol. Sci. 2025, 26, 5528. https://doi.org/10.3390/ijms26125528
Jiang L, Zhu M, Huang Y, Zhang Q. Genome-Wide Characterization and Expression Analysis of the Cysteine-Rich Polycomb-like Protein Gene Family in Response to Hormone Signaling in Apple (Malus domestica). International Journal of Molecular Sciences. 2025; 26(12):5528. https://doi.org/10.3390/ijms26125528
Chicago/Turabian StyleJiang, Le, Min Zhu, Ying Huang, and Quanyan Zhang. 2025. "Genome-Wide Characterization and Expression Analysis of the Cysteine-Rich Polycomb-like Protein Gene Family in Response to Hormone Signaling in Apple (Malus domestica)" International Journal of Molecular Sciences 26, no. 12: 5528. https://doi.org/10.3390/ijms26125528
APA StyleJiang, L., Zhu, M., Huang, Y., & Zhang, Q. (2025). Genome-Wide Characterization and Expression Analysis of the Cysteine-Rich Polycomb-like Protein Gene Family in Response to Hormone Signaling in Apple (Malus domestica). International Journal of Molecular Sciences, 26(12), 5528. https://doi.org/10.3390/ijms26125528





