Genome-Wide Association Study for Drought Tolerance in Cowpea (Vigna unguiculata (L.) Walp.) at Seedling Stage Using a Whole Genome Resequencing Approach
Abstract
1. Introduction
2. Results
2.1. Population Structure and Genetic Diversity
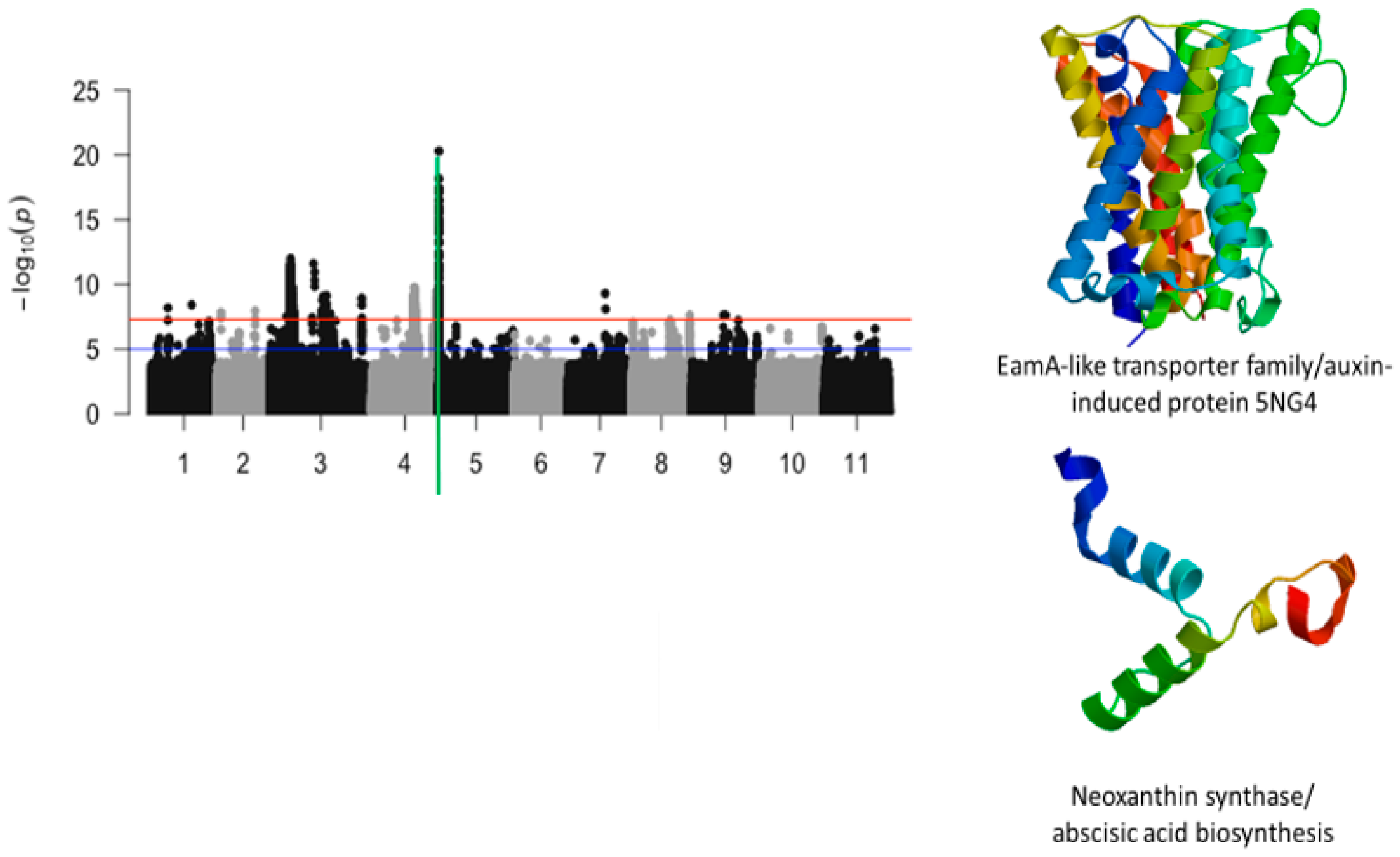
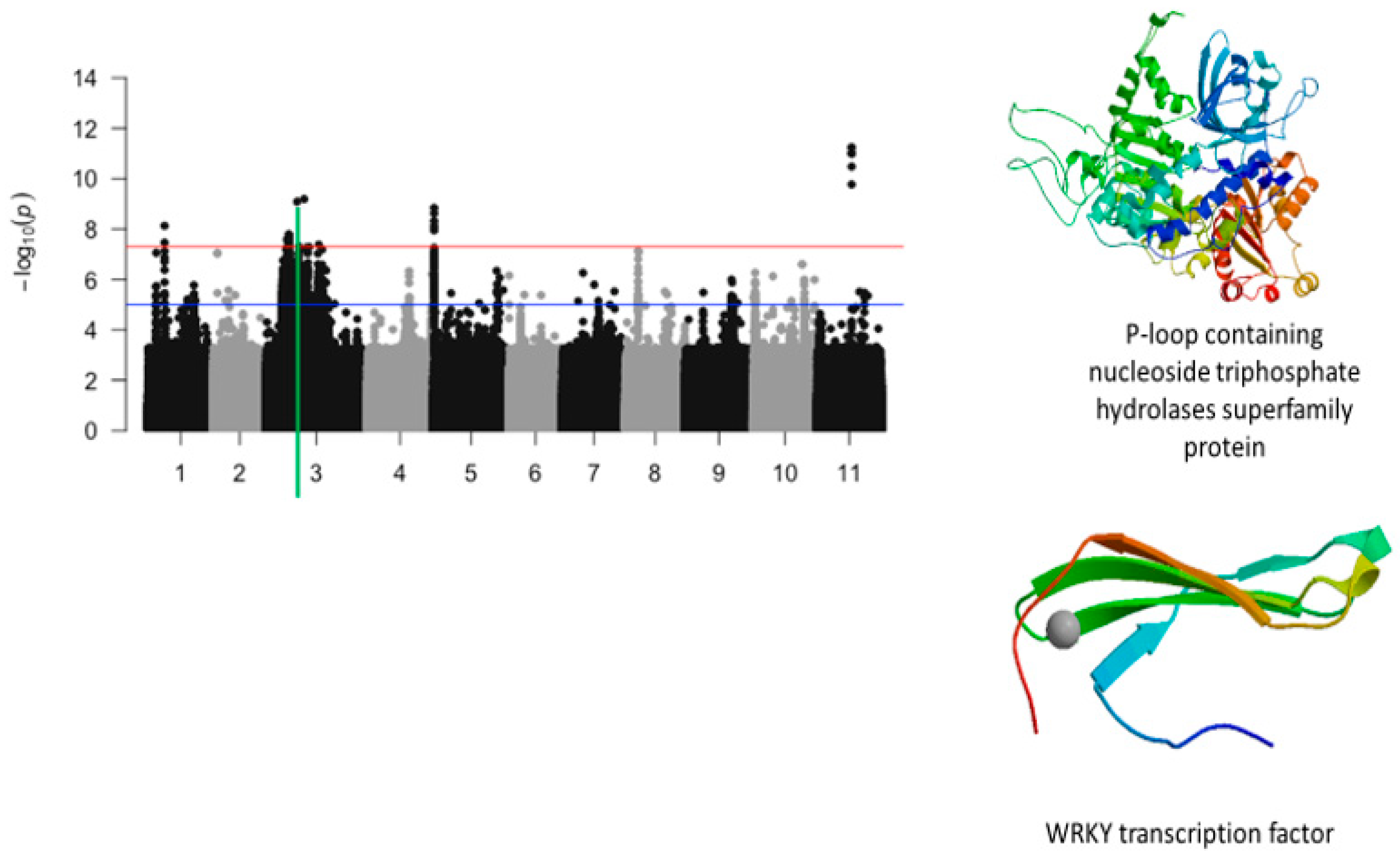
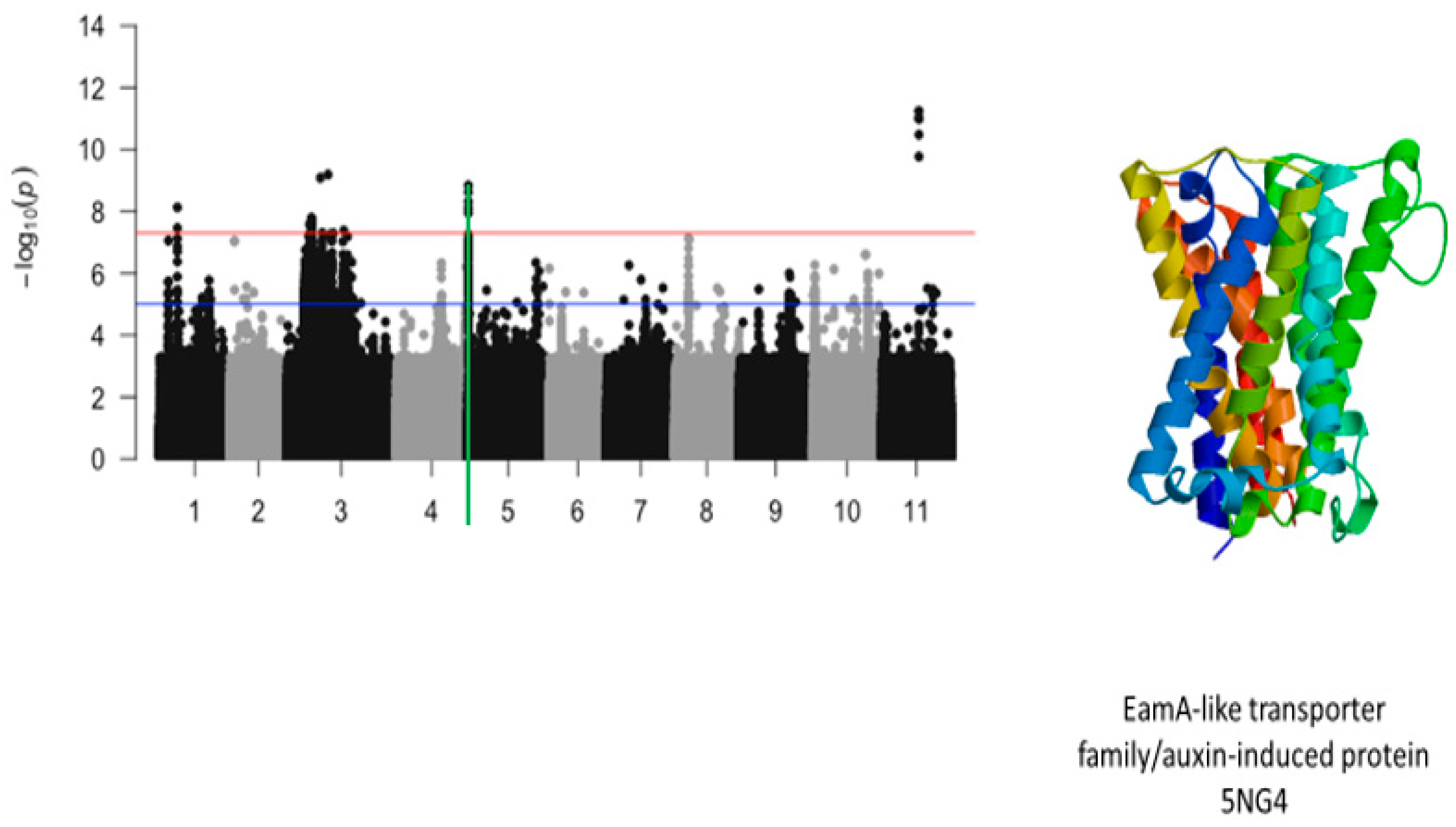
2.2. First Trifoliate Leaf Chlorosis Under Drought Stress
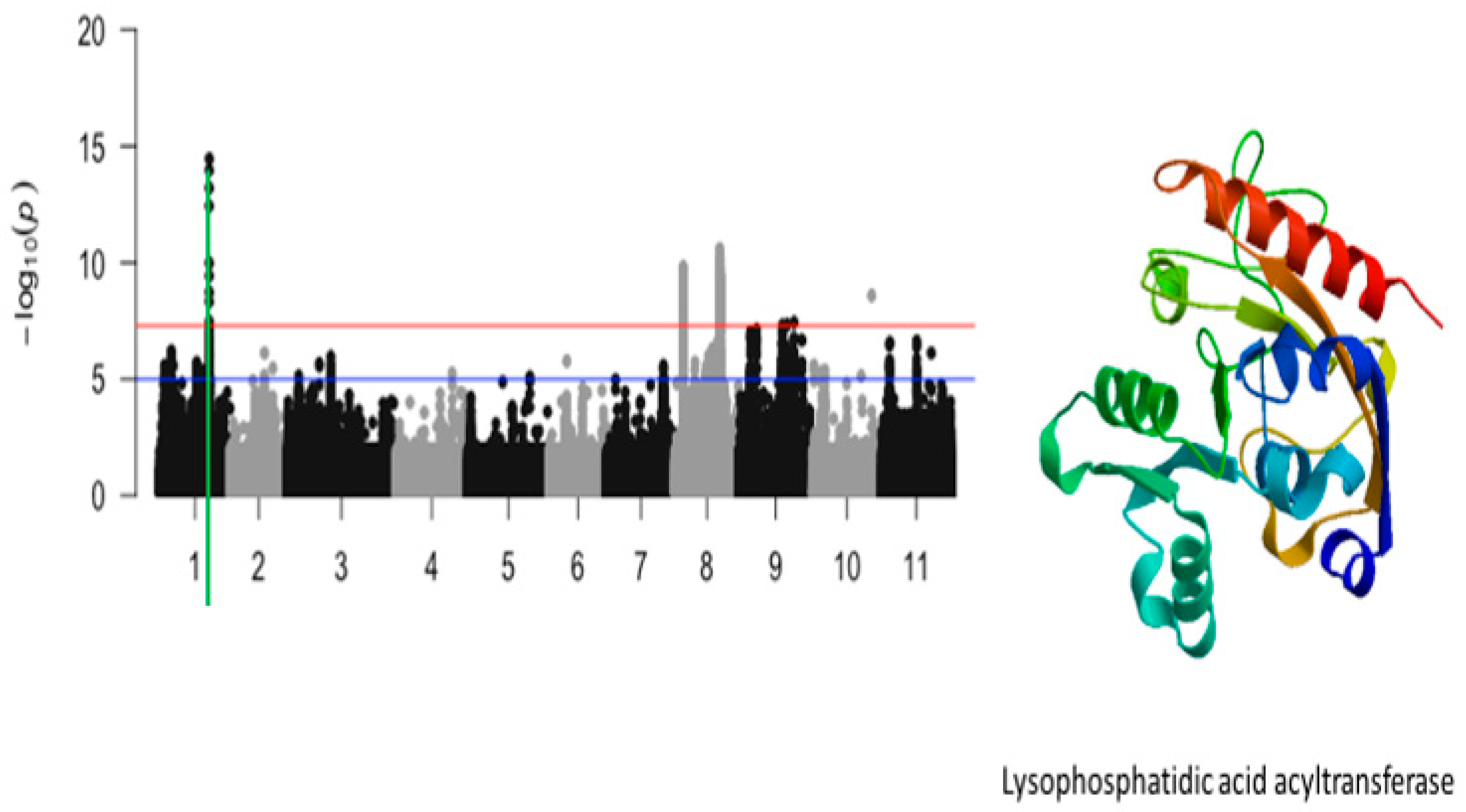
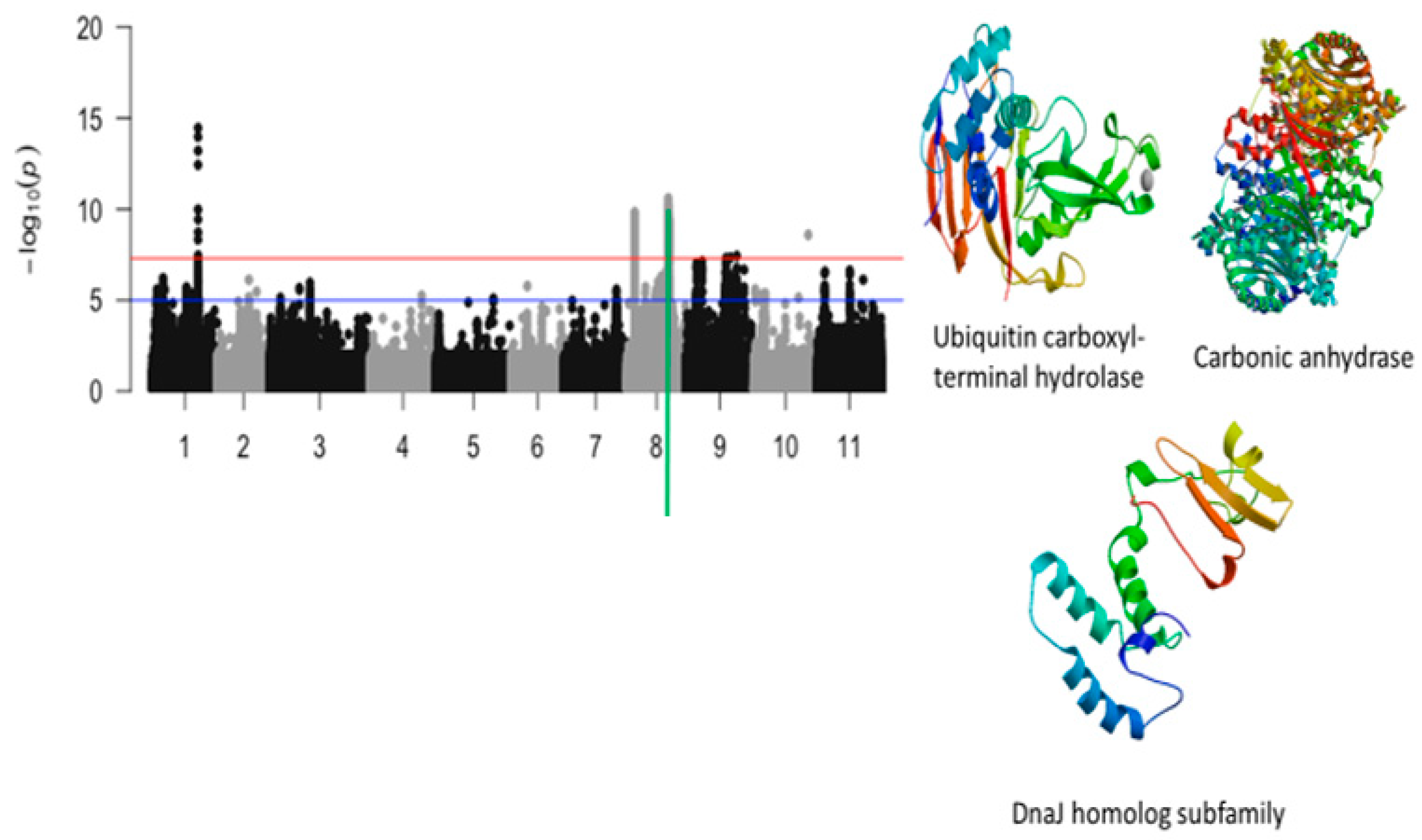
2.3. Unifoliate Leaf Chlorosis
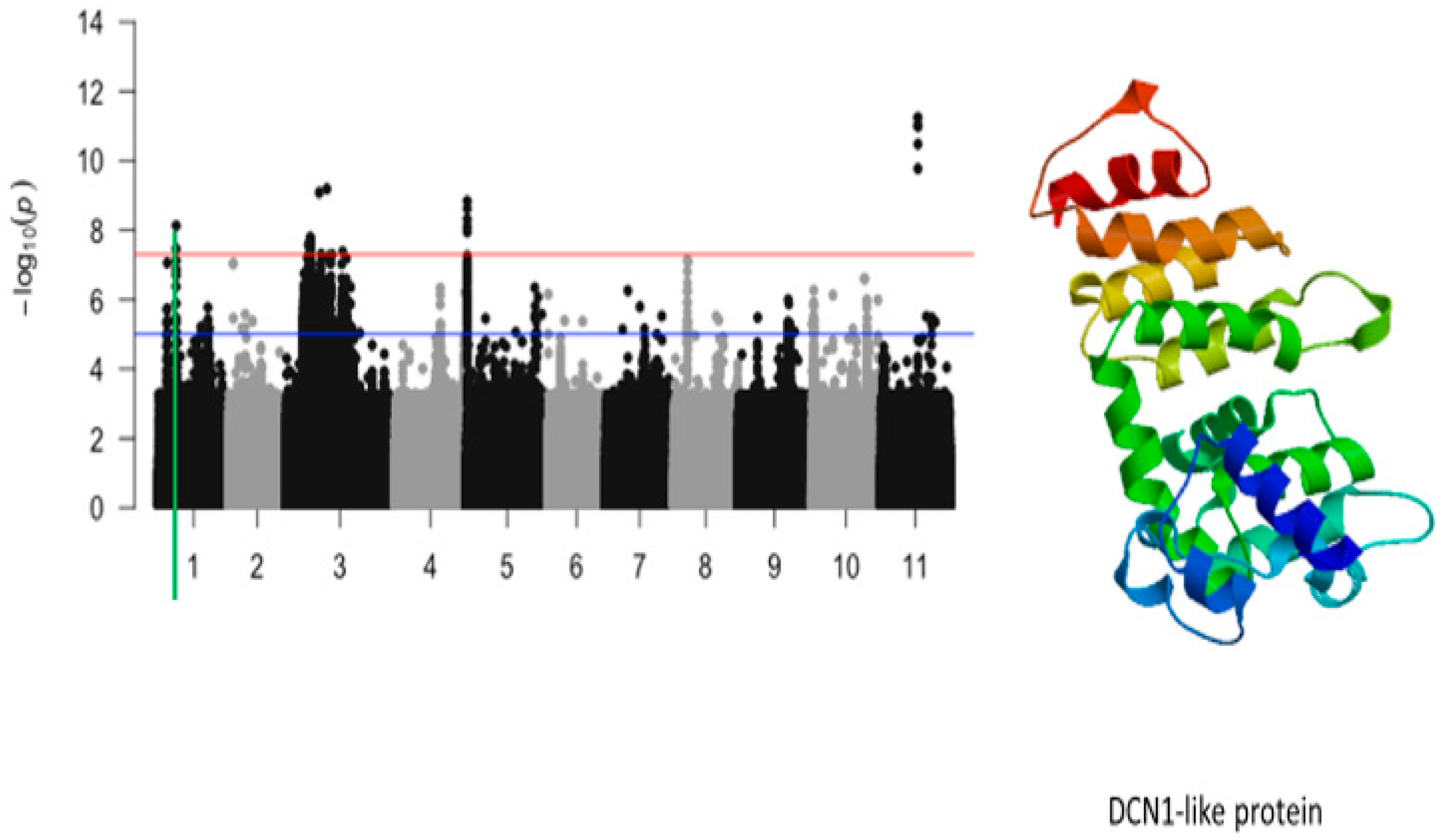
2.4. Plant Greenness Score
2.5. Protein Homologs
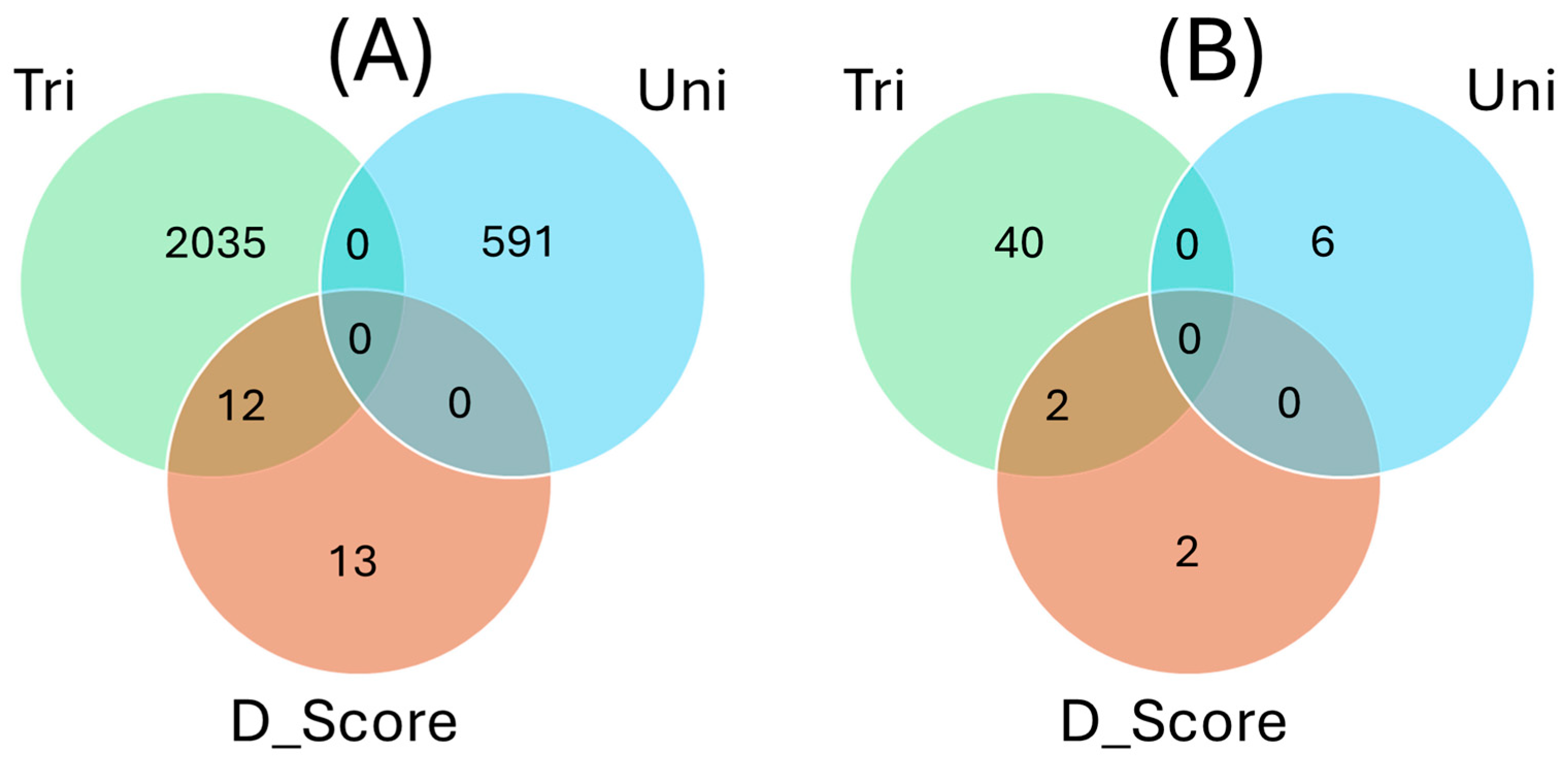
2.6. Overlapping SNPs and Functional Annotations
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Phenotyping
4.2. Genotyping
4.2.1. DNA Extraction, Library Preparation, and Whole-Genome Resequencing
4.2.2. SNP Calling, Mapping, and Filtering
4.3. Population Structure and Genetic Diversity Analysis
4.4. Genome Wide Association Study (GWAS)
4.5. Candidate Gene Search and Synteny Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tester, M.; Davenport, R. Na+ Tolerance and Na+ Transport in Higher Plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Lüking, I.; Yang, O. Plant Tolerance to Drought and Salinity: Stress Regulating Transcription Factors and Their Functional Significance in the Cellular Transcriptional Network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Cruz de Carvalho, M.H. Drought Stress and Reactive Oxygen Species: Production, Scavenging and Signaling. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Amatriaín, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O.; et al. Genome Resources for Climate-Resilient Cowpea, an Essential Crop for Food Security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef]
- Verdcourt, B. Studies in the Leguminosae-Papilionoïdeae for the’Flora of Tropical East Africa’: IV. Kew Bull. 1970, 24, 507–569. [Google Scholar] [CrossRef]
- Blackhurst, H.T.; Miller, J.C. Cowpea. In Hybridization of Crop Plants; American Society of Agronomy, Inc.: Madison, WI, USA, 1980; ISBN 978-0-89118-566-6. [Google Scholar]
- Burridge, J.D.; Schneider, H.M.; Huynh, B.L.; Roberts, P.A.; Bucksch, A.; Lynch, J.P. Genome-Wide Association Mapping and Agronomic Impact of Cowpea Root Architecture. Theor. Appl. Genet. 2016, 130, 419–431. [Google Scholar] [CrossRef]
- Fatokun, C.; Boukar, O.; Muranaka, S. Evaluation of Cowpea (Vigna unguiculata (L.) Walp.) Germplasm Lines for Tolerance to Drought. Plant Genet. Resour. 2012, 10, 171–176. [Google Scholar] [CrossRef]
- Timko, M.; Ehlers, J.; Roberts, P. Cowpea. In Pulses, Sugar and Tuber Crops; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Contour-ansel, D.; Torres-Franklin, M.L.; Cryz De Carvalho, M.H.; D’Arcy-Lameta, A.; Zuily-Fodil, Y. Glutathione Reductase in Leaves of Cowpea: Cloning of Two cDNAs, Expression and Enzymatic Activity under Progressive Drought Stress, Desiccation and Abscisic Acid Treatment. Ann. Bot. 2006, 98, 1279–1287. [Google Scholar] [CrossRef]
- Lucas, M.R.; Diop, N.-N.; Wanamaker, S.; Ehlers, J.D.; Roberts, P.A.; Close, T.J. Cowpea–Soybean Synteny Clarified through an Improved Genetic Map. Plant Genome J. 2011, 4, 218–225. [Google Scholar] [CrossRef]
- Muchero, W.; Ehlers, J.D.; Roberts, P.A. Restriction Site Polymorphism-Based Candidate Gene Mapping for Seedling Drought Tolerance in Cowpea [Vigna unguiculata (L.) Walp.]. Theor. Appl. Genet. 2010, 120, 509–518. [Google Scholar] [CrossRef]
- Muchero, W.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Genic SNP Markers and Legume Synteny Reveal Candidate Genes Underlying QTL for Macrophomina Phaseolina Resistance and Maturity in Cowpea [Vigna unguiculata (L) Walp.]. BMC Genomics 2011, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Jiang, C.; Huang, Z.; Torres-Jerez, I.; Chang, J.; Zhang, H.; Udvardi, M.; Liu, R.; Verdier, J. The Vigna unguiculata Gene Expression Atlas (VuGEA) from De Novo Assembly and Quantification of RNA-Seq Data Provides Insights into Seed Maturation Mechanisms. Plant J. 2016, 88, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Izzah, N.; Jayakodi, M. Genome-Wide SNP Identification and QTL Mapping for Black Rot Resistance in Cabbage. Plant Biol. 2015, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Thudi, M.; Khan, A.; Kumar, V. Whole Genome Re-Sequencing Reveals Genome-Wide Variations among Parental Lines of 16 Mapping Populations in Chickpea (Cicer arietinum L.). BMC Plant Biol. 2016, 16, 10. [Google Scholar] [CrossRef]
- Varshney, R.K.; Paulo, M.J.; Grando, S.; van Eeuwijk, F.A.; Keizer, L.C.P.; Guo, P.; Ceccarelli, S.; Kilian, A.; Baum, M.; Graner, A. Genome Wide Association Analyses for Drought Tolerance Related Traits in Barley (Hordeum vulgare L.). Field Crops Res. 2012, 126, 171–180. [Google Scholar] [CrossRef]
- Dhanapal, A.P.; Ray, J.D.; Singh, S.K.; Hoyos-Villegas, V.; Smith, J.R.; Purcell, L.C.; Andy King, C.; Cregan, P.B.; Song, Q.; Fritschi, F.B. Genome-Wide Association Study (GWAS) of Carbon Isotope Ratio (Δ13C) in Diverse Soybean [Glycine max (L.) Merr.] Genotypes. Theor. Appl. Genet. 2015, 128, 73–91. [Google Scholar] [CrossRef]
- Bac-Molenaar, J.A.; Vreugdenhil, D.; Granier, C.; Keurentjes, J.J.B. Genome-Wide Association Mapping of Growth Dynamics Detects Time-Specific and General Quantitative Trait Loci. J. Exp. Bot. 2015, 66, 5567–5580. [Google Scholar] [CrossRef]
- Pantalião, G.F.; Narciso, M.; Guimarães, C.; Castro, A.; Colombari, J.M.; Breseghello, F.; Rodrigues, L.; Vianello, R.P.; Borba, T.O.; Brondani, C. Genome Wide Association Study (GWAS) for Grain Yield in Rice Cultivated under Water Deficit. Genetica 2016, 144, 651–664. [Google Scholar] [CrossRef]
- Zhang, J.; Mason, A.S.; Wu, J.; Liu, S.; Zhang, X.; Luo, T.; Redden, R.; Batley, J.; Hu, L.; Yan, G. Identification of Putative Candidate Genes for Water Stress Tolerance in Canola (Brassica napus). Front. Plant Sci. 2015, 6, 1058. [Google Scholar] [CrossRef]
- Kang, Y.; Sakiroglu, M.; Krom, N.; Stanton-Geddes, J.; Wang, M.; Lee, Y.-C.; Young, N.D.; Udvardi, M. Genome-Wide Association of Drought-Related and Biomass Traits with HapMap SNPs in Medicago Truncatula. Plant Cell Environ. 2015, 38, 1997–2011. [Google Scholar] [CrossRef]
- Hoyos-Villegas, V. Identification of Genomic Regions and Development of Breeding Resources Associated with Drought Tolerance in Common Bean (Phaseolus vulgaris L.). Ph.D. Thesis, Michigan State University, East Lansing, MI, USA, 2015. [Google Scholar]
- Wang, N.; Wang, Z.; Liang, X.; Weng, J.; Lv, X.; Zhang, D.; Yang, J.; Yong, H.; Li, M.; Li, F.; et al. Identification of Loci Contributing to Maize Drought Tolerance in a Genome-Wide Association Study. Euphytica 2016, 210, 165–179. [Google Scholar] [CrossRef]
- Price, A.H. Believe It or Not, QTLs Are Accurate! Trends Plant Sci. 2006, 11, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, M.T.; Buckler, E.S.; Jannink, J.-L. Population Genetics of Genomics-Based Crop Improvement Methods. Trends Genet. 2011, 27, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Jalal, A.; Rauf, K.; Iqbal, B.; Khalil, R.; Mustafa, H.; Murad, M.; Khalil, F.; Khan, S.; Oliveira, C.E.d.S.; Filho, M.C.M.T. Engineering Legumes for Drought Stress Tolerance: Constraints, Accomplishments, and Future Prospects. S. Afr. J. Bot. 2023, 159, 482–491. [Google Scholar] [CrossRef]
- Sansa, O.O.; Ariyo, O.J.; Ayo-Vaughan, M.A.; Ekanem, U.O.; Ntukidem, S.O.; Abberton, M.T.; Oyatomi, O.A. Genetic Variability, Inter-Character Correlation, and Stability Performance in Cowpea for Drought Tolerance. J. Crop Improv. 2025, 39, 43–68. [Google Scholar] [CrossRef]
- Ibrahim, T.A.A.; Abdel-Ati, K.E.A.; Khaled, K.A.M.; Azoz, S.N.; Hassan, A.A. Physiological, Molecular and Anatomical Studies on Drought Tolerance in Cowpea. Egypt. J. Soil Sci. 2025, 65, 253–274. [Google Scholar] [CrossRef]
- Wu, X.; Sun, T.; Xu, W.; Sun, Y.; Wang, B.; Wang, Y.; Li, Y.; Wang, J.; Wu, X.; Lu, Z.; et al. Unraveling the Genetic Architecture of Two Complex, Stomata-Related Drought-Responsive Traits by High-Throughput Physiological Phenotyping and GWAS in Cowpea (Vigna unguiculata L. Walp). Front Genet 2021, 12, 743758. [Google Scholar] [CrossRef]
- Ding, M.; Tao, Y.; Hua, J.; Dong, Y.; Lu, S.; Qiang, J.; He, J. Genome-Wide Association Study Reveals Growth-Related SNPs and Candidate Genes in Largemouth Bass (Micropterus salmoides) Adapted to Hypertonic Environments. Int. J. Mol. Sci. 2025, 26, 1834. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, X.; Li, J.; Ning, X.; Xu, S.; Liu, P.; Guo, X.; Yuan, W.; Xie, B.; Wang, F.; et al. Identification of Elite Alleles and Candidate Genes for the Cotton Boll Opening Rate via a Genome-Wide Association Study. Int. J. Mol. Sci. 2025, 26, 2697. [Google Scholar] [CrossRef]
- Fang, W.-P.; Meinhardt, L.W.; Tan, H.-W.; Zhou, L.; Mischke, S.; Zhang, D. Varietal Identification of Tea (Camellia sinensis) Using Nanofluidic Array of Single Nucleotide Polymorphism (SNP) Markers. Hortic. Res. 2014, 1, 14035. [Google Scholar] [CrossRef]
- Singh, B.; Mai-Kodomi, Y.; Terao, T. A Simple Screening Method for Drought Tolerance in Cowpea. Indian J. Genet. Plant Breed. 1999, 59, 211–220. [Google Scholar]
- Iuchi, S.; Yamaguchi-Shinozaki, K.; Urao, T.; Terao, T.; Shinozaki, K. Novel Drought-Inducible Genes in the Highly Drought-Tolerant Cowpea: Cloning of cDNAs and Analysis of the Expression of the Corresponding Genes. Plant Cell Physiol. 1996, 37, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A Stress-Inducible Gene for 9-cis-Epoxycarotenoid Dioxygenase Involved in Abscisic Acid Biosynthesis under Water Stress in Drought-Tolerant Cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef] [PubMed]
- El Maarouf, H.; Zuily-Fodil, Y.; Gareil, M.; D’Arcy-Lameta, A.; Thu Pham-Thi, A. Enzymatic Activity and Gene Expression under Water Stress of Phospholipase D in Two Cultivars of Vigna unguiculata L.Walp. Differing in Drought Tolerance. Plant Mol. Biol. 1999, 39, 1257–1265. [Google Scholar] [CrossRef]
- de Paula, F.M.; Thi, A. Effects of Water Stress on the Biosynthesis and Degradation of Polyunsaturated Lipid Molecular Species in Leaves of Vigna unguiculata. Plant Physiol. Biochem. 1993, 31, 707–715. [Google Scholar]
- Marcel, G.C.F.; Matos, A.; D’Arcy-Lameta, A.; Kader, J.-C.; Zuily-Fodil, J.Y.; Pham-Thi, A. Two Novel Plant cDNAs Homologous to Animal Type-2 Phosphatidate Phosphatase Are Expressed in Cowpea Leaves and Are Differently Regulated by Water Deficits. Biochem Soc Trans 2000, 28, 915–917. [Google Scholar] [CrossRef]
- Sahsah, Y.; Campos, P.; Gareil, M.; Zuily-Fodil, Y.; Pham-Thi, A.T. Enzymatic Degradation of Polar Lipids in Vigna unguiculata Leaves and Influence of Drought Stress. Physiol. Plant. 1998, 104, 577–586. [Google Scholar] [CrossRef]
- Matos, A.R.; D’Arcy-Lameta, A.; França, M.; Pêtres, S.; Edelman, L.; Kader, J.-C.; Zuily-Fodil, Y.; Pham-Thi, A.T. A Novel Patatin-like Gene Stimulated by Drought Stress Encodes a Galactolipid Acyl Hydrolase. FEBS Lett. 2001, 491, 188–192. [Google Scholar] [CrossRef]
- Carvalho, M.; Lino-Neto, T.; Rosa, E.; Carnide, V. Cowpea: A Legume Crop for a Challenging Environment. J. Sci. Food Agric. 2017, 97, 4273–4284. [Google Scholar] [CrossRef]
- Diop, N.N.; Kidrič, M.; Repellin, A.; Gareil, M.; D’Arcy-Lameta, A.; Pham Thi, A.T.; Zuily-Fodil, Y. A Multicystatin Is Induced by Drought-Stress in Cowpea (Vigna unguiculata (L.) Walp.) Leaves. FEBS Lett. 2004, 577, 545–550. [Google Scholar] [CrossRef]
- D’Arcy-Lameta, A.; Ferrari-Iliou, R.; Contour-Ansel, D.; Pham-Thi, A.-T.; Zuily-Fodil, Y. Isolation and Characterization of Four Ascorbate Peroxidase cDNAs Responsive to Water Deficit in Cowpea Leaves. Ann. Bot. 2005, 97, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Gazendam, I.; Oelofse, D. Isolation of Cowpea Genes Conferring Drought Tolerance: Construction of a cDNA Drought Expression Library. Water SA 2009, 33, 387–392. [Google Scholar] [CrossRef]
- da Silva, H.A.P.; Galisa, P. de S.; Oliveira, R.S. da S.; Vidal, M.S.; Simões-Araújo, J.L. Gene Expression Induced by Abiotic Stress in Cowpea Nodules. Pesqui Agropecu Bras 2012, 47, 797–807. (In Spanish) [Google Scholar] [CrossRef]
- Barrera-Figueroa, B.; Gao, L. Identification and Comparative Analysis of Drought-Associated MicroRNAs in Two Cowpea Genotypes. BMC Plant Biol. 2011, 11, 127. [Google Scholar] [CrossRef]
- Shui, X.-R.; Chen, Z.-W.; Li, J.-X. MicroRNA Prediction and Its Function in Regulating Drought-Related Genes in Cowpea. Plant Sci. 2013, 210, 25–35. [Google Scholar] [CrossRef]
- Verbree, D.A.; Singh, B.B.; Payne, W.A. Genetics and Heritability of Shoot Drought Tolerance in Cowpea Seedlings. Crop Sci. 2015, 55, 146–153. [Google Scholar] [CrossRef]
- Huynh, B.-L.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Registration of a Cowpea [Vigna unguiculata (L.) Walp.] Multiparent Advanced Generation Intercross (MAGIC) Population. J. Plant Regist. 2019, 13, 281–286. [Google Scholar] [CrossRef]
- Ravelombola, W.; Shi, A.; Qin, J.; Weng, Y.; Bhattarai, G.; Zia, B.; Zhou, W.; Mou, B. Investigation on Various Aboveground Traits to Identify Drought Tolerance in Cowpea Seedlings. HortScience 2018, 53, 1757–1765. [Google Scholar] [CrossRef]
- Kisha, T.J.; Sneller, C.H.; Diers, B.W. Relationship between Genetic Distance among Parents and Genetic Variance in Populations of Soybean. Crop Sci. 1997, 37, 1317–1325. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten Years of Next-Generation Sequencing Technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.; et al. The Genome of Cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An Improved Ultrafast Tool for Short Read Alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A Package for the next Level of Genome-Wide Association Studies with Both Individuals and Markers in the Millions. GigaScience 2019, 8, giy154. [Google Scholar] [CrossRef] [PubMed]





| Traits | SNP | BP | CHR | Pval | LOD | Gene_ID | Functional_Annotation |
|---|---|---|---|---|---|---|---|
| Trifoliate leaf chlorosis | Vu01_10616309 | 10616309 | 1 | 6.54 × 10−9 | 8.18 | Vign01g055000.1 | Mn plant transporter |
| Vu01_25892353 | 25892353 | 1 | 3.74 × 10−9 | 8.43 | Vign01g09400.1 | NADH-cytochrome b5 reductase | |
| Vu02_24537084 | 24537084 | 2 | 1.12 × 10−8 | 7.95 | Vign02090500.1 | Retrotransposon | |
| Vu03_10803196 | 10803196 | 3 | 2.98 × 10−8 | 7.53 | Vign03g116500.1 | PB1 domain containing protein | |
| Vu03_12666912 | 12666912 | 3 | 2.04 × 10−8 | 7.69 | Vigun03g130400.1 | Protein phosphatase 2C | |
| Vu03_12897768 | 12897768 | 3 | 1.55 × 10−8 | 7.81 | Vign03g132300.1 | Pumilio-family RNA binding repeat | |
| Vu03_13212508 | 13212508 | 3 | 2.99 × 10−8 | 7.52 | Vign03g135100.1 | ATP-dependent DNA helicase 2 subunit 2 | |
| Vu03_13274473 | 13274473 | 3 | 1.89 × 10−8 | 7.72 | Vign03g135400.1 | RNA methylase-related | |
| Vu03_13295491 | 13295491 | 3 | 5.40 × 10−10 | 9.27 | Vign03g135700.1 | Vacuolar iron transporter | |
| Vu03_13297714 | 13297714 | 3 | 5.68 × 10−12 | 11.25 | Vign03g135800.1 | Vacuolar iron transporter | |
| Vu03_13302250 | 13302250 | 3 | 1.84 × 10−11 | 10.74 | Vign03g135900.1 | Vacuolar iron transporter | |
| Vu03_13352276 | 13352276 | 3 | 1.16 × 10−9 | 8.93 | Vigun03g136300.1 | EamA-like transporter family/Glucose-6-phosphate/phosphate and phosphoenolpyruvate/phosphate antiporter | |
| Vu03_13361294 | 13361294 | 3 | 5.04 × 10−10 | 9.30 | Vigun03g136400.1 | EamA-like transporter family/Glucose-6-phosphate/phosphate and phosphoenolpyruvate/phosphate antiporter | |
| Vu03_13376628 | 13376628 | 3 | 1.96 × 10−10 | 9.71 | Vigun03g136500.1 | EamA-like transporter family/Glucose-6-phosphate/phosphate and phosphoenolpyruvate/phosphate antiporter | |
| Vu03_13382599 | 13382599 | 3 | 2.55 × 10−10 | 9.59 | Vigun03g136600.1 | EamA-like transporter family/auxin-induced protein 5NG4 | |
| Vu03_13509429 | 13509429 | 3 | 5.66 × 10−11 | 10.25 | Vigun03g137500.1 | ABA responsive element binding factor | |
| Vu03_14318570 | 14318570 | 3 | 9.08 × 10−10 | 9.04 | Vigun03g142100.1 | Tetrahydroberberine oxidase | |
| Vu03_14743049 | 14743049 | 3 | 5.21 × 10−9 | 8.28 | Vigun03g144800.1 | WRKY TRANSCRIPTION FACTOR 28-RELATED | |
| Vu03_14763808 | 14763808 | 3 | 2.88 × 10−8 | 7.54 | Vigun03g144900.1 | O-acetyltransferase family protein | |
| Vu03_14806565 | 14806565 | 3 | 2.32 × 10−9 | 8.63 | Vigun03g145200.1 | Starch synthase | |
| Vu03_14815803 | 14815803 | 3 | 1.63 × 10−9 | 8.79 | Vigun03g145400.1 | Chlorophyll a/b binding protein | |
| Vu03_15222570 | 15222570 | 3 | 2.69 × 10−8 | 7.57 | Vigun03g148300.1 | 3-oxoacyl-synthase | |
| Vu03_36340055 | 36340055 | 3 | 2.93 × 10−9 | 8.53 | Vigun03g218100.1 | ABC-2 type transporter family protein | |
| Vu03_58980712 | 58980712 | 3 | 3.74 × 10−9 | 8.43 | Vigun03g384600.1 | H+-transporting ATPase | |
| Vu04_26966450 | 26966450 | 4 | 4.24 × 10−9 | 8.37 | Vigun04g109300.1 | CemA-like proton extrusion protein-related | |
| Vu04_27157237 | 27157237 | 4 | 6.15 × 10−9 | 8.21 | Vigun04g109500.1 | Protein FLOWERING LOCUS T (FT) | |
| Vu04_27241963 | 27241963 | 4 | 4.99 × 10−9 | 8.30 | Vigun04g109600.1 | TatD DNase family protein | |
| Vu04_27298716 | 27298716 | 4 | 5.99 × 10−9 | 8.22 | Vigun04g109700.1 | Aquaporin-like superfamily protein | |
| Vu04_27342140 | 27342140 | 4 | 2.73 × 10−9 | 8.56 | Vigun04g109800.1 | Nucleoside-triphosphatase | |
| Vu04_27505387 | 27505387 | 4 | 3.11 × 10−9 | 8.51 | Vigun04g110000.1 | rRNA-processing protein FCF | |
| Vu04_27528973 | 27528973 | 4 | 7.92 × 10−9 | 8.10 | Vigun04g110100.1 | Ubiquinone (electron-transporting coenzyme) biosynthesis protein | |
| Vu04_27714135 | 27714135 | 4 | 1.91 × 10−9 | 8.72 | Vigun04g110600.1 | No apical meristem (NAM) protein | |
| Vu04_27716250 | 27716250 | 4 | 4.48 × 10−9 | 8.35 | Vigun04g110700.1 | Tryptophan/tyrosine permease family | |
| Vu04_27778870 | 27778870 | 4 | 2.16 × 10−8 | 7.67 | Vigun04g110800.1 | Myb family transcription factor-related | |
| Vu04_27786623 | 27786623 | 4 | 8.27 × 10−10 | 9.08 | Vigun04g110900.1 | Pyruvate kinase | |
| Vu04_27797389 | 27797389 | 4 | 4.32 × 10−9 | 8.37 | Vigun04g111000.1 | Zinc finger protein | |
| Vu04_27830859 | 27830859 | 4 | 1.57 × 10−8 | 7.81 | Vigun04g111100.1 | CCR4-NOT transcription factor | |
| Vu04_27913211 | 27913211 | 4 | 1.59 × 10−8 | 7.80 | Vigun04g111200.1 | Glucan endo-1,3-beta-D-glucosidase | |
| Vu04_27913980 | 27913980 | 4 | 8.65 × 10−9 | 8.06 | Vigun04g111300.1 | GATA zinc finger | |
| Vu04_41785910 | 41785910 | 4 | 3.13 × 10−9 | 8.50 | Vigun04g193600.1 | Protein kinase superfamily | |
| Vu04_41800041 | 41800041 | 4 | 2.14 × 10−8 | 7.67 | Vigun04g193700.1 | NAD-dependent epimerase/dehydratase | |
| Vu04_41826262 | 41826262 | 4 | 7.69 × 10−9 | 8.11 | Vigun04g193800.1 | Translation initiation factor 3 subunit G | |
| Vu04_41832927 | 41832927 | 4 | 8.14 × 10−9 | 8.09 | Vigun04g194000.1 | Universal stress protein family | |
| Vu05_540561 | 540561 | 5 | 5.09 × 10−21 | 20.29 | Vigun05g006300.1 | EamA-like transporter family/auxin-induced protein 5NG4 | |
| Vu05_560665 | 560665 | 5 | 5.63 × 10−15 | 14.25 | Vigun05g006500.1 | Neoxanthin synthase/abscisic acid biosynthesis | |
| Vu07_23856082 | 23856082 | 7 | 5.11 × 10−10 | 9.29 | Vigun07g129600.1 | Protein tyrosine kinase | |
| Vu07_24143183 | 24143183 | 7 | 7.91 × 10−9 | 8.10 | Vigun07g131800.1 | Protoporphyrinogen oxidase/chloroplast precursor | |
| Vu08_37171764 | 37171764 | 8 | 2.43 × 10−8 | 7.61 | Vigun08g208700.1 | PPR repeat | |
| Unifoliate leaf chlorosis | Vu01_29544191 | 29544191 | 1 | 3.51 × 10−15 | 14.45 | Vigun01g119000.1 | Lysophosphatidic acid acyltransferase |
| Vu08_4931701 | 4931701 | 8 | 4.83 × 10−9 | 8.32 | Vigun08g046200.1 | Leucine-rich repeat | |
| Vu08_4945627 | 4945627 | 8 | 2.46 × 10−10 | 9.61 | Vigun08g046400.1 | Leucine-rich repeat | |
| Vu08_4952526 | 4952526 | 8 | 4.61 × 10−10 | 9.34 | Vigun08g046500.1 | Leucine-rich repeat | |
| Vu08_26752606 | 26752606 | 8 | 2.58 × 10−11 | 10.59 | Vigun08g107800.1 | Ubiquitin carboxyl-terminal hydrolase | |
| Vu08_26868733 | 26868733 | 8 | 1.49 × 10−9 | 8.83 | Vigun08g107900.1 | AT-hook DNA-binding family protein | |
| Vu08_26877485 | 26877485 | 8 | 8.31 × 10−11 | 10.08 | Vigun08g108100.1 | Carbonic anhydrase | |
| Vu08_26901689 | 26901689 | 8 | 9.11 × 10−11 | 10.04 | Vigun08g108400.1 | DnaJ homolog subfamily | |
| Vu10_35348050 | 35348050 | 10 | 2.58 × 10−9 | 8.59 | Vigun10g137100.1 | Leucine-rich repeat | |
| Plant greenness | Vu01_10616486 | 10616486 | 1 | 7.42 × 10−9 | 8.13 | Vigun01g054900.1 | DCN1-like protein |
| Vu03_13509429 | 13509429 | 3 | 2.63 × 10−8 | 7.58 | Vigun03g137600.1 | P-loop containing nucleoside triphosphate hydrolase superfamily protein | |
| Vu03_14725438 | 14725438 | 3 | 1.65 × 10−8 | 7.78 | Vigun03g144800.1 | WRKY transcription factor | |
| Vu05_541044 | 541044 | 5 | 2.40 × 10−9 | 8.62 | Vigun05g006300.1 | EamA-like transporter family/auxin-induced protein 5NG4 |
| Traits | Gene_ID | Functional_Annotations | Vun | Gma | Pvu | Mtr |
|---|---|---|---|---|---|---|
| Trifoliate leaf chlorosis | Vigun05g006300.1 | EamA-like transporter family/auxin-induced protein 5NG4 | 1 | 3 | 4 | 1 |
| Vigun05g006500.1 | Neoxanthin synthase/abscisic acid biosynthesis | 1 | 3 | 3 | 3 | |
| Vigun03g136600.1 | EamA-like transporter family/auxin-induced protein 5NG4 | 2 | 5 | 3 | 3 | |
| Vigun03g137500.1 | ABA responsive element binding factor | 0 | 1 | 1 | 0 | |
| Vigun03g135700.1 | Vacuolar iron transporter | 0 | 1 | 1 | 0 | |
| Vigun03g135800.1 | Vacuolar iron transporter | 4 | 9 | 4 | 8 | |
| Vigun03g135900.1 | Vacuolar iron transporter | 6 | 7 | 7 | 4 | |
| Vigun04g110600.1 | No apical meristem (NAM) protein | 1 | 4 | 2 | 0 | |
| Vigun04g110800.1 | Myb family transcription factor-related | 0 | 2 | 1 | 1 | |
| Uniifoliate leaf chlorosis | Vigun01g119000.1 | Lysophosphatidic acid acyltransferase | 1 | 4 | 2 | 2 |
| Vigun08g046200.1 | Leucine-rich repeat | 2 | 2 | 6 | 0 | |
| Vigun08g046400.1 | Leucine-rich repeat | 0 | 1 | 4 | 0 | |
| Vigun08g046500.1 | Leucine-rich repeat | 1 | 2 | 4 | 0 | |
| Vigun08g107800.1 | Ubiquitin carboxyl-terminal hydrolase | 0 | 0 | 1 | 0 | |
| Vigun08g108100.1 | Carbonic anhydrase/Carbonate dehydratase | 0 | 1 | 1 | 1 | |
| Vigun08g108400.1 | DnaJ homolog subfamily | 0 | 2 | 1 | 1 | |
| Vigun10g137100.1 | Leucine-rich repeat | 0 | 0 | 0 | 0 | |
| Plant greenness | Vigun01g054900.1 | DCN1-like protein | 0 | 2 | 1 | 1 |
| Vigun03g137600.1 | P-loop containing nucleoside triphosphate hydrolase superfamily protein | 0 | 1 | 0 | 0 | |
| Vigun03g144800.1 | WRKY transcription factor | 0 | 2 | 1 | 0 | |
| Vigun05g006300.1 | EamA-like transporter family/auxin-induced protein 5NG4 | 1 | 3 | 3 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravelombola, W.; Xiong, H.; Bhattarai, G.; Manley, A.; Cason, J.; Pham, H.; Zia, B.; Mou, B.; Shi, A. Genome-Wide Association Study for Drought Tolerance in Cowpea (Vigna unguiculata (L.) Walp.) at Seedling Stage Using a Whole Genome Resequencing Approach. Int. J. Mol. Sci. 2025, 26, 5478. https://doi.org/10.3390/ijms26125478
Ravelombola W, Xiong H, Bhattarai G, Manley A, Cason J, Pham H, Zia B, Mou B, Shi A. Genome-Wide Association Study for Drought Tolerance in Cowpea (Vigna unguiculata (L.) Walp.) at Seedling Stage Using a Whole Genome Resequencing Approach. International Journal of Molecular Sciences. 2025; 26(12):5478. https://doi.org/10.3390/ijms26125478
Chicago/Turabian StyleRavelombola, Waltram, Haizheng Xiong, Gehendra Bhattarai, Aurora Manley, John Cason, Hanh Pham, Bazgha Zia, Beiquan Mou, and Ainong Shi. 2025. "Genome-Wide Association Study for Drought Tolerance in Cowpea (Vigna unguiculata (L.) Walp.) at Seedling Stage Using a Whole Genome Resequencing Approach" International Journal of Molecular Sciences 26, no. 12: 5478. https://doi.org/10.3390/ijms26125478
APA StyleRavelombola, W., Xiong, H., Bhattarai, G., Manley, A., Cason, J., Pham, H., Zia, B., Mou, B., & Shi, A. (2025). Genome-Wide Association Study for Drought Tolerance in Cowpea (Vigna unguiculata (L.) Walp.) at Seedling Stage Using a Whole Genome Resequencing Approach. International Journal of Molecular Sciences, 26(12), 5478. https://doi.org/10.3390/ijms26125478






