RNA-Seq Uncovers Association of Endocrine-Disrupting Chemicals with Hub Genes and Transcription Factors in Aggressive Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. Discovery of Differentially Expressed Genes (DEGs) in RNA-Seq for PCa
2.2. RNA-Seq DEG Enrichment Analysis: GO, KEGG, and TFs
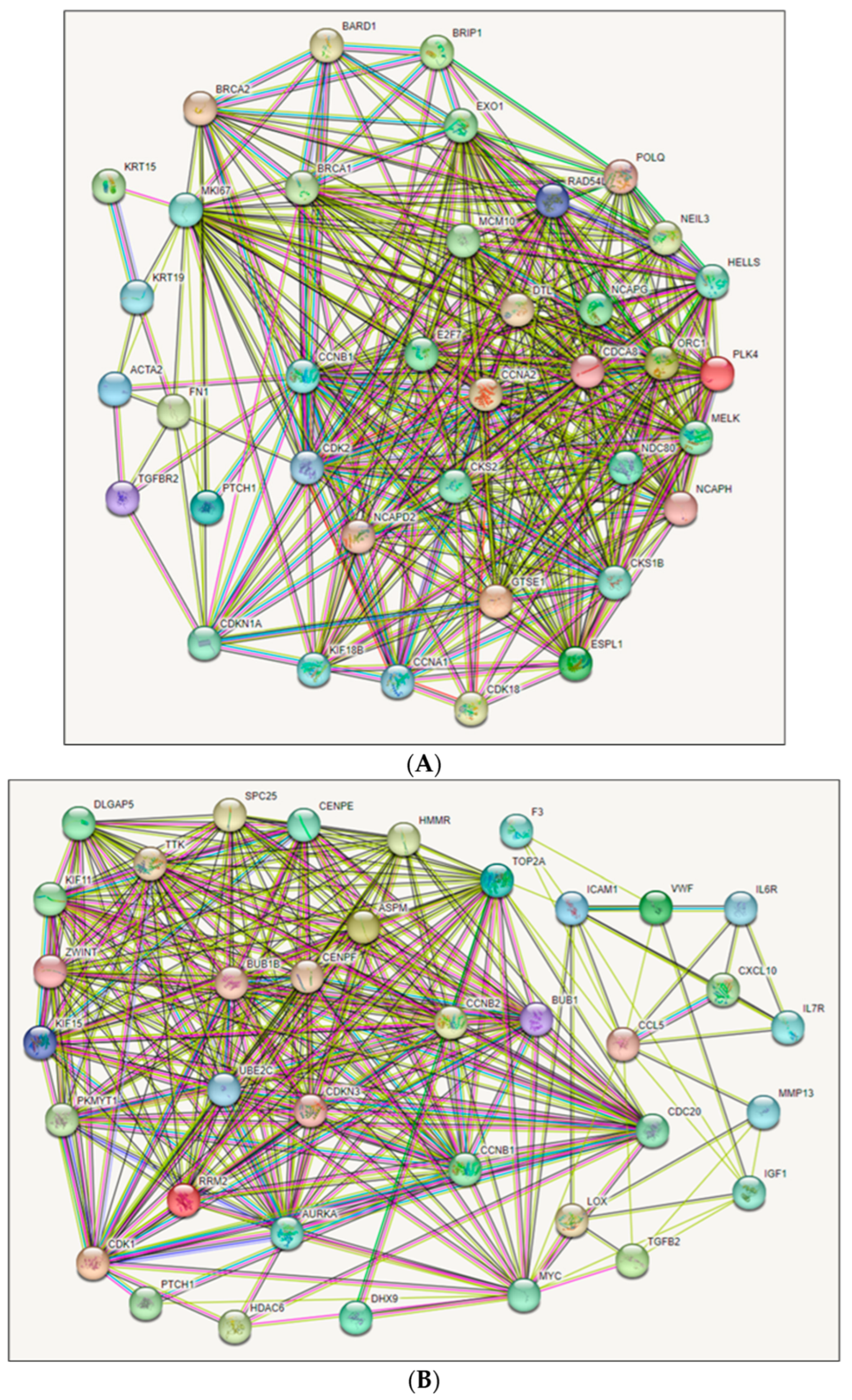
2.3. Protein–Protein Interaction (PPI) Networks and Module Selection Analysis
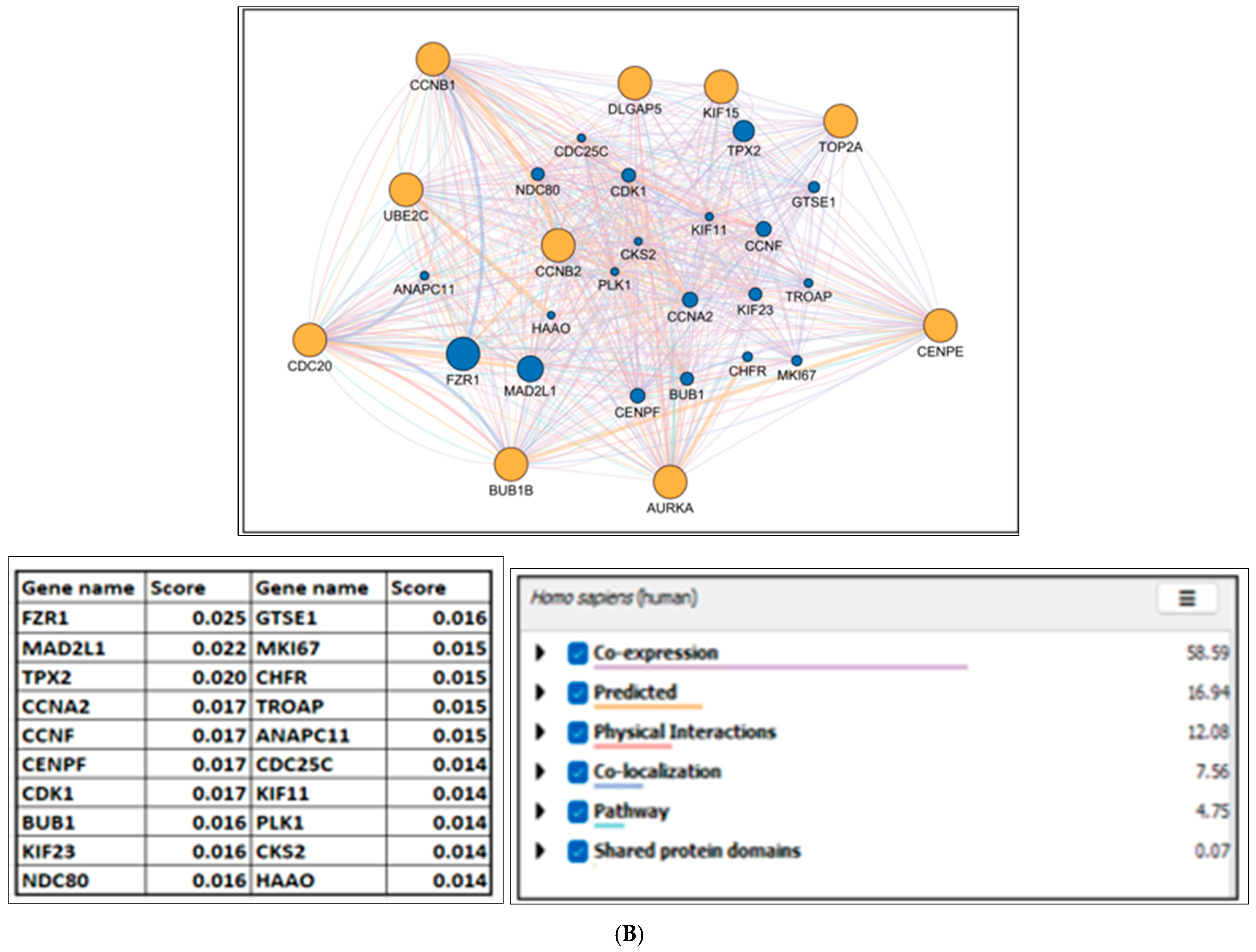
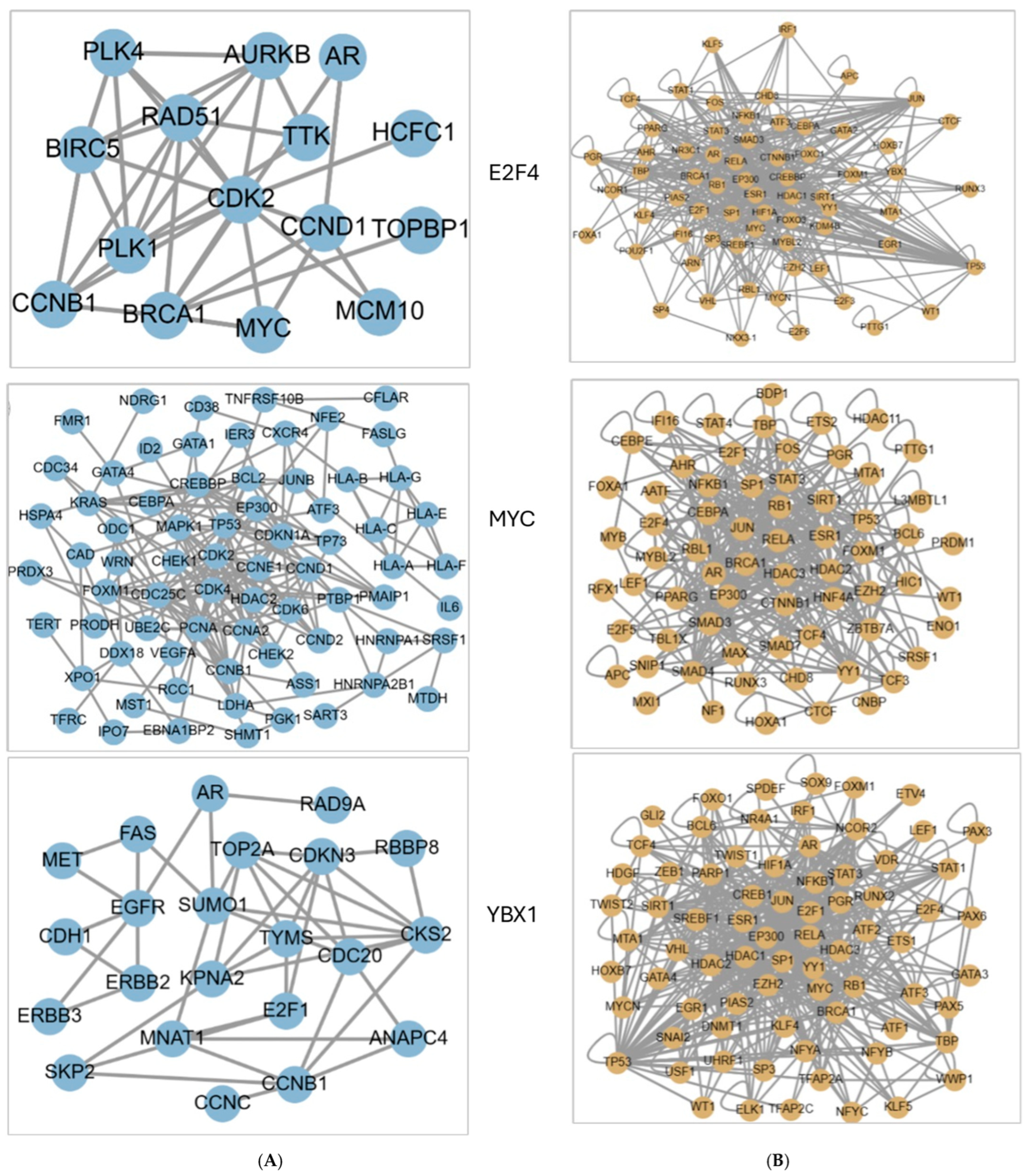
2.4. Hub Gene Identification
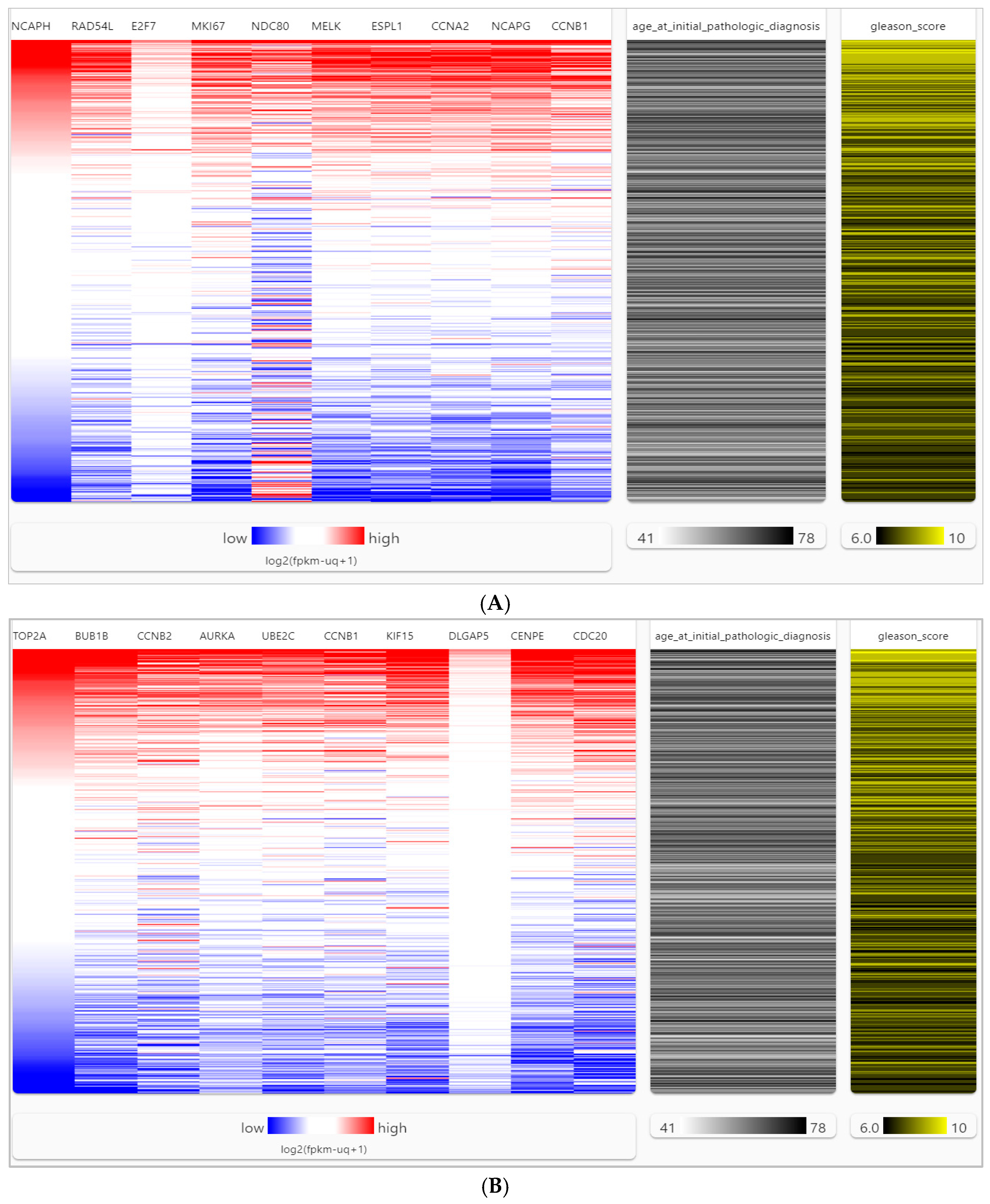
2.5. Genetic Alteration and Expression of Hub Genes
2.6. Hub Gene and Immune Cell Infiltration Estimations
2.7. GlueGO and GluePedia
2.8. Hub Gene Redundancy and Dependency Analysis
2.9. Enrichr and Hub Gene Enrichment Analysis
2.10. Regulatory TFs and Their Associated Network with Hub Genes
2.11. Comparison of Phenotypes for Hub Genes and TFs
3. Discussion
4. Materials and Methods
4.1. Omics Data Acquisition
4.2. Data Processing
4.3. Statistical Analysis
4.4. Identification of DEGs and Pathway Enrichment Analysis
4.5. Biological System Analysis and Module Network Mining
4.6. Genetic Alteration in Hub Genes
4.7. Visualizing the Heatmap and Hub Gene Expressions
4.8. Hub Genes’ Association with Immune Cell Infiltration
4.9. ClueGO and CluePedia: Functional Enrichment Analysis
4.10. Redundancy Analyses of Hub Genes
4.11. Hub Gene Enrichment Analysis—Enrichr
4.12. TF Association Network with Hub Genes
4.13. Causal Association and Protein Interactions and Phenotypes with DEGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADT | Androgen Deprivation Therapy |
| AR | Androgen Receptor |
| BP | Biological Process |
| cBioPortal | cBio Cancer Genomics Portal |
| CC | Cellular Component |
| ChEA | Chip Enrichment Analysis |
| CT | Computed Tomography |
| DAVID | Database for Annotation, Visualization, and Integrated Discovery |
| DEGs | Differentially Expressed Genes |
| DHT | Dihydrotestosterone |
| DOIDs | Disease Ontology Identifiers |
| DNMC | Degree, Density of Maximum Neighborhood Component |
| E2F4 | E2F Transcription Factor 4 |
| EDCs | Endocrine-Disrupting Chemicals |
| EPC | Edge Percolated Component |
| FDR | False Discovery Rate |
| FPKM | Fragments Per Kilobase of transcript per Million |
| GDC | Genomic Data Commons |
| GO | Gene Ontology |
| GEO | Expression Omnibus database |
| GISTIC | Genomic Identification of Significant Targets in Cancer |
| GTEx | Genotype-Tissue Expression |
| ICGC | International Cancer Genome Consortium |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KM | Kaplan–Meier |
| MCC | Maximal Clique Centrality |
| MCODE | Molecular Complex Detection |
| MF | Molecular Function |
| MNC | Maximum Neighborhood Component |
| MRI | Magnetic Resonance Imaging |
| MSK | Memorial Sloan Kettering Cancer Center |
| MYC | MYC Proto-Oncogene, BHLH Transcription Factor |
| NFY | Nuclear transcription factor Y |
| NCBI | National Center for Biotechnology Information |
| NHANES | National Health and Nutrition Examination Survey |
| PCa | Prostate Cancer |
| PEI | Pathway Enrichment Analysis |
| PPI | Protein–Protein Interaction |
| PSA | Prostate-Specific Antigen |
| SIGNOR | SIGnaling Network Open Resource |
| STRING | Retrieval of Interacting Genes/Proteins |
| TCGA | The Cancer Genome Atlas |
| TFs | Transcription Factors |
| TIMER 2.0 | Tumor Immunity Evaluation Resource 2.0 |
| TRRUST | Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining |
| UCSC_TFBS | University of California, Santa Cruz—transcription factor binding sites |
| UCSC-Xena | University of California Santa Cruz—Xena |
| YBX1 | Y box binding protein 1 |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Viale, P.H. The American Cancer Society’s Facts & Figures: 2022 Edition. J. Adv. Pract. Oncol. 2020, 11, 135–136. [Google Scholar] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Packer, J.R.; Maitland, N.J. The molecular and cellular origin of human prostate cancer. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 1238–1260. [Google Scholar] [CrossRef]
- Tagai, E.K.; Miller, S.M.; Kutikov, A.; Diefenbach, M.A.; Gor, R.A.; Al-Saleem, T.; Chen, D.Y.; Fleszar, S.; Roy, G. Prostate Cancer Patients’ Understanding of the Gleason Scoring System: Implications for Shared Decision-Making. J. Cancer Educ. 2018, 34, 441–445. [Google Scholar] [CrossRef]
- Messner, E.A.; Steele, T.M.; Tsamouri, M.M.; Hejazi, N.; Gao, A.C.; Mudryj, M.; Ghosh, P.M. The androgen receptor in prostate cancer: Effect of structure, ligands and spliced variants on therapy. Biomedicines 2020, 8, 422. [Google Scholar] [CrossRef]
- Centenera, M.M.; Selth, L.A.; Ebrahimie, E.; Butler, L.M.; Tilley, W.D. New Opportunities for Targeting the Androgen Receptor in Prostate Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a030478. [Google Scholar] [CrossRef]
- Cai, C.; He, H.H.; Chen, S.; Coleman, I.; Wang, H.; Fang, Z.; Chen, S.; Nelson, P.S.; Liu, X.S.; Brown, M.; et al. Androgen Receptor Gene Expression in Prostate Cancer Is Directly Suppressed by the Androgen Receptor Through Recruitment of Lysine-Specific Demethylase 1. Cancer Cell 2011, 20, 457–471. [Google Scholar] [CrossRef]
- Saltman, A.; Zegar, J.; Haj-Hamed, M.; Verma, S.; Sidana, A. Prostate cancer biomarkers and multiparametric MRI: Is there a role for both in prostate cancer management? Ther. Adv. Urol. 2021, 13, 1756287221997186. [Google Scholar] [CrossRef]
- Wang, R.; Xiao, Y.; Pan, M.; Chen, Z.; Yang, P. Integrative Analysis of Bulk RNA-Seq and Single-Cell RNA-Seq Unveils the Characteristics of the Immune Microenvironment and Prognosis Signature in Prostate Cancer. J. Oncol. 2022, 2022, 1–28. [Google Scholar] [CrossRef]
- Draisma, G.; Etzioni, R.; Tsodikov, A.; Mariotto, A.; Wever, E.; Gulati, R.; Feuer, E.; de Koning, H. Lead Time and Overdiagnosis in Prostate-Specific Antigen Screening: Importance of Methods and Context. J. Natl. Cancer Inst. 2009, 101, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Catalona, W.J. Prostate Cancer Screening. Med. Clin. N. Am. 2018, 102, 199–214. [Google Scholar] [CrossRef]
- Moyer, V.A. Screening for prostate cancer: U.S. preventive services task force recommendation statement. Ann. Intern. Med. 2012, 157, 120–134. [Google Scholar] [CrossRef]
- Gu, P.; Yang, D.; Zhu, J.; Zhang, M.; He, X. Bioinformatics analysis identified hub genes in prostate cancer tumorigenesis and metastasis. Math. Biosci. Eng. MBE 2021, 18, 3180–3196. [Google Scholar] [CrossRef]
- Endo, T.; Uzawa, K.; Suzuki, H.; Tanzawa, H.; Ichikawa, T. Characteristic gene expression profiles of benign prostatic hypertrophy and prostate cancer. Int. J. Oncol. 2009, 35, 499–509. [Google Scholar] [PubMed]
- Sun, J.; Li, S.; Wang, F.; Fan, C.; Wang, J. Identification of key pathways and genes in pten mutation prostate cancer by bioinformatics analysis. BMC Med. Genet. 2019, 20, 191. [Google Scholar] [CrossRef]
- Song, F.; Zhang, Y.; Pan, Z.; Hu, X.; Yi, Y.; Zheng, X.; Wei, H.; Huang, P. Identification of novel key genes associated with the metastasis of prostate cancer based on bioinformatics prediction and validation. Cancer Cell Int. 2021, 21, 559. [Google Scholar] [CrossRef]
- Liu, K.; Chen, Y.; Feng, P.; Wang, Y.; Sun, M.; Song, T.; Tan, J.; Li, C.; Liu, S.; Kong, Q.; et al. Identification of Pathologic and Prognostic Genes in Prostate Cancer Based on Database Mining. Front. Genet. 2022, 13, 854531. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Ding, Z. Identification of key genes in prostate cancer gene expression profile by bioinformatics. Andrologia 2019, 51, e13169. [Google Scholar] [CrossRef]
- Song, Z.; Chao, F.; Zhuo, Z.; Ma, Z.; Li, W.; Chen, G. Identification of hub genes in prostate cancer using robust rank aggregation and weighted gene co-expression network analysis. Aging 2019, 11, 4736–4756. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Yan, K.; Lin, J.; Zheng, Z.; Bi, J. Identification of core genes associated with prostate cancer progression and outcome via bioinformatics analysis in multiple databases. PeerJ 2020, 2020, e8786. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Song, Y.; Deng, S. Combined analysis and validation for DNA methylation and gene expression profiles associated with prostate cancer. Cancer Cell Int. 2019, 19, 50. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Jin, X.; Wang, K. Integrated Bioinformatics Analysis of Potential Biomarkers for Prostate Cancer. Pathol. Oncol. Res. 2017, 25, 455–460. [Google Scholar] [CrossRef]
- He, Z.; Duan, X.; Zeng, G. Identification of potential biomarkers and pivotal biological pathways for prostate cancer using bioinformatics analysis methods. PeerJ 2019, 2019, e7872. [Google Scholar] [CrossRef]
- Song, Z.; Huang, Y.; Zhao, Y.; Ruan, H.; Yang, H.; Cao, Q.; Liu, D.; Zhang, X.; Chen, K. The Identification of Potential Biomarkers and Biological Pathways in Prostate Cancer. J. Cancer 2019, 10, 1398–1408. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Dai, Y.; Zhuang, T.; Yue, X.; Chen, Y.; Wang, X.; Duan, X.; Ping, Y.; Xie, Y.; Cao, Y.; et al. Identification and Validation of Three Hub Genes Involved in Cell Proliferation and Prognosis of Castration-Resistant Prostate Cancer. Oxidative Med. Cell. Longev. 2022, 2022, 8761112. [Google Scholar] [CrossRef]
- Roy, D.; Morgan, M.; Yoo, C.; Deoraj, A.; Roy, S.; Yadav, V.; Garoub, M.; Assaggaf, H.; Doke, M. Integrated Bioinformatics, environmental epidemiologic and genomic approaches to identify environmental and molecular links between endometriosis and breast cancer. Int. J. Mol. Sci. 2015, 16, 25285–25322. [Google Scholar] [CrossRef]
- Deoraj, A.; Yoo, C.; Roy, D. Integrated bioinformatics biostatistics and molecular epidemiologic approaches to study how the environment and genes work together to affect the development of complex chronic diseases. In Gene-Environment Interaction Analysis: Methods in Bioinformatics and Computational Biology; Anno, S., Ed.; Pan Stanford Publishing Pte. Ltd.: New York, NY, USA, 2016; pp. 151–191. [Google Scholar]
- Prins, G.S. Endocrine Disruptors and Prostate Cancer Risk. Endocr. Relat. Cancer 2008, 15, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Tarapore, P.; Ying, J.; Ouyang, B.; Burke, B.; Bracken, B.; Ho, S.-M. Exposure to Bisphenol a Correlates with Early-Onset Prostate Cancer and Promotes Centrosome Amplification and Anchorage-Independent Growth in Vitro. PLoS ONE 2014, 9, e90332. [Google Scholar] [CrossRef]
- Golden, R.; Gandy, J.; Vollmer, G. A Review of the Endocrine Activity of Parabens and Implications for Potential Risks to Human Health. Crit. Rev. Toxicol. 2005, 35, 435–458. [Google Scholar] [CrossRef]
- Ho, S.M.; Rao, R.; To, S.; Schoch, E.; Tarapore, P. Bisphenol A and Its Analogues Disrupt Centrosome Cycle and Microtubule Dynamics in Prostate Cancer. Endocr. Relat. Cancer 2017, 24, 83–96. [Google Scholar] [CrossRef]
- De Falco, M.; Laforgia, V. Combined Effects of Different Endocrine-Disrupting Chemicals (EDCs) on Prostate Gland. Int. J. Environ. Res. Public Health 2021, 18, 9772. [Google Scholar] [CrossRef]
- Alwadi, D.; Felty, Q.; Roy, D.; Yoo, C.; Deoraj, A. Environmental Phenol and Paraben Exposure Risks and Their Potential Influence on the Gene Expression Involved in the Prognosis of Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 3679. [Google Scholar] [CrossRef]
- Alwadi, D.; Felty, Q.; Yoo, C.; Roy, D.; Deoraj, A. Endocrine Disrupting Chemicals Influence Hub Genes Associated with Aggressive Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 3191. [Google Scholar] [CrossRef]
- Jarred, R.A.; Keikha, M.; Dowling, C.; McPherson, S.J.; Clare, A.M.; Husband, A.J.; Pedersen, J.S.; Frydenberg, M.; Risbridger, G.P. Induction of Apoptosis in Low to Moderate-Grade Human Prostate Carcinoma by Red Clover-derived Dietary Isoflavones. Cancer Epidemiol. Biomark. Prev. 2002, 11, 1689–1696. [Google Scholar]
- Doultsinos, D.; Mills, I.G. Derivation and application of molecular signatures to prostate cancer: Opportunities and challenges. Cancers 2021, 13, 495. [Google Scholar] [CrossRef]
- Lakshman, M.; Xu, L.; Ananthanarayanan, V.; Cooper, J.; Takimoto, C.H.; Helenowski, I.; Pelling, J.C.; Bergan, R.C. Dietary genistein inhibits metastasis of human prostate cancer in mice. Cancer Res. 2008, 68, 2024–2032. [Google Scholar] [CrossRef]
- Artacho-Cordón, F.; Fernández, M.F.; Frederiksen, H.; Iribarne-Durán, L.M.; Jiménez-Díaz, I.; Vela-Soria, F.; Andersson, A.M.; Martin-Olmedo, P.; Peinado, F.M.; Olea, N.; et al. Environmental Phenols and Parabens in Adipose Tissue from Hospitalized Adults in Southern Spain. Environ. Int. 2018, 119, 203–211. [Google Scholar] [CrossRef]
- Tai, K.-Y.; Shiah, S.-G.; Shieh, Y.-S.; Kao, Y.-R.; Chi, C.-Y.; Huang, E.; Lee, H.-S.; Chang, L.-C.; Yang, P.-C.; Wu, C.-W. DNA methylation and histone modification regulate silencing of epithelial cell adhesion molecule for tumor invasion and progression. Oncogene 2007, 26, 3989–3997. [Google Scholar] [CrossRef]
- Bonkhoff, H. Estrogen receptor signaling in prostate cancer: Implications for carcinogenesis and tumor progression. Prostate 2018, 78, 2–10. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, J.; Zhang, J. Expression of P53 and Its Mechanism in Prostate Cancer. Oncol. Lett. 2018, 16, 378–382. [Google Scholar] [CrossRef]
- Habrowska-Górczyńska, D.E.; Kozieł, M.J.; Kowalska, K.; Piastowska-Ciesielska, A.W. FOXO3A and Its Regulators in Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 12530. [Google Scholar] [CrossRef]
- Carabet, L.A.; Lallous, N.; Leblanc, E.; Ban, F.; Morin, H.; Lawn, S.; Ghaidi, F.; Lee, J.; Mills, I.G.; Gleave, M.E.; et al. Computeraided drug discovery of Myc-Max inhibitors as potential therapeutics for prostate cancer. Eur. J. Med. Chem. 2018, 160, 108–119. [Google Scholar] [CrossRef]
- Qiu, X.; Boufaied, N.; Hallal, T.; Feit, A.; de Polo, A.; Luoma, A.M.; Larocque, J.; Zadra, G.; Xie, Y.; Gu, S.; et al. MYC drives aggressive prostate cancer by disrupting transcriptional pause release at androgen receptor targets. Nat. Commun. 2021, 13, 2559. [Google Scholar] [CrossRef]
- Xie, B.; Wang, S.; Jiang, N.; Li, J.J. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett. 2019, 443, 56–66. [Google Scholar] [CrossRef]
- Feng, T.; Wei, D.; Li, Q.; Yang, X.; Han, Y.; Luo, Y.; Jiang, Y. Four Novel Prognostic Genes Related to Prostate Cancer Identified Using Co-Expression Structure Network Analysis. Front. Genet. 2021, 12, 584164. [Google Scholar] [CrossRef]
- Qing, Y.; Wang, Y.; Hu, C.; Zhang, H.; Huang, Y.; Zhang, Z.; Ma, T.; Zhang, S.; Li, K. Evaluation of Notch Family Genes’ Expression and Prognostic Value in Prostate Cancer. Transl. Androl. Urol. 2022, 11, 627–642. [Google Scholar] [CrossRef]
- Brouwer-Visser, J.; Cheng, W.-Y.; Bauer-Mehren, A.; Maisel, D.; Lechner, K.; Andersson, E.; Dudley, J.T.; Milletti, F. Regulatory T-Cell Genes Drive Altered Immune Microenvironment in Adult Solid Cancers and Allow for Immune Contextual Patient Subtyping. Cancer Epidemiol. Biomark. Prev. 2018, 27, 103–112. [Google Scholar] [CrossRef]
- Wang, X.; Simpson, E.R.; Brown, K.A. P53: Protection against Tumor Growth beyond Effects on Cell Cycle and Apoptosis. Cancer Res. 2016, 76, 1668. [Google Scholar] [CrossRef]
- Zeng, X.; Shi, G.; He, Q.; Zhu, P. Screening and Predicted Value of Potential Biomarkers for Breast Cancer Using Bioinformatics Analysis. Sci. Rep. 2021, 11, 20799. [Google Scholar] [CrossRef]
- Gao, J.; Xia, R.; Chen, J.; Gao, J.; Luo, X.; Ke, C.; Ren, C.; Li, J.; Mi, Y. Inhibition of Esophageal-Carcinoma Cell Proliferation by Genistein via Suppression of JAK1/2-STAT3 and AKT/MDM2/P53 Signaling Pathways. Aging 2020, 12, 6240–6259. [Google Scholar] [CrossRef]
- Engeland, K. Cell Cycle Arrest through Indirect Transcriptional Repression by P53: I Have a Dream. Cell Death Differ. 2017, 25, 114–132. [Google Scholar] [CrossRef]
- Kumar, A.; Coleman, I.; Morrissey, C.; Zhang, X.; True, L.D.; Gulati, R.; Etzioni, R.; Bolouri, H.; Montgomery, B.; White, T.; et al. Substantial Interindividual and Limited Intraindividual Genomic Diversity among Tumors from Men with Metastatic Prostate Cancer. Nat. Med. 2016, 22, 369–378. [Google Scholar] [CrossRef]
- Ribeiro, F.R.; Jerónimo, C.; Henrique, R.; Fonseca, D.; Oliveira, J.; Lothe, R.A.; Teixeira, M.R. 8Q Gain Is an Independent Predictor of Poor Survival in Diagnostic Needle Biopsies from Prostate Cancer Suspects. Clin. Cancer Res. 2006, 12, 3961–3970. [Google Scholar] [CrossRef]
- DuPree, E.L.; Mazumder, S.; Almasan, A. Genotoxic Stress Induces Expression of E2F4, Leading to Its Association with P130 in Prostate Carcinoma Cells. Cancer Res. 2004, 64, 4390–4393. [Google Scholar] [CrossRef]
- Taylor, W.R.; Schonthal, A.H.; Galante, J.; Stark, G.R. p130/E2F4 binds to and represses the cdc2 promoter in response to p53. J. Biol. Chem. 2001, 276, 1998–2006. [Google Scholar] [CrossRef]
- Yang, J.; Song, K.; Krebs, T.L.; Jackson, M.W.; Danielpour, D. RB/E2F4 and Smad2/3 Link Survivin to TGF-β-Induced Apoptosis and Tumor Progression. Oncogene 2008, 27, 5326–5338. [Google Scholar] [CrossRef]
- Shiota, M.; Fujimoto, N.; Imada, K.; Yokomizo, A.; Itsumi, M.; Takeuchi, A.; Kuruma, H.; Inokuchi, J.; Tatsugami, K.; Uchiumi, T.; et al. Potential Role for YB-1 in Castration-Resistant Prostate Cancer and Resistance to Enzalutamide through the Androgen Receptor V7. J. Natl. Cancer Inst. 2016, 108, djw005. [Google Scholar] [CrossRef]
- Montes, M.; Becerra, S.; Sánchez-Álvarez, M.; Suñé, C. Functional Coupling of Transcription and Splicing. Gene 2012, 501, 104–117. [Google Scholar] [CrossRef]
- Wyatt, A.W.; Gleave, M.E. Targeting the Adaptive Molecular Landscape of Castration-resistant Prostate Cancer. EMBO Mol. Med. 2015, 7, 878–894. [Google Scholar] [CrossRef]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Edgar, R. NCBI GEO: Mining tens of millions of expression profiles—Database and tools update. Nucleic Acids Res. 2007, 35, D760–D765. [Google Scholar] [CrossRef] [PubMed]
- Poluri, R.T.; Paquette, V.; Allain, É.P.; Lafront, C.; Joly-Beauparlant, C.; Weidmann, C.; Droit, A.; Guillemette, C.; Pelletier, M.; Audet-Walsh, É. KLF5 and NFYA Factors as Novel Regulators of Prostate Cancer Cell Metabolism. Endocr.-Relat. Cancer 2021, 28, 257–271. [Google Scholar] [CrossRef]
- Poluri, R.T.K.; Beauparlant, C.J.; Droit, A.; Audet-Walsh, É. RNA sequencing data of human prostate cancer cells treated with androgens. Data Brief 2019, 25, 104372. [Google Scholar] [CrossRef]
- Jividen, K.; Kedzierska, K.Z.; Yang, C.; Szlachta, K.; Ratan, A.; Paschal, B.M. Genomic analysis of DNA repair genes and androgen signaling in prostate cancer. BMC Cancer 2018, 18, 960. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Jividen, K.; Kamata, T.; Dworak, N.; Oostdyk, L.; Remlein, B.; Pourfarjam, Y.; Kim, I.; Du, K.; Abbas, T.; et al. Androgen signaling uses a writer and a reader of ADP-ribosylation to regulate protein complex assembly. Nat. Commun. 2021, 12, 2705. [Google Scholar] [CrossRef]
- Nyquist, M.D.; Corella, A.; Mohamad, O.; Coleman, I.; Kaipainen, A.; Kuppers, D.A.; Lucas, J.M.; Paddison, P.J.; Plymate, S.R.; Nelson, P.S.; et al. Molecular determinants of response to high-dose androgen therapy in prostate cancer. JCI Insight 2019, 4, e129715. [Google Scholar] [CrossRef]
- Brzezinka, K.; Nevedomskaya, E.; Lesche, R.; Haegebarth, A.; Ter Laak, A.; Fernández-Montalván, A.E.; Eberspaecher, U.; Werbeck, N.D.; Moenning, U.; Siegel, S.; et al. Characterization of the menin-MLL interaction as therapeutic cancer target. Cancers 2020, 12, 201. [Google Scholar] [CrossRef]
- Cato, L.; Neeb, A.; Sharp, A.; Buzón, V.; Ficarro, S.B.; Yang, L.; Muhle-Goll, C.; Kuznik, N.C.; Riisnaes, R.; Rodrigues, D.N.; et al. Development of bag-1L as a therapeutic target in androgen receptor-dependent prostate cancer. eLife 2023, 6, e27159. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Willmann, D.; McMillan, J.; Forne, I.; Metzger, P.; Gerhardt, S.; Petroll, K.; Von Maessenhausen, A.; Urban, S.; Schott, A.; et al. Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat. Struct. Mol. Biol. 2016, 23, 132–139. [Google Scholar] [CrossRef]
- Lin, S.; Wei, H.; Maeder, D.; Franklin, R.B.; Feng, P. Profiling of zinc-altered gene expression in human prostate normal vs. cancer cells: A time course study. J. Nutr. Biochem. 2009, 20, 1000–1012. [Google Scholar] [CrossRef]
- Zhang, H.; Gordon, R.; Li, W.; Yang, X.; Pattanayak, A.; Fowler, G.; Zhang, L.; Catalona, W.J.; Ding, Y.; Xu, L.; et al. Genistein treatment duration effects biomarkers of cell motility in human prostate. PLoS ONE 2019, 14, e0214078. [Google Scholar] [CrossRef] [PubMed]
- Rooney, J.P.; Chorley, B.; Kleinstreuer, N.; Corton, J.C. Identification of Androgen Receptor Modulators in a Prostate Cancer Cell Line Microarray Compendium. Toxicol. Sci. 2018, 166, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Firlej, V.; Soyeux, P.; Nourieh, M.; Huet, E.; Semprez, F.; Allory, Y.; Londono-Vallejo, A.; de la Taille, A.; Vacherot, F.; Destouches, D. Overexpression of Nucleolin and Associated Genes in Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 4491. [Google Scholar] [CrossRef]
- Teslow, E.A.; Bao, B.; Dyson, G.; Legendre, C.; Mitrea, C.; Sakr, W.; Carpten, J.D.; Powell, I.; Bollig-Fischer, A. Exogenous IL-6 induces mRNA splice variant MBD2_v2 to promote stemness in TP53 wild-type, African American PCa cells. Mol. Oncol. 2018, 12, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Wang, S.; Xu, Y.; Wang, W.; Soares, F.; Ahmed, M.; Su, P.; Wang, T.; Orouji, E.; Xu, X.; et al. MYC reshapes CTCF-mediated chromatin architecture in prostate cancer. Nat. Commun. 2023, 14, 1787. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Rahman, M.H.; Rana, H.K.; Peng, S.; Hu, X.; Chen, C.; Quinn, J.M.W.; Moni, M.A. Bioinformatics and machine learning methodologies to identify the effects of central nervous system disorders on glioblastoma progression. Brief. Bioinform. 2021, 22, bbaa365. [Google Scholar] [CrossRef]
- Seifert, E. OriginPro 9.1: Scientific data analysis and graphing software-software review. J. Chem. Inf. Model. 2014, 54, 1552. [Google Scholar] [CrossRef]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. (2007–2015). Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 27 November 2023).
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Du, P.; Kibbe, W.A.; Lin, S.M. lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008, 24, 1547–1548. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; Cherry, J.M.; Ashburner, M.; Ball, C.A.; Blake, J.A.; Butler, H.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Omranian, S.; Nikoloski, Z.; Grimm, D.G. Computational identification of protein complexes from network interactions: Present state, challenges, and the way forward. Comput. Struct. Biotechnol. J. 2022, 20, 2699–2712. [Google Scholar] [CrossRef] [PubMed]
- Bandettini, W.P.; Kellman, P.; Mancini, C.; Booker, O.J.; Vasu, S.; Leung, S.W.; Wilson, J.R.; Shanbhag, S.M.; Chen, M.Y.; Arai, A.E. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: A clinical validation study. J. Cardiovasc. Magn. Reson. 2012, 14, 83. [Google Scholar] [CrossRef]
- Chin, C.; Chen, S.; Wu, H.; Ho, C.; Ko, M.; Lin, C. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Cao, Z.; Zhang, S. An integrative and comparative study of pan-cancer transcriptomes reveals distinct cancer common and specific signatures. Sci. Rep. 2016, 6, 33398. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Li, B.; Severson, E.; Pignon, J.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Hahn, W.C. Abstract IA17: Defining cancer dependency maps. Cancer Res. 2020, 80, IA17. [Google Scholar] [CrossRef]
- Dwane, L.; Behan, F.M.; Gonçalves, E.; Lightfoot, H.; Yang, W.; van der Meer, D.; Shepherd, R.; Pignatelli, M.; Iorio, F.; Garnett, M.J. Project Score database: A resource for investigating cancer cell dependencies and prioritizing therapeutic targets. Nucleic Acids Res. 2021, 49, D1365–D1372. [Google Scholar] [CrossRef]
- Redman, M.; King, A.; Watson, C.; King, D. What is CRISPR/Cas9? Arch. Dis. Child. Educ. Pract. Ed. 2016, 101, 213–215. [Google Scholar] [CrossRef]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA Interference: Biology, Mechanism, and Applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. ENRICHR: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with ENRICHR. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Han, H.; Shim, H.; Shin, D.; Shim, J.E.; Ko, Y.; Shin, J.; Kim, H.; Cho, A.; Kim, E.; Lee, T.; et al. TRRUST: A reference database of human transcriptional regulatory interactions. Sci. Rep. 2015, 5, 11432. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Cho, J.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Blom, U.M.; Wang, P.I.; Shim, J.E.; Marcotte, E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011, 21, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Razick, S.; Magklaras, G.; Donaldson, I.M. iRefIndex: A consolidated protein interaction database with provenance. BMC Bioinform. 2008, 9, 405. [Google Scholar] [CrossRef]
- Yang, H.; Robinson, P.N.; Wang, K. Phenolyzer: Phenotype-based prioritization of candidate genes for human diseases. Nat. Methods 2015, 12, 841–843. [Google Scholar] [CrossRef]
- Surdo, P.L.; Iannuccelli, M.; Contino, S.; Castagnoli, L.; Licata, L.; Cesareni, G.; Perfetto, L. SIGNOR 3.0, the SIGnaling network open resource 3.0: 2022 update. Nucleic Acids Res. 2023, 51, D631–D637. [Google Scholar]
- Perfetto, L.; Briganti, L.; Calderone, A.; Perpetuini, A.C.; Iannuccelli, M.; Langone, F.; Licata, L.; Marinkovic, M.; Mattioni, A.; Pavlidou, T.; et al. SIGNOR: A database of causal relationships between biological entities. Nucleic Acids Res. 2016, 44, D548–D554. [Google Scholar] [CrossRef]
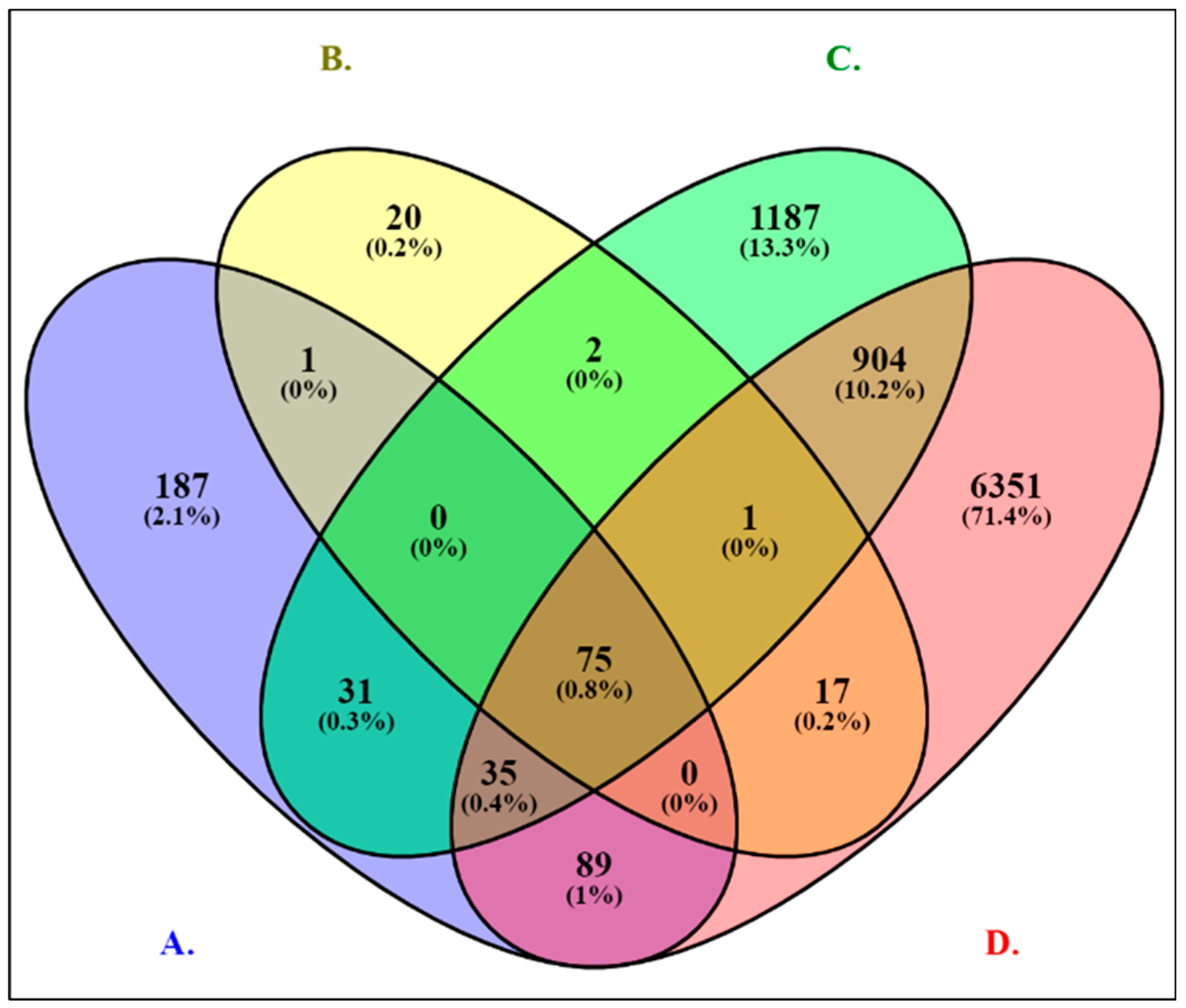


| Category | # | TFs | # of Genes | p-Value | FDR 2 |
|---|---|---|---|---|---|
| Upregulated UCSC_TFBS 1 | 1 | MYC | 28 | 6.48 × 10−5 | 0.011 |
| 2 | NFY | 21 | 1.03 × 10−3 | 0.086 | |
| 3 | STAT | 33 | 5.52 × 10−3 | 0.222 | |
| 4 | CEBP | 31 | 7.13 × 10−3 | 0.222 | |
| 5 | YY1 | 34 | 9.82 × 10−3 | 0.243 | |
| 6 | EVI1 | 17 | 1.16 × 10−2 | 0.266 | |
| 7 | HSF1 | 22 | 1.13 × 10−2 | 0.266 | |
| 8 | CDPCR3HD | 26 | 1.40 × 10−2 | 0.266 | |
| 9 | STAT5A | 26 | 1.68 × 10−2 | 0.266 | |
| 10 | PBX1 | 19 | 1.58 × 10−2 | 0.269 | |
| Downregulated UCSC_TFBS | 1 | MYC | 22 | 1.60 × 10−2 | 1.00 |
| 2 | NFY | 19 | 2.70 × 10−2 | 1.00 | |
| 3 | NFAT | 23 | 2.90 × 10−2 | 1.00 | |
| 4 | TATA | 20 | 3.10 × 10−2 | 1.00 | |
| 5 | IRF7 | 28 | 6.20 × 10−2 | 1.00 | |
| 6 | MEF2 | 18 | 6.60 × 10−2 | 1.00 | |
| 7 | FREAC3 | 18 | 9.10 × 10−2 | 1.00 | |
| 8 | ISRE | 23 | 9.80 × 10−2 | 1.00 | |
| 9 | CDP | 19 | 6.80 × 10−2 | 1.00 | |
| 10 | NF1 | 18 | 9.90 × 10−2 | 1.00 |
| MCC | DMNC | MNC | Degree | EPC | MCC ꓵ DMNC ꓵ MNC ꓵ Degree ꓵ EPC |
|---|---|---|---|---|---|
| NCAPH | NCAPH | NCAPH | NCAPH | NCAPH | NCAPH |
| RAD54L | RAD54L | RAD54L | RAD54L | RAD54L | RAD54L |
| E2F7 | E2F7 | E2F7 | E2F7 | E2F7 | E2F7 |
| MKI67 | MKI67 | MKI67 | MKI67 | MKI67 | MKI67 |
| NDC80 | NDC80 | NDC80 | NDC80 | NDC80 | NDC80 |
| MELK | MELK | MELK | MELK | MELK | MELK |
| ESPL1 | ESPL1 | ESPL1 | ESPL1 | ESPL1 | ESPL1 |
| CCNA2 | CCNA2 | CCNA2 | CCNA2 | CCNA2 | CCNA2 |
| CKS2 | GTSE1 | ASPM | CKS2 | GTSE1 | |
| ORC1 | CHFR | FZR1 | ORC1 | CHFR | |
| PLK1 | DTL | DTL | ANAPC11 | DTL | |
| MCM10 | DTL | MCM10 | PLK1 | MCM10 | |
| HELLS | HELLS | ANAPC11 | HELLS | HELLS | |
| NCAPG | CCNB1 | NCAPG | NCAPG | NCAPG | NCAPG |
| CCNB1 | NCAPG | CCNB1 | CCNB1 | CCNB1 | CCNB1 |
| PLK4 | ANAPC11 | PLK4 | PLK4 | FZR1 | |
| HAAO | EXO1 | EXO1 | FZR1 | SMC4 | |
| HAAO | SPC25 | FOXP4 | FZR1 | EXO1 | |
| MCM10 | DTL | FOXP4 | PLK1 | MCM10 | |
| CKS2 | GTSE1 | ASPM | CKS2 | GTSE1 |
| MCC | DMNC | MNC | Degree | EPC | MCC ꓵ DMNC ꓵ MNC ꓵ Degree ꓵ EPC |
|---|---|---|---|---|---|
| TOP2A | TOP2A | TOP2A | TOP2A | TOP2A | TOP2A |
| CCNB1 | CDC20 | ASPM | GTSE1 | ASPM | |
| CDC20 | BUB1B | CDC20 | CDC20 | CDC20 | CDC20 |
| BUB1B | HMMR | BUB1B | BUB1B | BUB1B | BUB1B |
| HMMR | TTK | HMMR | HMMR | GTSE1 | |
| RRM2 | TTK | RRM2 | RRM2 | RRM2 | |
| CDK1 | BUB1 | GTSE1 | CDK1 | CDK1 | |
| TTK | CENPF | TTK | TTK | GTSE1 | |
| BUB1 | CDKN3 | BUB1 | BUB1 | BUB1 | BUB1 |
| CENPF | PKMYT1 | CENPF | CENPF | CENPF | CENPE |
| CDKN3 | CCNB2 | CDKN3 | CDKN3 | CDKN3 | |
| CCNB2 | KIF15 | CCNB2 | CCNB2 | CCNB2 | CCNB2 |
| KIF15 | AURKA | KIF15 | KIF15 | KIF15 | KIF15 |
| AURKA | KIF11 | AURKA | AURKA | AURKA | AURKA |
| KIF11 | DLGAP5 | KIF11 | KIF11 | KIF11 | |
| DLGAP5 | ZWINT | DLGAP5 | DLGAP5 | DLGAP5 | DLGAP5 |
| ZWINT | CENPE | ZWINT | GTSE1 | ZWINT | |
| CENPE | SPC25 | CENPE | CENPE | CENPE | |
| UBE2C | UBE2C | UBE2C | UBE2C | UBE2C | UBE2C |
| ASPM | GTSE1 | ASPM | ASPM | GTSE1 |
| Group | Cell Lines | |||||||
|---|---|---|---|---|---|---|---|---|
| # | GEO Profile | Platform | Annotation Platform | Total | PCa | Control | References | |
| Cell lines treated with R1881 treatment | 1 | GSE70466 | GPL16791 | Illumina HiSeq 2500 | 6 | 3 | 3 | [63] |
| 2 | GSE151290 | GPL16791 | Illumina HiSeq 2500 | 16 | 8 | 8 | [64] | |
| 3 | GSE128749 | GPL11154 | Illumina HiSeq 2000 | 11 | 5 | 6 | [65] | |
| 4 | GSE120660 | GPL16791 | Illumina HiSeq 2500 | 21 | 12 | 9 | [66] | |
| 5 | GSE135879 | GPL16791 | Illumina HiSeq 2500 | 12 | 6 | 6 | [67] | |
| 6 | GSE136272 | GPL16791 | Illumina HiSeq 2500 | 12 | 6 | 6 | [68] | |
| Cell lines with EDC exposure | 7 | GSE218556 | GPL24676 | Illumina NovaSeq 6000 | 6 | 3 | 3 | [69] |
| 8 | GSE64529 | GPL11154 | Illumina HiSeq 2000 | 6 | 3 | 3 | [70] | |
| 9 | GSE5590 | GPL2986 | Illumina HiSeq 2500 | 6 | 3 | 3 | [71] | |
| 10 | GSE128339 | GPL8842 | Illumina NovaSeq 6000 | 34 | 24 | 10 | [72] | |
| 11 | GSE109021 | GPL10558 | Illumina HiSeq 2000 | 18 | 15 | 3 | [73] | |
| Cell lines with Gleason scores and Cells with MYC overexpression | 12 | GSE200879 | GPL32170 | Illumina HiSeq 2500 | 124 | 115 | 9 | [74] |
| 13 | GSE103512 | GPL13158 | Illumina NovaSeq 6000 | 57 | 50 | 7 | [49] | |
| Cell lines between Age and Race | 14 | GSE104131 | GPL16791 | Illumina HiSeq 2500 | 29 | 16 | 13 | [75] |
| 15 | GSE200167 | GPL24676 | Illumina NovaSeq 6000 | 6 | 3 | 3 | [76] | |
| Total Samples | 364 | 272 | 92 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alwadi, D.; Felty, Q.; Doke, M.; Roy, D.; Yoo, C.; Deoraj, A. RNA-Seq Uncovers Association of Endocrine-Disrupting Chemicals with Hub Genes and Transcription Factors in Aggressive Prostate Cancer. Int. J. Mol. Sci. 2025, 26, 5463. https://doi.org/10.3390/ijms26125463
Alwadi D, Felty Q, Doke M, Roy D, Yoo C, Deoraj A. RNA-Seq Uncovers Association of Endocrine-Disrupting Chemicals with Hub Genes and Transcription Factors in Aggressive Prostate Cancer. International Journal of Molecular Sciences. 2025; 26(12):5463. https://doi.org/10.3390/ijms26125463
Chicago/Turabian StyleAlwadi, Diaaidden, Quentin Felty, Mayur Doke, Deodutta Roy, Changwon Yoo, and Alok Deoraj. 2025. "RNA-Seq Uncovers Association of Endocrine-Disrupting Chemicals with Hub Genes and Transcription Factors in Aggressive Prostate Cancer" International Journal of Molecular Sciences 26, no. 12: 5463. https://doi.org/10.3390/ijms26125463
APA StyleAlwadi, D., Felty, Q., Doke, M., Roy, D., Yoo, C., & Deoraj, A. (2025). RNA-Seq Uncovers Association of Endocrine-Disrupting Chemicals with Hub Genes and Transcription Factors in Aggressive Prostate Cancer. International Journal of Molecular Sciences, 26(12), 5463. https://doi.org/10.3390/ijms26125463







