Repurposing Biomolecules from Aerva javanica Against DDX3X in LAML: A Computer-Aided Therapeutic Approach
Abstract
1. Introduction
2. Results
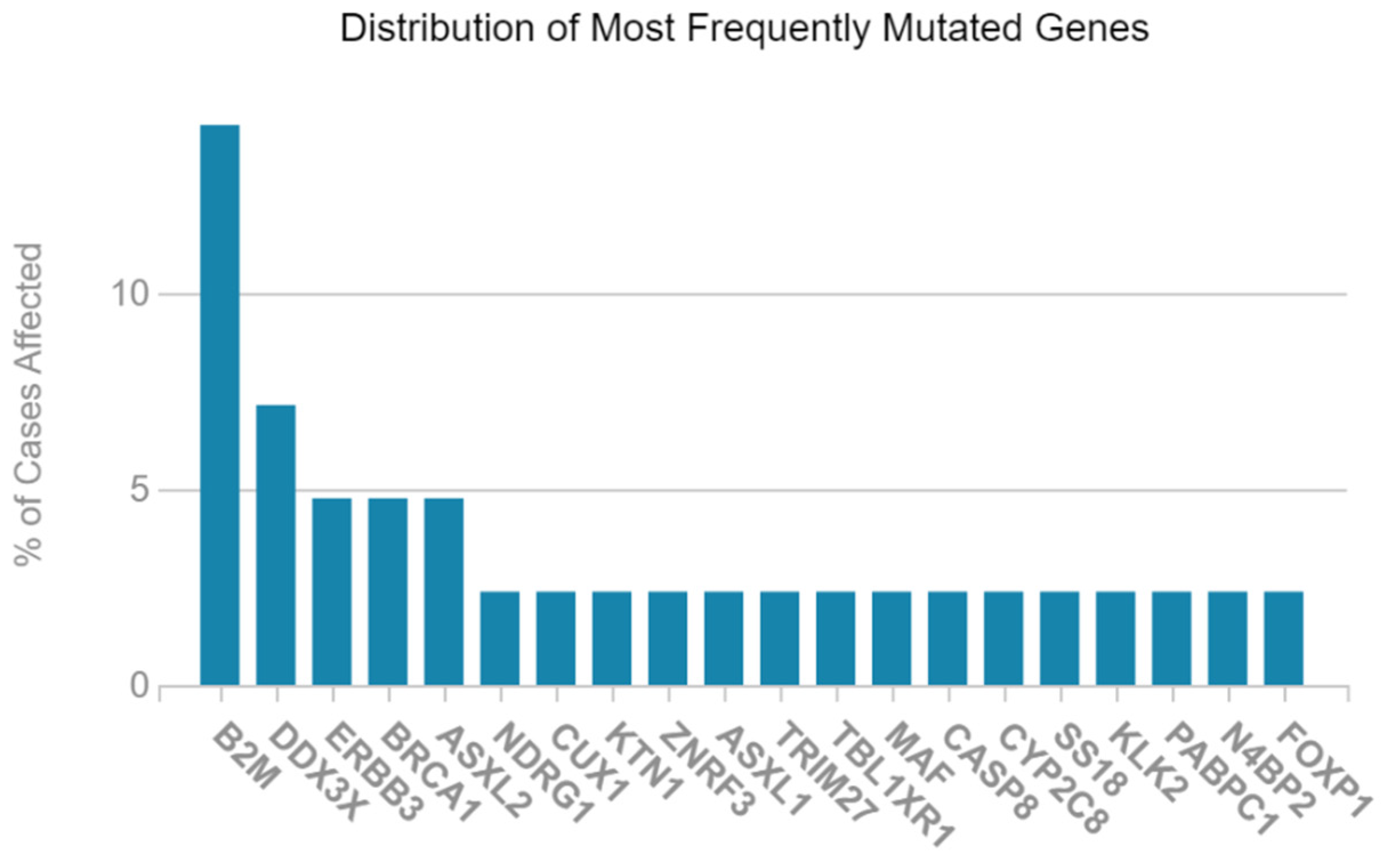
2.1. Mutated Genes in Acute Myeloid Leukemia (LAML)
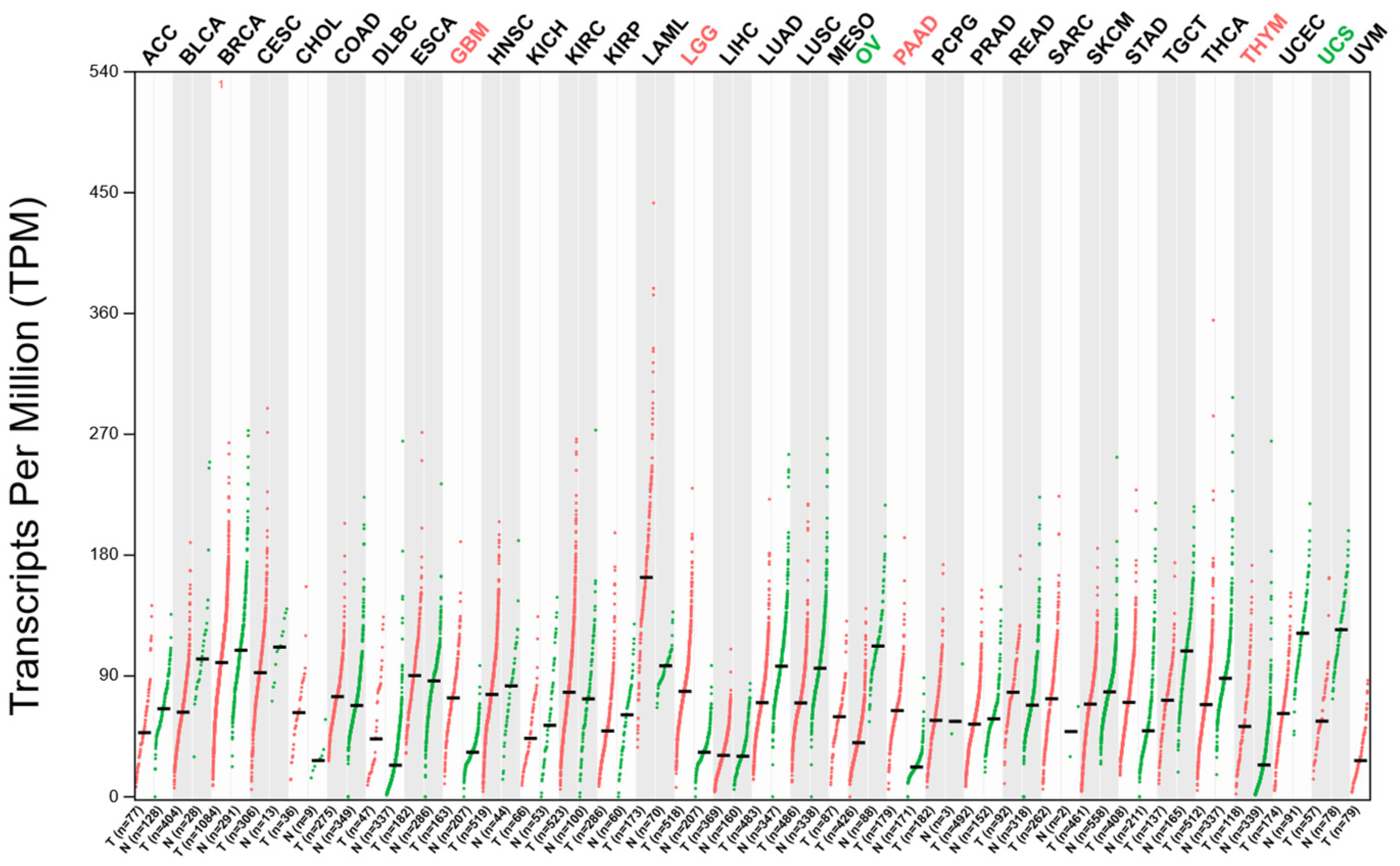
2.2. The Expression Analysis of DDX3X
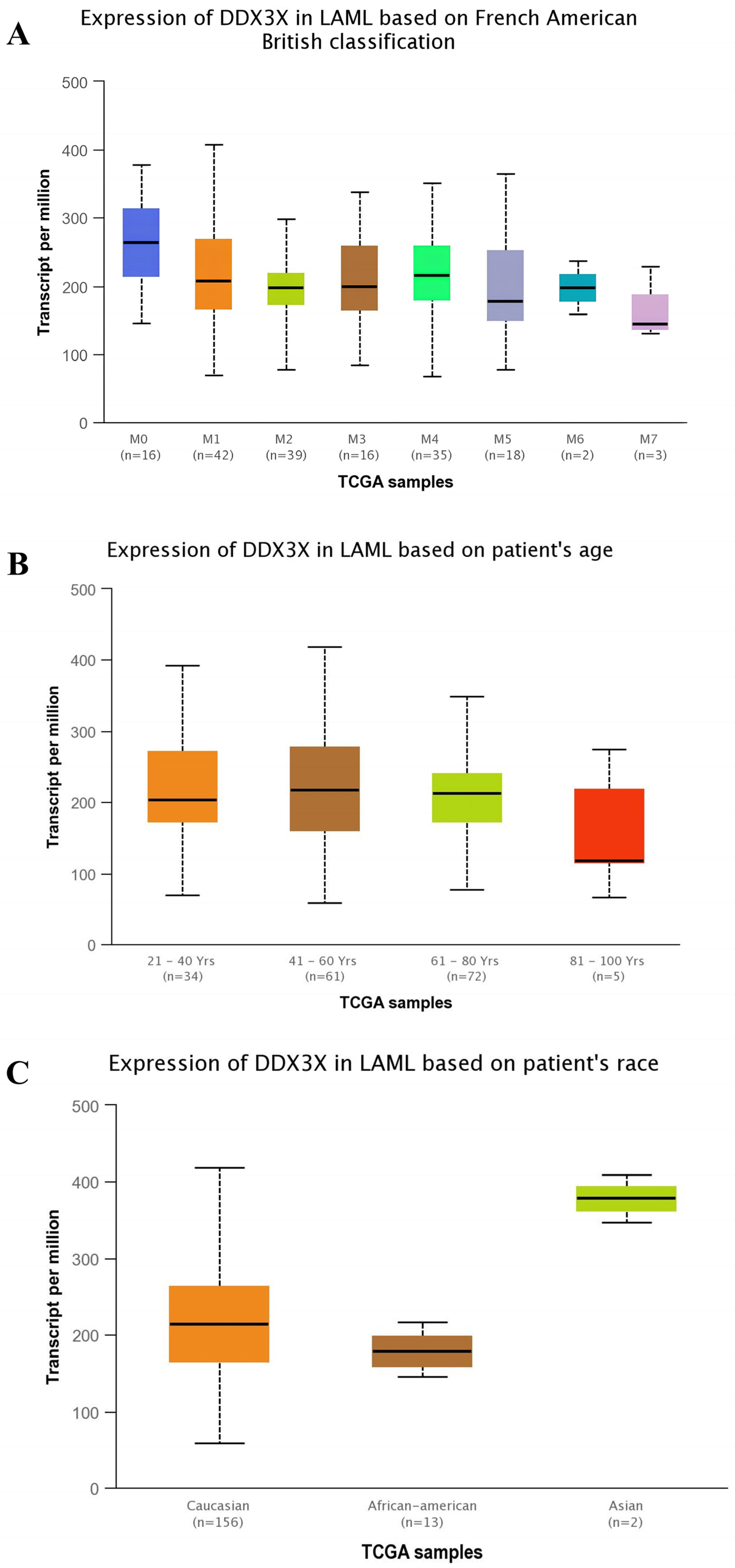
2.3. Expression Profiling of DDX3X at Various Stages
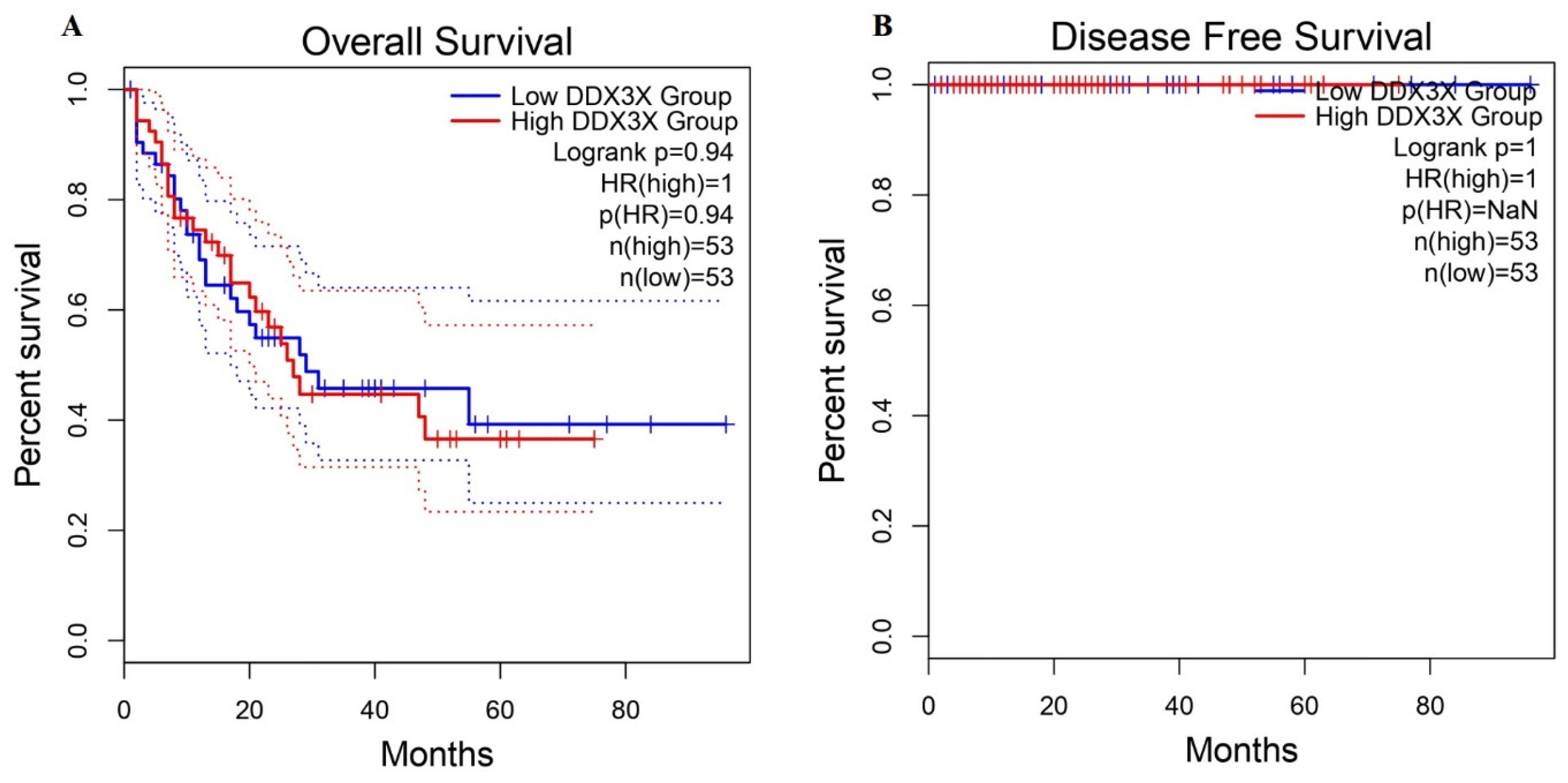
2.4. Survival Analysis of DDX3X
2.5. Principal Component Analysis (PCA) of DDX3X
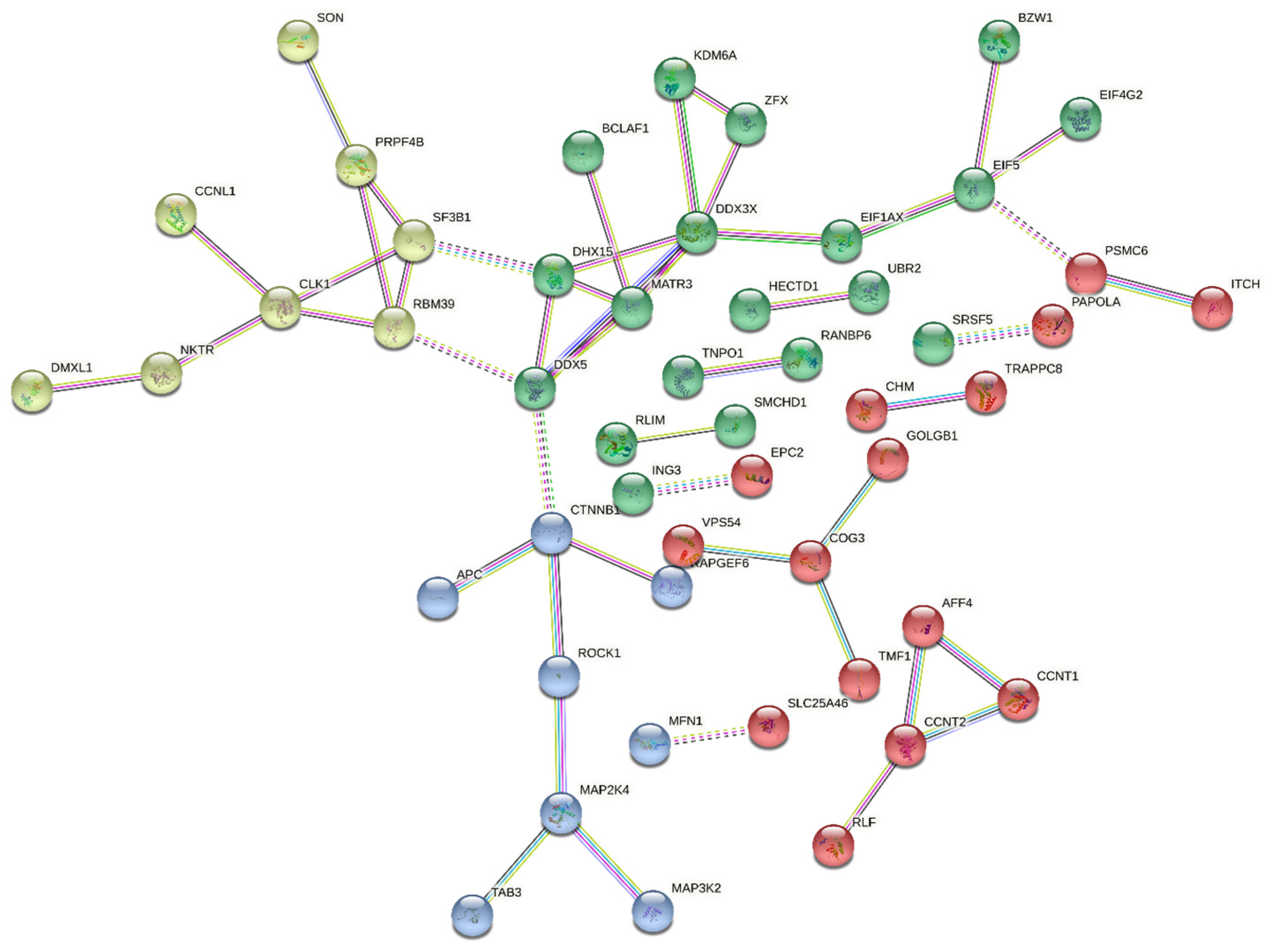
2.6. Interactions of DDX3X with Other Proteins
2.7. A. javanica Compounds and Their Pharmacokinetic Screening
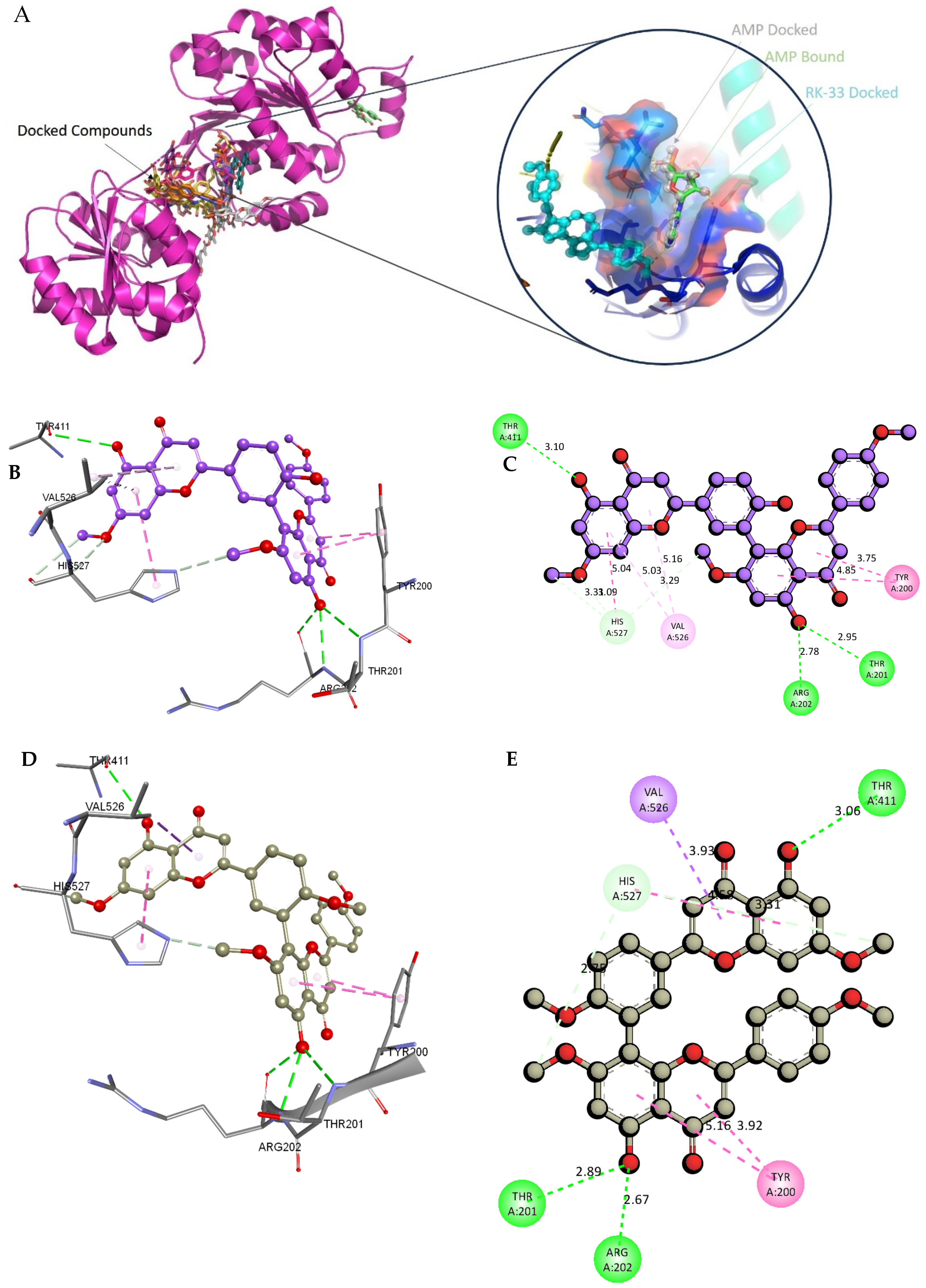
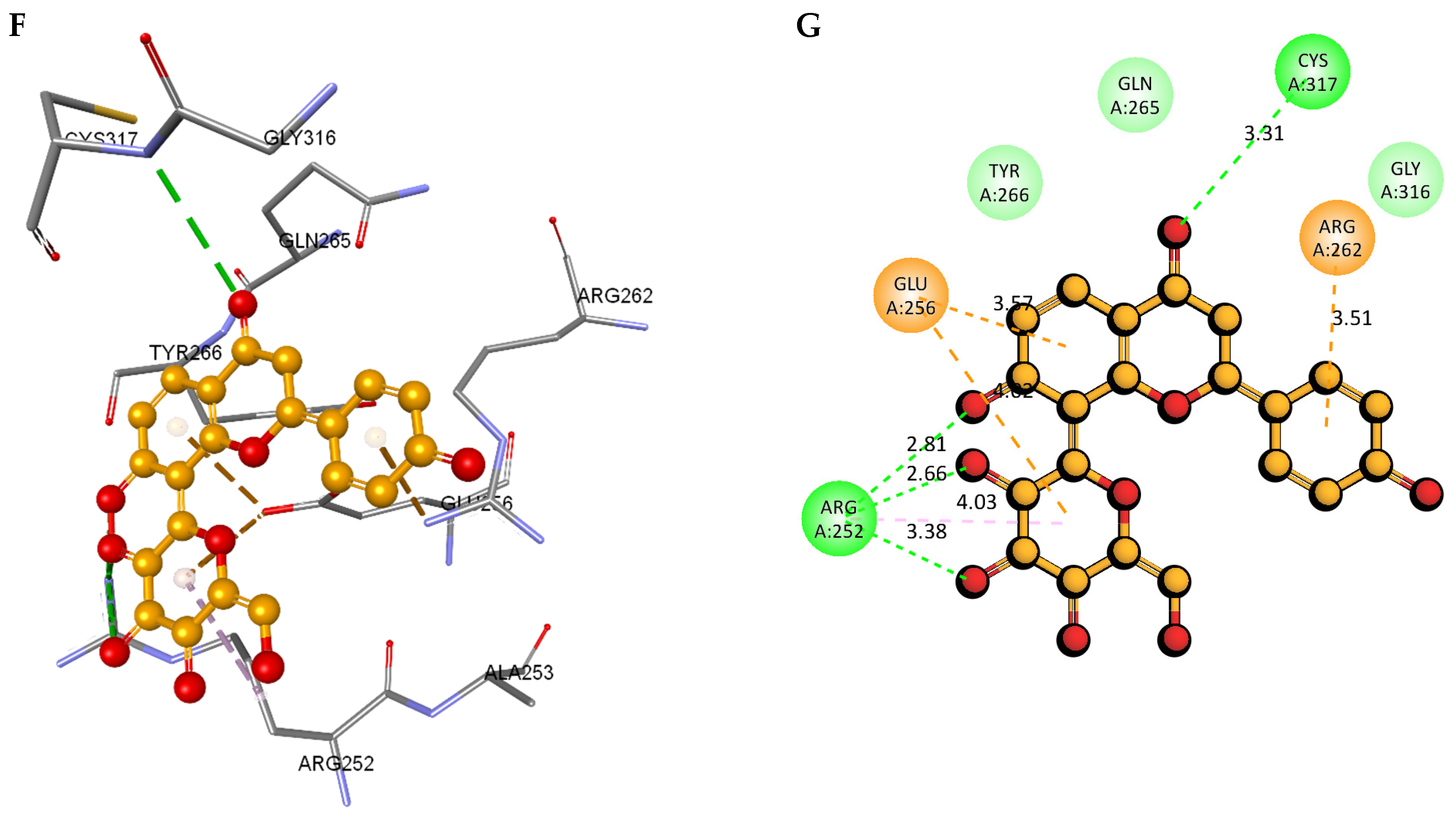
2.8. Molecular Docking and Intra-Molecular Interaction Analysis of DDX3X and the A. javanica Compounds
3. Discussions
4. Materials and Methods
4.1. The Data Sources and Bioinformatics Tools
4.2. The Identification of Highly Mutated Genes in Acute Myeloid Leukemia (LAML)
4.3. The Pan-Cancer Expression Analysis
4.4. Stage-Specific Expression Profiling
4.5. Survival Analysis
4.6. The Co-Expression Analysis
4.7. The Principal Component Analysis (PCA)
4.8. The Protein–Protein Interaction Network
4.9. Pharmacokinetic Screening of A. javanica Compounds
4.10. The Molecular Docking Analysis
4.11. Interaction Profiling and Visualization
4.12. Validation of the Docking Protocol Using RK-33
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HIA | human intestinal absorption |
| Caco-2 permeability | Caco-2 cell monolayer permeability |
| BBB | blood–brain barrier |
| PPB | plasma protein binding |
References
- Khwaja, A.; Bjorkholm, M.; Gale, R.E.; Levine, R.L.; Jordan, C.T.; Ehninger, G.; Bloomfield, C.D.; Estey, E.; Burnett, A.; Linch, D.C.; et al. Acute myeloid leukaemia. Nat. Rev. Dis. Primers 2016, 2, 16010. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zhang, J. Global leukemia burden and trends: A comprehensive analysis of temporal and spatial variations from 1990–2021 using GBD (Global Burden of Disease) data. BMC Public Health 2025, 25, 262. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Jani, C. Mapping incidence and mortality of leukemia and its subtypes in 21 world regions in last three decades and projections to 2030. Ann. Hematol. 2022, 101, 1523–1534. [Google Scholar] [CrossRef] [PubMed]
- Bawazir, A.; Al-Zamel, N.; Amen, A.; Akiel, M.A.; Alhawiti, N.M.; Alshehri, A. The burden of leukemia in the Kingdom of Saudi Arabia: 15 years period (1999–2013). BMC Cancer 2019, 19, 703. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, G.; Cai, X.; Liu, Y.; Qian, B.; Li, D. Global, regional, and national burden of acute myeloid leukemia, 1990–2021: A systematic analysis for the global burden of disease study 2021. Biomark. Res. 2024, 12, 101. [Google Scholar] [CrossRef]
- Alsobhi, E.M.; Farahat, F.; Daghistani, M.; Awad, K.; Al-Zahran, O.; Al-Saiari, A.; Koshak, F. Overall survival of adult acute myeloid leukemia based on cytogenetic and molecular abnormalities during 5 years in a single center study. Saudi Med. J. 2019, 40, 1171. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Cortes, J.E. Mutations in AML: Prognostic and therapeutic implications. Hematol. Am. Soc. Hematol. Educ. Program 2016, 2016, 348–355. [Google Scholar] [CrossRef]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 38. [Google Scholar] [CrossRef]
- Ali, M.A.M. DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021, 296, 198352. [Google Scholar] [CrossRef]
- Ryan, C.S.; Schröder, M. The human DEAD-box helicase DDX3X as a regulator of mRNA translation. Front. Cell Dev. Biol. 2022, 10, 1033684. [Google Scholar] [CrossRef]
- He, Y.-N.; Han, X.R.; Wang, D.; Hou, J.L.; Hou, X.M. Dual mode of DDX3X as an atp-dependent RNA helicase and atp-independent nucleic acid chaperone. Biochem. Biophys. Res. Commun. 2024, 714, 149964. [Google Scholar] [CrossRef] [PubMed]
- Floor, S.N.; Condon, K.J.; Sharma, D.; Jankowsky, E.; Doudna, J.A. Autoinhibitory interdomain interactions and subfamily-specific extensions redefine the catalytic core of the human DEAD-box protein DDX3. J. Biol. Chem. 2016, 291, 2412–2421. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, K.C.; Schroeder, T.; Gu, S.; Revalde, J.L.; Floor, S.N. A novel reporter for helicase activity in translation uncovers DDX3X interactions. RNA 2024, 30, 1041–1057. [Google Scholar] [CrossRef] [PubMed]
- Gadek, M.; Sherr, E.H.; Floor, S.N. The variant landscape and function of DDX3X in cancer and neurodevelopmental disorders. Trends Mol. Med. 2023, 29, 726–739. [Google Scholar] [CrossRef]
- Lin, T.C. DDX3X Multifunctionally Modulates Tumor Progression and Serves as a Prognostic Indicator to Predict Cancer Outcomes. Int. J. Mol. Sci. 2019, 21, 281. [Google Scholar] [CrossRef]
- Wang, Y.; Bao, Y.; Zhang, S.; Wang, Z. Splicing dysregulation in cancer: From mechanistic understanding to a new class of therapeutic targets. Sci. Chin. Life Sci. 2020, 63, 469–484. [Google Scholar] [CrossRef]
- Vesuna, F.; Akhrymuk, I.; Smith, A.; Winnard, P.T., Jr.; Lin, S.C.; Panny, L.; Scharpf, R.; Kehn-Hall, K.; Raman, V. RK-33, a small molecule inhibitor of host RNA helicase DDX3, suppresses multiple variants of SARS-CoV-2. Front. Microbiol. 2022, 13, 959577. [Google Scholar] [CrossRef]
- Nasim, N.; Sandeep, I.S.; Mohanty, S. Plant-derived natural products for drug discovery: Current approaches and prospects. Nucleus 2022, 65, 399–411. [Google Scholar] [CrossRef]
- Movaliya, V.; Zaveri, M. A review on the Pashanbheda plant Aerva javanica. Int. J. Pharm. Sci. Rev. Res. 2014, 25, 268–275. [Google Scholar]
- Thotathil, V.; Sidiq, N.; Fakhroo, A.; Sreerama, L. Phytochemical Analysis of Anastatica hierochuntica and Aerva javanica Grown in Qatar: Their Biological Activities and Identification of Some Active Ingredients. Molecules 2023, 28, 3364. [Google Scholar] [CrossRef]
- Jensen, M.A.; Ferretti, V.; Grossman, R.L.; Staudt, L.M. The NCI Genomic Data Commons as an engine for precision medicine. Blood J. Am. Soc. Hematol. 2017, 130, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cui, B.; Zhou, Y.; Wang, X.; Wu, W.; Wang, Z.; Dai, Z.; Cheng, Q.; Yang, K. B2M overexpression correlates with malignancy and immune signatures in human gliomas. Sci. Rep. 2021, 11, 5045. [Google Scholar] [CrossRef]
- Middha, S.; Yaeger, R.; Shia, J.; Stadler, Z.K.; King, S.; Guercio, S.; Paroder, V.; Bates, D.D.B.; Rana, S.; Diaz, L.A., Jr.; et al. Majority of B2M-mutant and-deficient colorectal carcinomas achieve clinical benefit from immune checkpoint inhibitor therapy and are microsatellite instability-high. JCO Precis. Oncol. 2019, 3, 1–14. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Driscoll, J.J.; Rixe, O. Overall survival: Still the gold standard: Why overall survival remains the definitive end point in cancer clinical trials. Cancer J. 2009, 15, 401–405. [Google Scholar] [CrossRef]
- Koster, J.; Volckmann, R.; Zwijnenburg, D.; Molenaar, P.; Versteeg, R. R2: Genomics analysis and visualization platform. Cancer Res. 2019, 79, 2490. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Högbom, M.; Collins, R.; van den Berg, S.; Jenvert, R.M.; Karlberg, T.; Kotenyova, T.; Flores, A.; Karlsson Hedestam, G.B.; Schiavone, L.H. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J. Mol. Biol. 2007, 372, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.-C.; Chen, Y.F.; Lin, S.R.; Yang, J.M. iGEMDOCK: A graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform. 2011, 12, S33. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.N.; Atkinson, S.C.; Audsley, M.D.; Heaton, S.M.; Jans, D.A.; Borg, N.A. RK-33 is a broad-spectrum antiviral agent that targets DEAD-box RNA helicase DDX3X. Cells 2020, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Bol, G.M.; Vesuna, F.; Xie, M.; Zeng, J.; Aziz, K.; Gandhi, N.; Levine, A.; Irving, A.; Korz, D.; Tantravedi, S.; et al. Targeting DDX 3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 2015, 7, 648–669. [Google Scholar] [CrossRef]
- Phung, B.; Cieśla, M.; Sanna, A.; Guzzi, N.; Beneventi, G.; Cao Thi Ngoc, P.; Lauss, M.; Cabrita, R.; Cordero, E.; Bosch, A.; et al. The X-linked DDX3X RNA helicase dictates translation reprogramming and metastasis in melanoma. Cell Rep. 2019, 27, 3573–3586.e7. [Google Scholar] [CrossRef]
- Palusiak, M.; Grabowski, S.J. Methoxy group as an acceptor of proton in hydrogen bonds. J. Mol. Struct. 2002, 642, 97–104. [Google Scholar] [CrossRef]
- De Almeida, W.B.; O’Malley, P.J. The effect of methoxy group rotation and hydrogen bonding on the redox properties of ubiquinone. Comput. Theor. Chem. 2015, 1069, 11–17. [Google Scholar] [CrossRef]

| CID | HIA | Caco-2 Permeability | Pgp Inhibitor | BBB Penetration | PPB | CYP3A4 Inhibitor |
|---|---|---|---|---|---|---|
| 5281643 | 0.720165 | 0.024945 | 0.019489 | 0.36468 | 0.34701 | 0.005815 |
| 5282149 | 0.813789 | 0.04693 | 0.015533 | 0.52433 | 0.3447 | 0.0056 |
| 5320646 | 0.834688 | 0.038304 | 0.779624 | 0.070213 | 1.064644 | 0.102586 |
| 5490003 | 0.915642 | 0.06874 | 0.943979 | 0.364681 | 0.970795 | 0.243789 |
| 15559724 | 0.916661 | 0.067603 | 0.902655 | 0.229925 | 1.035477 | 0.287866 |
| 74819331 | 0.720941 | 0.06123 | 0.028965 | 0.681398 | 0.402518 | 0.084012 |
| 74978256 | 0.296633 | 0.005106 | 0.438269 | 0.160238 | 0.878509 | 0.270137 |
| 91885208 | 0.764231 | 0.033519 | 0.019925 | 0.370717 | 0.480753 | 0.00434 |
| 162817595 | 0.576042 | 0.026792 | 0.874571 | 0.034886 | 1.014972 | 0.088901 |
| 162817597 | 0.200472 | 0.003026 | 0.020575 | 0.1995 | 0.375597 | 0.011781 |
| 162998749 | 0.376207 | 0.006379 | 0.022332 | 0.161656 | 0.399283 | 0.004699 |
| 163189397 | 0.381535 | 0.010827 | 0.895205 | 0.02035 | 0.891346 | 0.06044 |
| 46184988 | 0.97246 | 0.772927 | 0.321354 | 0.894446 | 0.879919 | 0.849846 |
| 6083 | 0.133681 | 0.006179 | 0.019417 | 0.272034 | 0.157684 | 0.008052 |
| CID | MW | LogP(o/w) | TPSA | HBAs | HBDs | Lipinski Violations |
|---|---|---|---|---|---|---|
| 5281643 | 464.379 | −0.62847 | 210.51 | 12 | 8 | Not accepted |
| 5282149 | 448.38 | −0.63566 | 190.28 | 11 | 7 | Not accepted |
| 5320646 | 566.518 | 5.267783 | 159.8 | 10 | 4 | Not accepted |
| 5490003 | 594.572 | 5.576282 | 137.8 | 10 | 2 | Not accepted |
| 15559724 | 580.545 | 5.608342 | 148.8 | 10 | 3 | Not accepted |
| 74819331 | 418.398 | −0.43163 | 156.91 | 9 | 6 | Accepted |
| 74978256 | 770.693 | 1.493971 | 284.73 | 18 | 9 | Not accepted |
| 91885208 | 478.406 | −0.53838 | 199.51 | 12 | 7 | Not accepted |
| 162817595 | 624.551 | 2.556964 | 225.81 | 14 | 7 | Not accepted |
| 162817597 | 624.548 | −1.07282 | 258.43 | 16 | 9 | Not accepted |
| 162998749 | 552.485 | −1.25717 | 250.97 | 14 | 10 | Not accepted |
| 163189397 | 870.81 | 2.335287 | 289.03 | 20 | 7 | Not accepted |
| 46184988 | 428.452 | 2.046065 | 96.95 | 9 | 0 | Accepted |
| 6083 | 347.224 | −2.69664 | 186.07 | 10 | 5 | Accepted |
| CID | AMES Toxicity | Mutagenicity | Carcinogens | Hepatotoxicity | hERG Inhibitor | LD50 (mg/kg) |
|---|---|---|---|---|---|---|
| 5281643 | 0.777389 | Inactive | 0.627358 | Inactive | 0.015333 | 5000 |
| 5282149 | 0.848903 | Inactive | 0.684536 | Inactive | 0.008304 | 5000 |
| 5320646 | 0.640871 | Inactive | 0.672932 | Inactive | 0.1671 | 4000 |
| 5490003 | 0.836254 | Inactive | 0.551992 | Inactive | 0.317555 | 4000 |
| 15559724 | 0.702942 | Inactive | 0.556323 | Inactive | 0.255199 | 4000 |
| 74819331 | 0.326982 | Active | 0.456241 | Inactive | 0.008861 | 2000 |
| 74978256 | 0.210756 | Inactive | 0.25206 | Inactive | 0.088186 | 5000 |
| 91885208 | 0.779684 | Inactive | 0.589165 | Inactive | 0.00742 | 5000 |
| 162817595 | 0.625209 | Active | 0.40003 | Inactive | 0.170601 | 5000 |
| 162817597 | 0.27347 | Inactive | 0.284445 | Inactive | 0.048891 | 5000 |
| 162998749 | 0.533229 | Inactive | 0.401097 | Inactive | 0.00953 | 5000 |
| 163189397 | 0.50833 | Inactive | 0.232467 | Inactive | 0.115662 | 5000 |
| 46184988 | 0.507371 | Active | 0.621935 | Inactive | 0.004847 | 360 |
| 6083 | 0.289806 | Inactive | 0.286742 | Inactive | 0.074652 | 11,250 |
| CID | Two-Dimensional Structure | vDW | Binding Energy | Intra-Molecular Interactions |
|---|---|---|---|---|
| RK-33 Docked |  | −81.6631 | −94.781 | Carbon–Hydrogen Bonds: HIS227, PRO203, THR201. Pi–Pi T-shaped: HIS527. Alkyl: ARG202. Pi–Alkyl: PRO205, ARG202. |
| 74978256 |  | −135.631 | −159.906 | Conventional Hydrogen Bonds: TYR200, THR201, GLN207. Carbon–Hydrogen Bonds: GLY229, TYR200. Pi–Anion: GLU285, TYR200. Pi-Donor Hydrogen Bonds: THR201, ARG202. Pi–Sigma: TYR200. Pi–Alkyl: ARG202. |
| 163189397 |  | −128.61 | −155.06 | Attractive Charge: ARG202, GLU524. Conventional Hydrogen Bonds: THR201, ARG202. Carbon Hydrogen Bonds: SER520, THR201. Pi-Donor Hydrogen Bonds: THR202. Pi–Sigma: THR201. Pi–Pi T-Shaped: HIS527. Pi–Alkyl: ARG202. |
| 74819331 |  | −129.001 | −147.727 | Conventional Hydrogen Bonds: ARG252, CYS317. Pi–Cation: ARG262. Pi–Anion: GLU256. Pi–Alkyl: ARG252. |
| 5490003 |  | −107.721 | −120.524 | Conventional Hydrogen Bonds: THR201, ARG202, THR411. Carbon–Hydrogen Bonds: HIS527. Pi–Sigma: VAL526. Pi–Pi Stacked: TYR200. Pi–Pi T-Shaped: HIS527. |
| 15559724 |  | −107.546 | −119.038 | Conventional Hydrogen Bonds: THR201, ARG202, THR411. Carbon Hydrogen Bonds: HIS527. Pi–Pi Stacked: TYR200. Pi–Pi T-Shaped: HIS527. Pi–Alkyl: VAL526. |
| 162817595 | 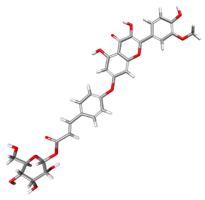 | −97.1523 | −116.779 | Conventional Hydrogen Bonds: GLY227, GLY229, LYS230, VAL526. Carbon Hydrogen Bonds: GLY530, GLN281, SER228. Pi–Pi Stacked: HIS527. Alkyl: VAL405. Pi–Alkyl: PRO205. |
| 162817597 | 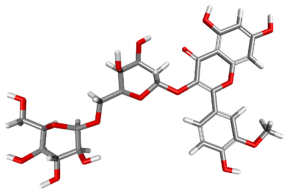 | −73.4756 | −115.757 | Conventional Hydrogen Bonds: THR411, SER412, GLY504, ARG531, ALA502, LEU505, GLY530. Carbon–Hydrogen Bonds: MET167. Amide–Pi Stacked: GKY227. P–Akyl: PRO205. |
| AMP Docked | 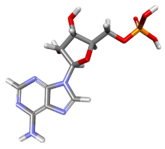 | −77.5459 | −115.411 | Conventional Hydrogen Bonds: TYR200, GLN207, GLY227, SER228, GLY229, LYS230, THR231, ARG202. Pi–Pi Stacked: TYR200. Pi–Pi Alkyl: ALA232. Attractive Charges: LYS230. |
| 5281643 |  | −72.3283 | −112.611 | Conventional Hydrogen Bonds: GLY504, ARG531, PRO203, ALA502, LEU505, GLY227. Carbon–Hydrogen Bonds: THR204, PRO205, ARG503. Pi–Cation: MET167. Pi–Pi Stacked: HIS527. Pi–Alkyl: PRO205. |
| 5320646 | 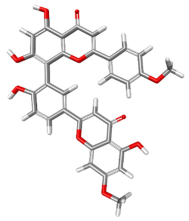 | −87.5561 | −112.148 | Conventional Hydrogen Bonds: ARG199, THE201, GLN207, LYS288, ARG199, ARG202, GLN207. Pi–Sigma: ARG199. Pi–Pi Stacked: TYR200. Alkyl: LYS288. Pi–Alkyl: ARG199. |
| 5282149 |  | −68.5132 | −108.82 | Conventional Hydrogen Bonds: LYS208, GLY504, HIS527, ARG531LEU505. Pi–Alkyl: PRO205, ARG503. |
| 91885208 |  | −73.5348 | −106.394 | Conventional Hydrogen Bonds: VAL206, SER228, THR411, ARG531, ARG534, THR226, GLY530, LEU505, SER228. Alkyl: VAL168, PRO205, VAL206, VAL405. Pi–Alkyl: PRO205. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asiri, A.; Alrehaily, A.; Al Ali, A.; Abu-Alghayth, M.H.; Tasleem, M. Repurposing Biomolecules from Aerva javanica Against DDX3X in LAML: A Computer-Aided Therapeutic Approach. Int. J. Mol. Sci. 2025, 26, 5445. https://doi.org/10.3390/ijms26125445
Asiri A, Alrehaily A, Al Ali A, Abu-Alghayth MH, Tasleem M. Repurposing Biomolecules from Aerva javanica Against DDX3X in LAML: A Computer-Aided Therapeutic Approach. International Journal of Molecular Sciences. 2025; 26(12):5445. https://doi.org/10.3390/ijms26125445
Chicago/Turabian StyleAsiri, Abdulaziz, Abdulwahed Alrehaily, Amer Al Ali, Mohammed H. Abu-Alghayth, and Munazzah Tasleem. 2025. "Repurposing Biomolecules from Aerva javanica Against DDX3X in LAML: A Computer-Aided Therapeutic Approach" International Journal of Molecular Sciences 26, no. 12: 5445. https://doi.org/10.3390/ijms26125445
APA StyleAsiri, A., Alrehaily, A., Al Ali, A., Abu-Alghayth, M. H., & Tasleem, M. (2025). Repurposing Biomolecules from Aerva javanica Against DDX3X in LAML: A Computer-Aided Therapeutic Approach. International Journal of Molecular Sciences, 26(12), 5445. https://doi.org/10.3390/ijms26125445








