Phage–Antibiotic Synergy Enhances Biofilm Eradication and Survival in a Zebrafish Model of Pseudomonas aeruginosa Infection
Abstract
1. Introduction
2. Results
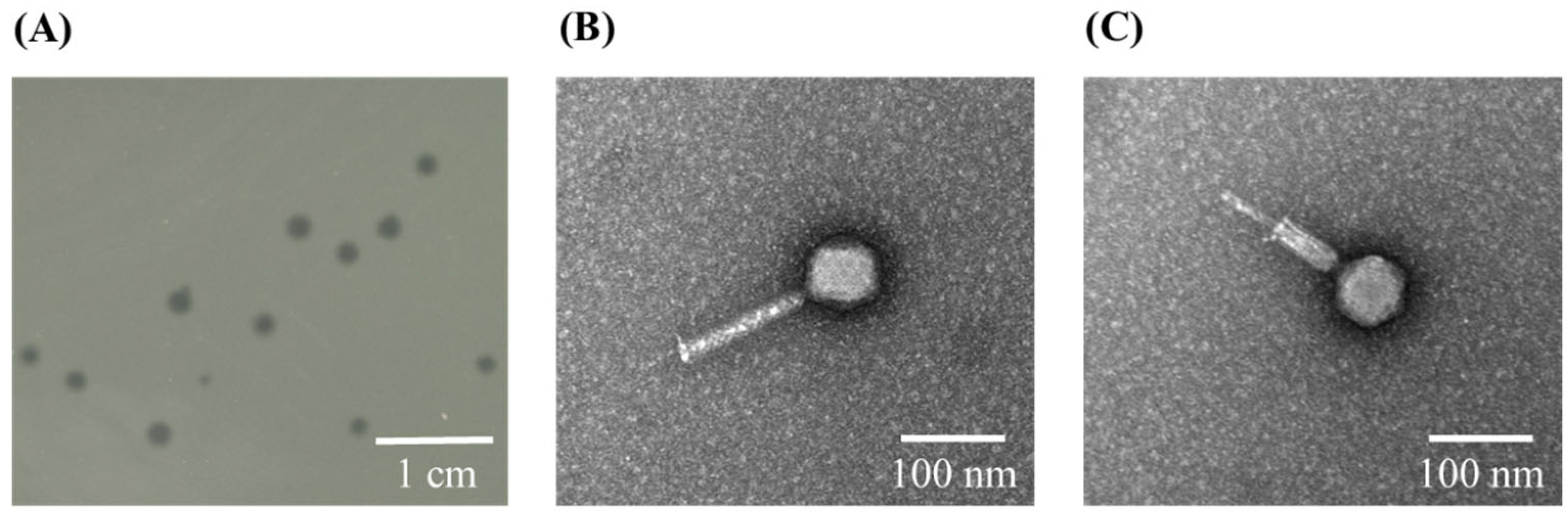
2.1. Isolation, Purification, and Morphology of P. aeruginosa Phage phiLCL12
2.2. Host Typing and Sensitivity to phiLCL12
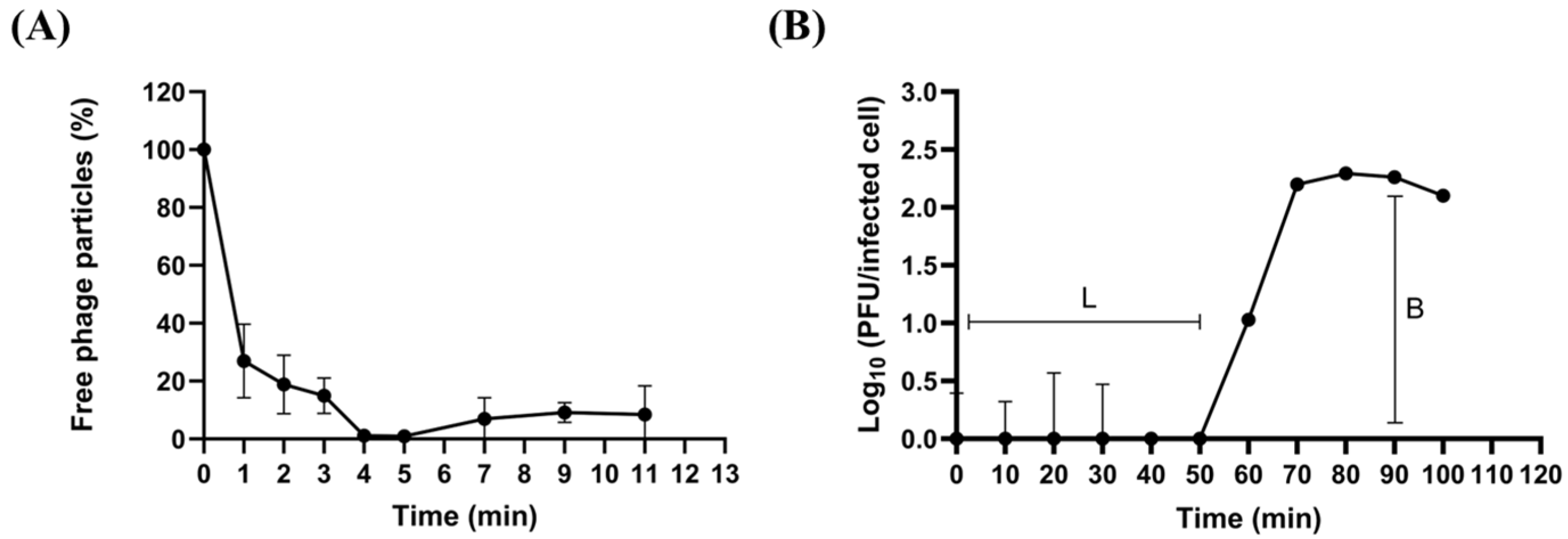
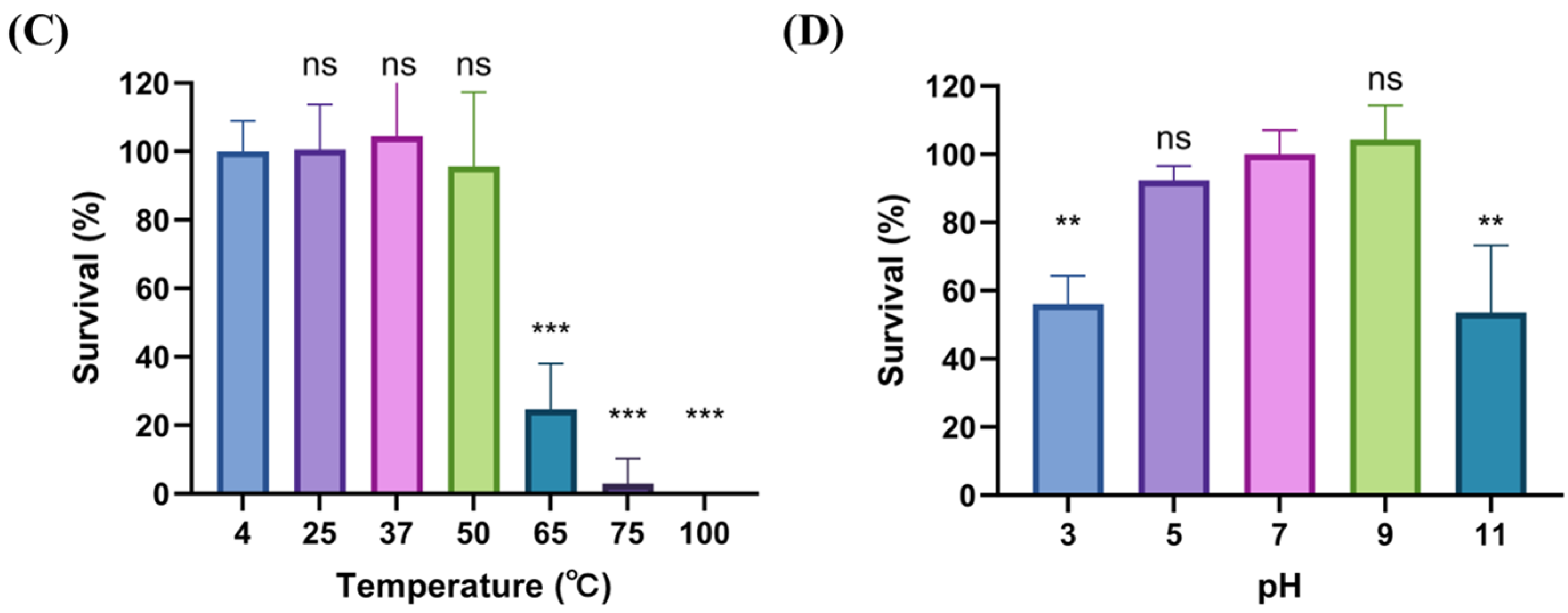
2.3. Biological Characteristics of phiLCL12
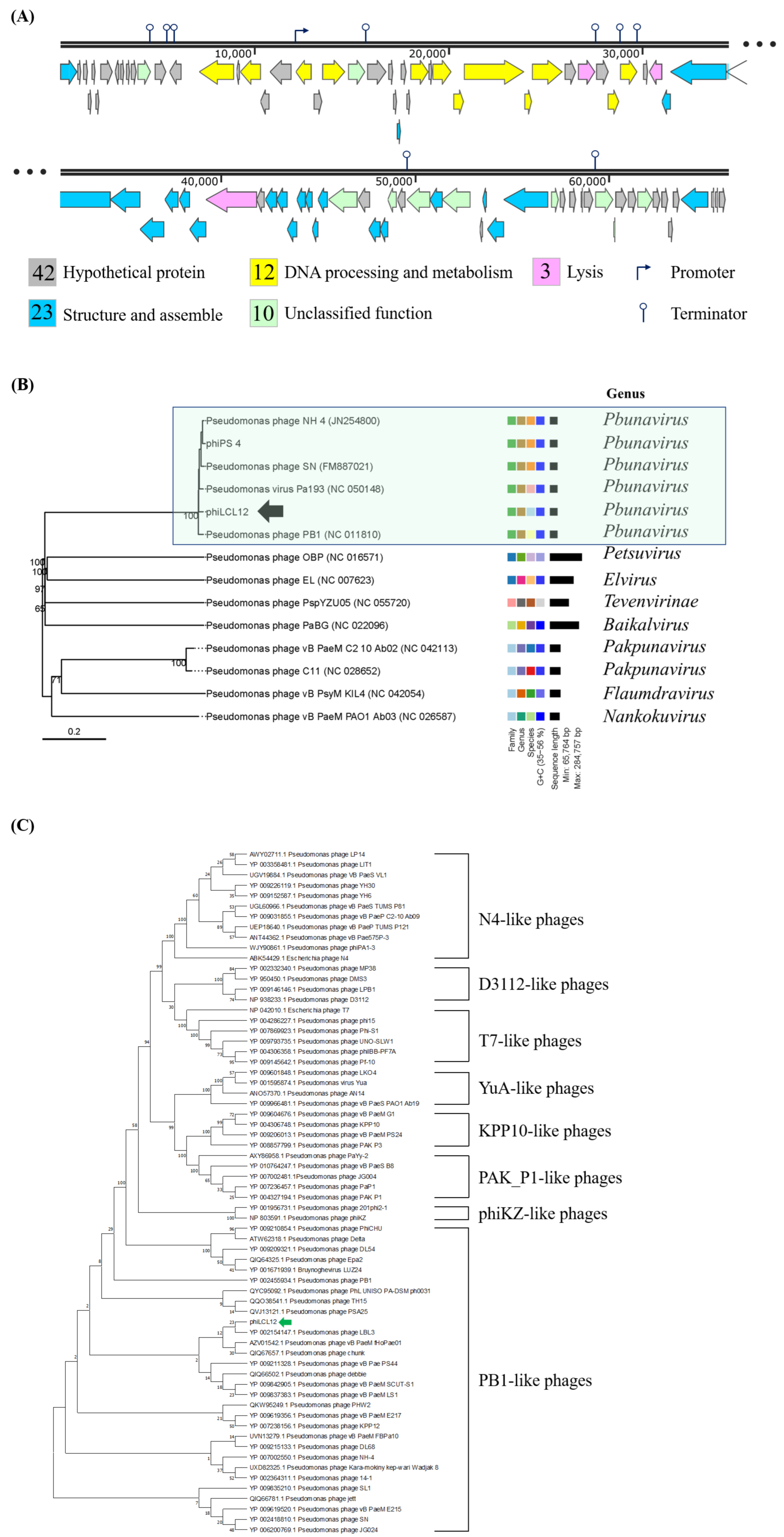
2.4. Genome Annotation and Phylogenetic Analysis
2.5. Structural Protein Analysis
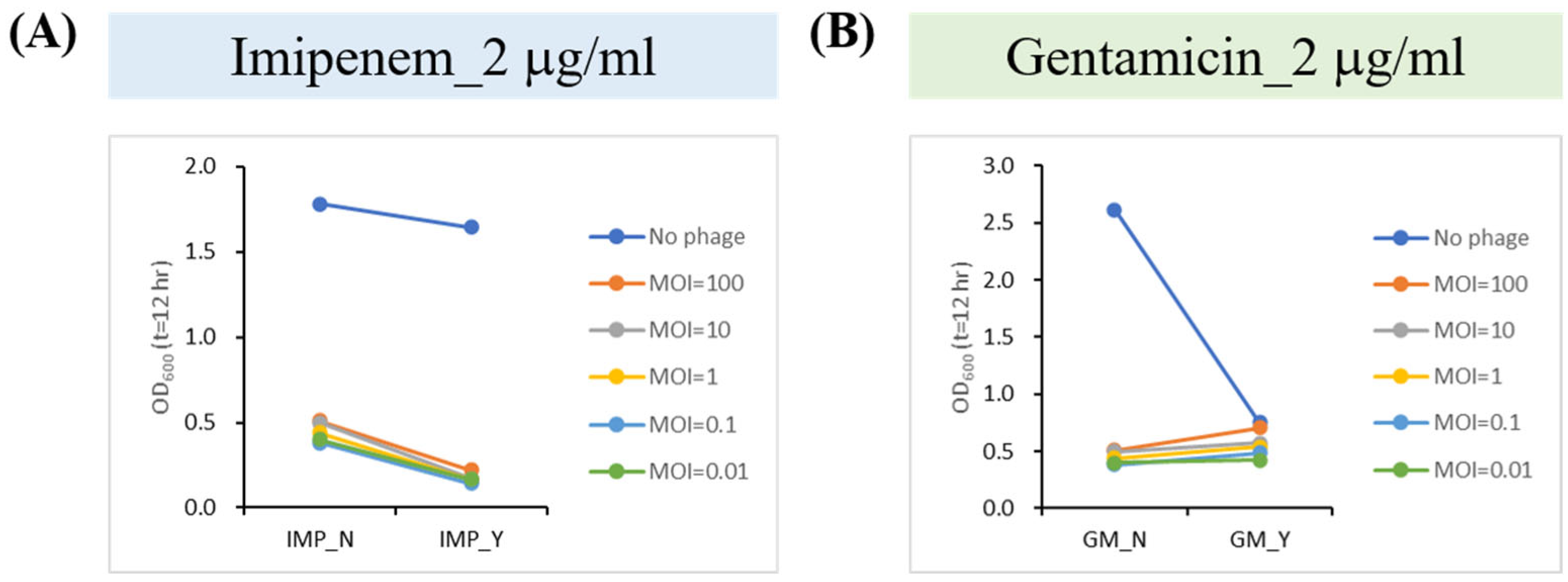
2.6. Phage–Antibiotic Synergy (PAS) Inhibits Bacterial Growth
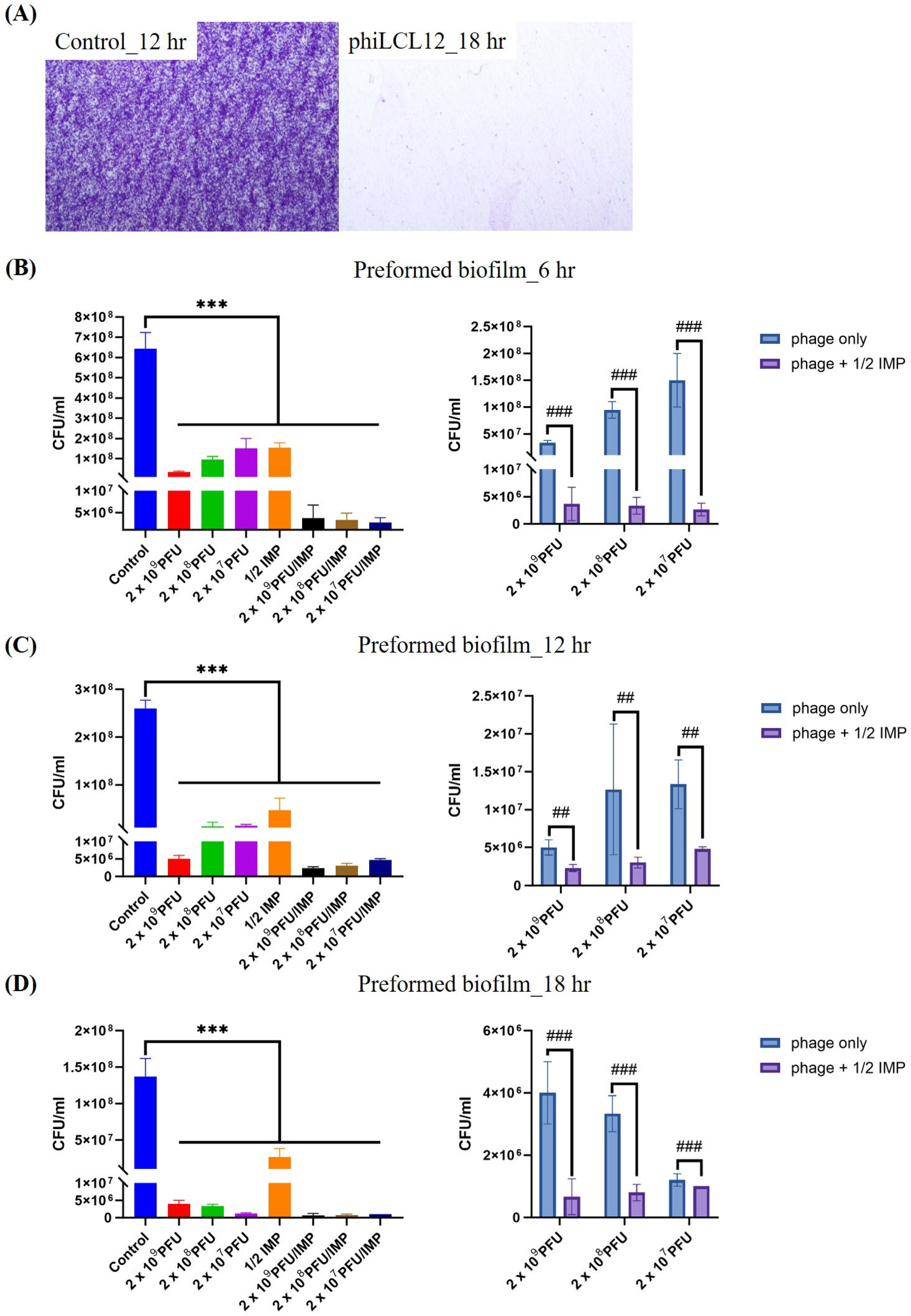
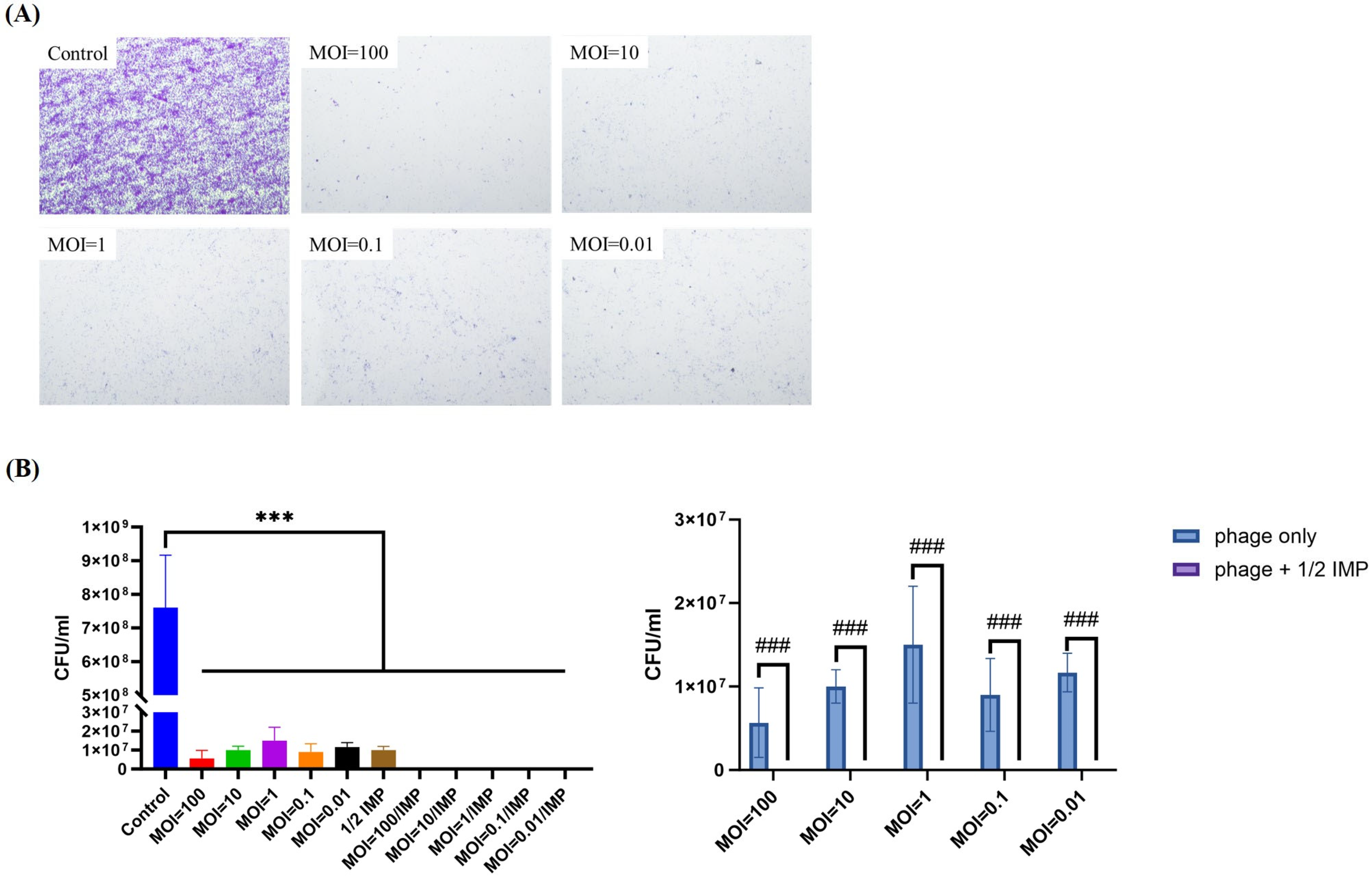
2.7. PAS Outperforms Phage Alone in Eradicating and Inhibiting P. aeruginosa Biofilms
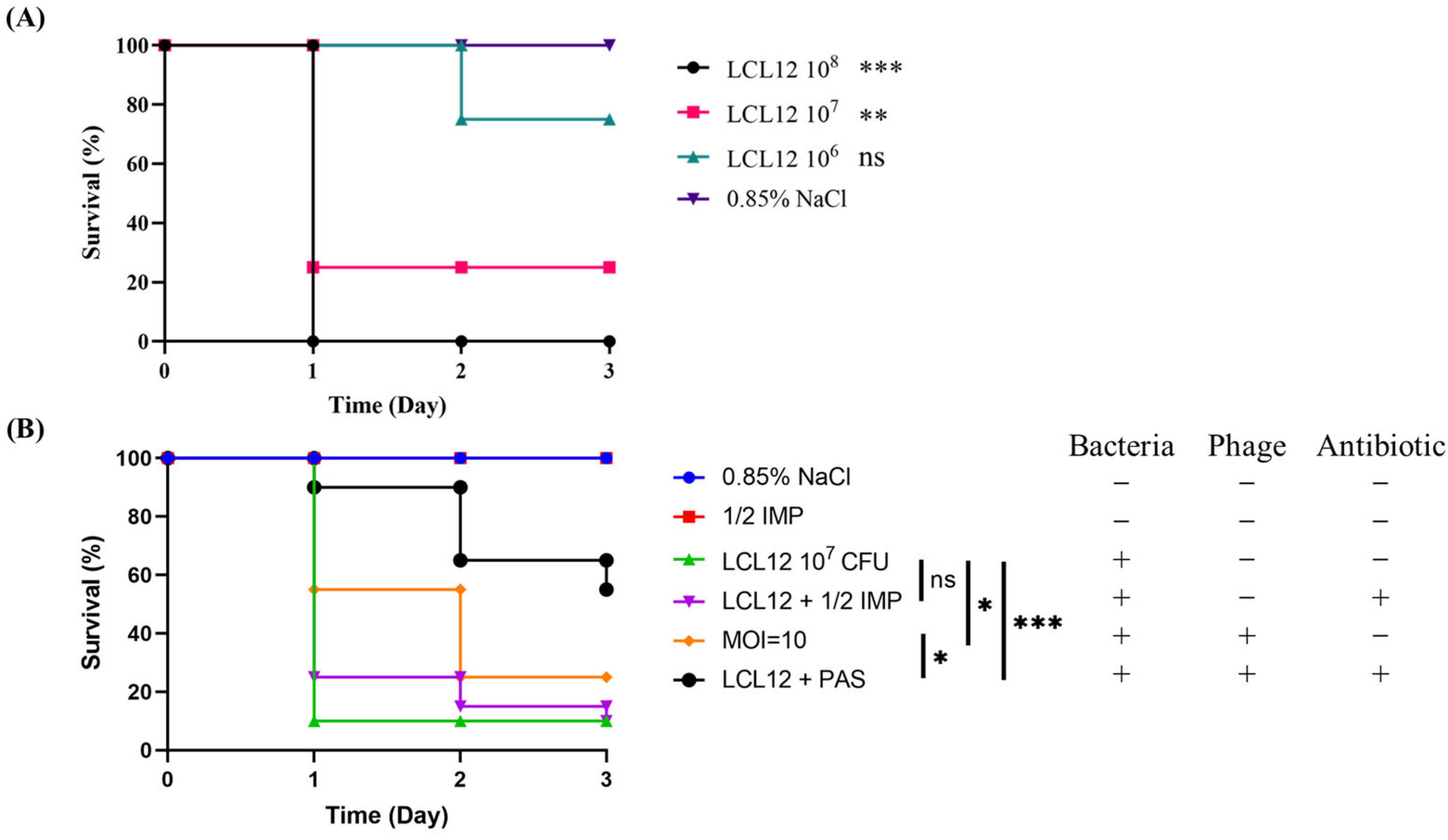
2.8. In Vivo Assessment of PAS Efficacy in a Zebrafish Model
3. Discussion
4. Materials and Methods
4.1. Bacteria Collection and Culture
4.2. Isolation and Purification of Bacteriophage
4.3. Transmission Electron Microscopy (TEM) Analysis
4.4. Host Range Test
4.5. Adsorption, One-Step Growth Curve, and Stability Assay
4.6. Bacterial Lysis
4.7. Phage DNA Extraction
4.8. Phage DNA Sequencing and Genome Analysis
4.9. Phage Structure Protein Analysis
4.10. Phage–Antibiotic Synergy Assay
4.11. Biofilm Eradication and Inhibition
4.12. In Vivo Assessment of PAS Efficacy Against P. aeruginosa LCL12 Infection
4.13. Statistical Analysis
4.14. Nucleotide Sequence Accession Number
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wheater, D.W.F.; Mara, D.D.; Jawad, L.; Oragui, J. Pseudomonas aeruginosa and Escherichia coli in sewage and fresh water. Water Res. 1980, 14, 713–721. [Google Scholar] [CrossRef]
- Walker, J.; Moore, G. Pseudomonas aeruginosa in hospital water systems: Biofilms, guidelines, and practicalities. J. Hosp. Infect. 2015, 89, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Schärer, V.; Meier, M.T.; Schuepbach, R.A.; Zinkernagel, A.S.; Boumasmoud, M.; Chakrakodi, B.; Brugger, S.D.; Fröhlich, M.R.; Wolfensberger, A.; Sax, H.; et al. An intensive care unit outbreak with multi-drug-resistant Pseudomonas aeruginosa—Spotlight on sinks. J. Hosp. Infect. 2023, 139, 161–167. [Google Scholar] [CrossRef]
- Mahmoud, S.F.; Fayez, M.; Swelum, A.A.; Alswat, A.S.; Alkafafy, M.; Alzahrani, O.M.; Alsunaini, S.J.; Almuslem, A.; Al Amer, A.S.; Yusuf, S. Genetic Diversity, Biofilm Formation, and Antibiotic Resistance of Pseudomonas aeruginosa Isolated from Cow, Camel, and Mare with Clinical Endometritis. Vet. Sci. 2022, 9, 239. [Google Scholar] [CrossRef]
- Kour, A.; Jaglan, S.; Sharma, S.; Sharma, S. A new strategy to treat Pseudomonas aeruginosa infected burn wounds: Antimicrobial cocktails as potent topical therapy. Med. Hypotheses 2023, 180, 111167. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Silva Filho, L.V.; Ferreira Fde, A.; Reis, F.J.; Britto, M.C.; Levy, C.E.; Clark, O.; Ribeiro, J.D. Pseudomonas aeruginosa infection in patients with cystic fibrosis: Scientific evidence regarding clinical impact, diagnosis, and treatment. J. Bras. Pneumol. 2013, 39, 495–512. [Google Scholar] [CrossRef]
- Tredget, E.E.; Shankowsky, H.A.; Joffe, A.M.; Inkson, T.I.; Volpel, K.; Paranchych, W.; Kibsey, P.C.; Alton, J.D.; Burke, J.F. Epidemiology of infections with Pseudomonas aeruginosa in burn patients: The role of hydrotherapy. Clin. Infect. Dis. 1992, 15, 941–949. [Google Scholar] [CrossRef]
- Yin, R.; Cheng, J.; Wang, J.; Li, P.; Lin, J. Treatment of Pseudomonas aeruginosa infectious biofilms: Challenges and strategies. Front. Microbiol. 2022, 13, 955286. [Google Scholar] [CrossRef]
- Kadirvelu, L.; Sivaramalingam, S.S.; Jothivel, D.; Chithiraiselvan, D.D.; Karaiyagowder Govindarajan, D.; Kandaswamy, K. A review on antimicrobial strategies in mitigating biofilm-associated infections on medical implants. Curr. Res. Microb. Sci. 2024, 6, 100231. [Google Scholar] [CrossRef]
- Omwenga, E.O.; Awuor, S.O. The Bacterial Biofilms: Formation, Impacts, and Possible Management Targets in the Healthcare System. Can. J. Infect. Dis. Med. Microbiol. 2024, 2024, 1542576. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, K.; Meshram, S. Biofilm Formation in Chronic Infections: A Comprehensive Review of Pathogenesis, Clinical Implications, and Novel Therapeutic Approaches. Cureus 2024, 16, e70629. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.-S.; Harnod, D.; Hsueh, P.-R. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2022, 12, 823684. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Wise, M.G.; Hsieh, T.-C.; Lu, H.-C.; Chen, W.-T.; Cheng, M.-H.; Siddiqui, F.; Young, K.; Motyl, M.R.; Sahm, D.F. Temporal and geographical prevalence of carbapenem-resistant Pseudomonas aeruginosa and the in vitro activity of ceftolozane/tazobactam and comparators in Taiwan—SMART 2012–2021. J. Glob. Antimicrob. Resist. 2023, 34, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.-S.; Lee, Y.-L.; Liu, P.-Y.; Lu, M.-C.; Ko, W.-C.; Hsueh, P.-R. Multicenter surveillance of antimicrobial susceptibilities and resistance mechanisms among Enterobacterales species and non-fermenting Gram-negative bacteria from different infection sources in Taiwan from 2016 to 2018. J. Microbiol. Immunol. Infect. 2022, 55, 463–473. [Google Scholar] [CrossRef]
- Ayobami, O.; Brinkwirth, S.; Eckmanns, T.; Markwart, R. Antibiotic resistance in hospital-acquired ESKAPE-E infections in low- and lower-middle-income countries: A systematic review and meta-analysis. Emerg. Microbes Infect. 2022, 11, 443–451. [Google Scholar] [CrossRef]
- Naddaf, M. 40 million deaths by 2050: Toll of drug-resistant infections to rise by 70. Nature 2024, 633, 747–748. [Google Scholar] [CrossRef]
- Chegini, Z.; Khoshbayan, A.; Vesal, S.; Moradabadi, A.; Hashemi, A.; Shariati, A. Bacteriophage therapy for inhibition of multi drug-resistant uropathogenic bacteria: A narrative review. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 30. [Google Scholar] [CrossRef]
- Hetta, H.F.; Rashed, Z.I.; Ramadan, Y.N.; Al-Kadmy, I.M.S.; Kassem, S.M.; Ata, H.S.; Nageeb, W.M. Phage Therapy, a Salvage Treatment for Multidrug-Resistant Bacteria Causing Infective Endocarditis. Biomedicines 2023, 11, 2860. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Li, X.; He, Y.; Wang, Z.; Wei, J.; Hu, T.; Si, J.; Tao, G.; Zhang, L.; Xie, L.; Abdalla, A.E.; et al. A combination therapy of Phages and Antibiotics: Two is better than one. Int. J. Biol. Sci. 2021, 17, 3573–3582. [Google Scholar] [CrossRef] [PubMed]
- Łusiak-Szelachowska, M.; Międzybrodzki, R.; Drulis-Kawa, Z.; Cater, K.; Knežević, P.; Winogradow, C.; Amaro, K.; Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Rękas, J.; et al. Bacteriophages and antibiotic interactions in clinical practice: What we have learned so far. J. Biomed. Sci. 2022, 29, 23. [Google Scholar] [CrossRef] [PubMed]
- Segall, A.M.; Roach, D.R.; Strathdee, S.A. Stronger together? Perspectives on phage-antibiotic synergy in clinical applications of phage therapy. Curr. Opin. Microbiol. 2019, 51, 46–50. [Google Scholar] [CrossRef]
- Siddell, S.G.; Smith, D.B.; Adriaenssens, E.; Alfenas-Zerbini, P.; Dutilh, B.E.; Garcia, M.L.; Junglen, S.; Krupovic, M.; Kuhn, J.H.; Lambert, A.J.; et al. Virus taxonomy and the role of the International Committee on Taxonomy of Viruses (ICTV). J. Gen. Virol. 2023, 104, 001840. [Google Scholar] [CrossRef]
- Akrami, S.; Ekrami, A.; Jahangirimehr, F.; Yousefi Avarvand, A. High prevalence of multidrug-resistant Pseudomonas aeruginosa carrying integron and exoA, exoS, and exoU genes isolated from burn patients in Ahvaz, southwest Iran: A retrospective study. Health Sci. Rep. 2024, 7, e2164. [Google Scholar] [CrossRef]
- Firouzi-Dalvand, L.; Pooladi, M.; Nowroozi, J.; Akhvan-Sepahi, A.; Hooshiyar, M. Presence of exoU and exoS Genes in Pseudomonas aeruginosa Isolated from Urinary Tract Infections. Infect. Epidemiol. Med. 2016, 2, 8–11. [Google Scholar] [CrossRef]
- Nicas, T.I.; Frank, D.W.; Stenzel, P.; Lile, J.D.; Iglewski, B.H. Role of exoenzyme S in chronic Pseudomonas aeruginosa lung infections. Eur. J. Clin. Microbiol. 1985, 4, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, V.; Yom, K.; Ozer, E.A.; Pura, O.; Hughes, A.; Murphy, K.R.; Cudzilo, L.; Mitchell, D.; Hauser, A.R. Environmental reservoirs for exoS+ and exoU+ strains of Pseudomonas aeruginosa. Env. Microbiol. Rep. 2018, 10, 485–492. [Google Scholar] [CrossRef]
- Wdowiak, M.; Paczesny, J.; Raza, S. Enhancing the Stability of Bacteriophages Using Physical, Chemical, and Nano-Based Approaches: A Review. Pharmaceutics 2022, 14, 1936. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder—An open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef] [PubMed]
- Kleinheinz, K.A.; Joensen, K.G.; Larsen, M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 2014, 4, e27943. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Rajandas, H.; Parimannan, S.; Manickam, R.; Marimuthu, K.; Petersen, B.; Clokie, M.R.J.; Millard, A.; Sicheritz-Pontén, T. PhageLeads: Rapid Assessment of Phage Therapeutic Suitability Using an Ensemble Machine Learning Approach. Viruses 2022, 14, 342. [Google Scholar] [CrossRef] [PubMed]
- Naville, M.; Ghuillot-Gaudeffroy, A.; Marchais, A.; Gautheret, D. ARNold: A web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 2011, 8, 11–13. [Google Scholar] [CrossRef]
- Sampaio, M.; Rocha, M.; Oliveira, H.; Dias, O. Predicting promoters in phage genomes using PhagePromoter. Bioinformatics 2019, 35, 5301–5302. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.Z.; Yuan, W.G.; Shang, J.; Shi, Y.H.; Yang, L.L.; Liu, M.; Zhu, P.; Jin, T.; Sun, Y.; Yuan, L.H. Virus classification for viral genomic fragments using PhaGCN2. Brief. Bioinform. 2023, 24, bbac505. [Google Scholar] [CrossRef]
- Gu Liu, C.; Green Sabrina, I.; Min, L.; Clark Justin, R.; Salazar Keiko, C.; Terwilliger Austen, L.; Kaplan Heidi, B.; Trautner Barbara, W.; Ramig Robert, F.; Maresso Anthony, W. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 2020, 11, e01462-20. [Google Scholar] [CrossRef]
- Kamal, F.; Dennis, J.J. Burkholderia cepacia complex Phage-Antibiotic Synergy (PAS): Antibiotics stimulate lytic phage activity. Appl. Environ. Microbiol. 2015, 81, 1132–1138. [Google Scholar] [CrossRef]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef]
- Feltman, H.; Schulert, G.; Khan, S.; Jain, M.; Peterson, L.; Hauser, A.R. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 2001, 147 Pt 10, 2659–2669. [Google Scholar] [CrossRef]
- Zhuo, C.; Wang, L.; Xiao, S.-N.; Li, H.-Y.; Qiu, G.-X.; Zhong, N.-S. Clinical significance of virulence-related genes of type III secretion system of Pseudomonas aeruginosa. Zhonghua Shao Shang Za Zhi Zhonghua Shaoshang Zazhi Chin. J. Burns 2010, 26, 354–359. [Google Scholar]
- Horna, G.; Amaro, C.; Palacios, A.; Guerra, H.; Ruiz, J. High frequency of the exoU+/exoS+ genotype associated with multidrug-resistant “high-risk clones” of Pseudomonas aeruginosa clinical isolates from Peruvian hospitals. Sci. Rep. 2019, 9, 10874. [Google Scholar] [CrossRef] [PubMed]
- Yousefi-Avarvand, A.; Khashei, R.; Sedigh Ebrahim-Saraie, H.; Emami, A.H.; Zomorodian, K.; Motamedifar, M. The Frequency of Exotoxin A and Exoenzymes S and U Genes Among Clinical Isolates of Pseudomonas aeruginosa in Shiraz, Iran. Int. J. Mol. Cell. Med. 2015, 4, 167–173. [Google Scholar] [PubMed]
- Song, Y.; Mu, Y.; Wong, N.-K.; Yue, Z.; Li, J.; Yuan, M.; Zhu, X.; Hu, J.; Zhang, G.; Wei, D.; et al. Emergence of hypervirulent Pseudomonas aeruginosa pathotypically armed with co-expressed T3SS effectors ExoS and ExoU. hLife 2023, 1, 44–56. [Google Scholar] [CrossRef]
- Ceyssens, P.-J.; Miroshnikov, K.; Mattheus, W.; Krylov, V.N.; Robben, J.; Noben, J.-P.; Vanderschraeghe, S.; Sykilinda, N.N.; Kropinski, A.M.; Volckaert, G.; et al. Comparative analysis of the widespread and conserved PB1-like viruses infecting Pseudomonas aeruginosa. Environ. Microbiol. 2009, 11, 2874–2883. [Google Scholar] [CrossRef]
- Garbe, J.; Wesche, A.; Bunk, B.; Kazmierczak, M.; Selezska, K.; Rohde, C.; Sikorski, J.; Rohde, M.; Jahn, D.; Schobert, M. Characterization of JG024, a Pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol. 2010, 10, 301. [Google Scholar] [CrossRef]
- Sharma, S.; Datta, S.; Chatterjee, S.; Dutta, M.; Samanta, J.; Vairale, M.G.; Gupta, R.; Veer, V.; Dwivedi, S.K. Isolation and characterization of a lytic bacteriophage against Pseudomonas aeruginosa. Sci. Rep. 2021, 11, 19393. [Google Scholar] [CrossRef]
- Rezk, N.; Abdelsattar, A.S.; Elzoghby, D.; Agwa, M.M.; Abdelmoteleb, M.; Aly, R.G.; Fayez, M.S.; Essam, K.; Zaki, B.M.; El-Shibiny, A. Bacteriophage as a potential therapy to control antibiotic-resistant Pseudomonas aeruginosa infection through topical application onto a full-thickness wound in a rat model. J. Genet. Eng. Biotechnol. 2022, 20, 133. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, Z.; Mao, Z.; Tang, M.; Chen, H.; Qian, C.; Zeng, W.; Zhou, T.; Wu, Q. Quorum sensing gene lasR promotes phage vB_Pae_PLY infection in Pseudomonas aeruginosa. BMC Microbiol. 2024, 24, 207. [Google Scholar] [CrossRef]
- Cui, J.; Shi, X.; Wang, X.; Sun, H.; Yan, Y.; Zhao, F.; Zhang, C.; Liu, W.; Zou, L.; Han, L.; et al. Characterization of a lytic Pseudomonas aeruginosa phage vB_PaeP_ASP23 and functional analysis of its lysin LysASP and holin HolASP. Front. Microbiol. 2023, 14, 1093668. [Google Scholar] [CrossRef]
- Schrader, H.S.; Schrader, J.O.; Walker, J.J.; Wolf, T.A.; Nickerson, K.W.; Kokjohn, T.A. Bacteriophage infection and multiplication occur in Pseudomonas aeruginosa starved for 5 years. Can. J. Microbiol. 1997, 43, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Chegini, Z.; Khoshbayan, A.; Taati Moghadam, M.; Farahani, I.; Jazireian, P.; Shariati, A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Ratner, L. Biomarkers and Preclinical Models for Adult T-Cell Leukemia-Lymphoma Treatment. Front. Microbiol. 2019, 10, 2109. [Google Scholar] [CrossRef] [PubMed]
- Engeman, E.; Freyberger, H.R.; Corey, B.W.; Ward, A.M.; He, Y.; Nikolich, M.P.; Filippov, A.A.; Tyner, S.D.; Jacobs, A.C. Synergistic Killing and Re-Sensitization of Pseudomonas aeruginosa to Antibiotics by Phage-Antibiotic Combination Treatment. Pharmaceuticals 2021, 14, 184. [Google Scholar] [CrossRef]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front. Cell. Infect. Microbiol. 2017, 7, 418. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.F.; Krisch, H.M. Phage-Antibiotic Synergy (PAS): Beta-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef]
- Uchiyama, J.; Shigehisa, R.; Nasukawa, T.; Mizukami, K.; Takemura-Uchiyama, I.; Ujihara, T.; Murakami, H.; Imanishi, I.; Nishifuji, K.; Sakaguchi, M.; et al. Piperacillin and ceftazidime produce the strongest synergistic phage—Antibiotic effect in Pseudomonas aeruginosa. Arch. Virol. 2018, 163, 1941–1948. [Google Scholar] [CrossRef]
- Kim, M.; Jo, Y.; Hwang, Y.J.; Hong, H.W.; Hong, S.S.; Park, K.; Myung, H. Phage-Antibiotic Synergy via Delayed Lysis. Appl. Environ. Microbiol. 2018, 84, e02085-18. [Google Scholar] [CrossRef]
- Gawish, A.A.; Mohamed, N.A.; El-shennawy, G.A.; Mohamed, H.A. An investigation of type 3 secretion toxins encoding-genes of Pseudomonas aeruginosa isolates in a University Hospital in Egypt. J. Microbiol. Infect. Dis. 2013, 3, 116–122. [Google Scholar] [CrossRef]
- Paranos, P.; Pournaras, S.; Meletiadis, J. A single-layer spot assay for easy, fast, and high-throughput quantitation of phages against multidrug-resistant Gram-negative pathogens. J. Clin. Microbiol. 2024, 62, e0074324. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Altaf, M.M.; Ahmad, I. In vitro Biofilm Development and Enhanced Rhizosphere Colonization of Triticum aestivum by Fluorescent pseudomonas sp. J. Pure Appl. Microbiol. 2019, 13, 1441–1449. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Lee, Y.-P.; Lin, N.-T.; Yang, H.-H.; Teh, S.-H.; Lin, L.-C. Therapeutic effect and anti-biofilm ability assessment of a novel phage, phiPA1-3, against carbapenem-resistant Pseudomonas aeruginosa. Virus Res. 2023, 335, 199178. [Google Scholar] [CrossRef]

| Sources | phiLCL12 a | exoS b | exoU c | Phenotype | |
|---|---|---|---|---|---|
| P. aeruginosa | |||||
| PA001 | clinical | +++ | + | − | Invasion |
| LCL12 | clinical | ++++ | − | + | Cytotoxic |
| LCL13 | clinical | ++++ | − | + | Cytotoxic |
| LCL14 | clinical | ++++ | − | + | Cytotoxic |
| PS-1 | bile | +++ | + | − | Invasion |
| PS-2 | pus | +++ | − | + | Cytotoxic |
| PS-3 | urine | − | + | − | Invasion |
| PS-4 | pus | ++++ | − | + | Cytotoxic |
| PS-5 | sputum | ++ | − | + | Cytotoxic |
| PS-6 | sputum | +++ | − | + | Cytotoxic |
| PS-7 | sputum | + | + | − | Invasion |
| PA10 | clinical | ++ | + | − | Invasion |
| PA13 | clinical | − | + | − | Invasion |
| PA18 | clinical | ++++ | + | + | Invasion/Cytotoxic |
| PA20 | clinical | ++++ | + | − | Invasion |
| PA22 | clinical | + | + | − | Invasion |
| PA25 | clinical | ++ | + | − | Invasion |
| PA75 | clinical | ++ | + | − | Invasion |
| PA76 | clinical | +++ | + | − | Invasion |
| PA77 | clinical | ++ | − | + | Cytotoxic |
| PA78 | clinical | ++ | + | − | Invasion |
| PA79 | clinical | +++ | − | + | Cytotoxic |
| PA80 | clinical | ++ | + | − | Invasion |
| PA81 | clinical | + | + | − | Invasion |
| PA82 | clinical | ++ | − | + | Cytotoxic |
| PA83 | clinical | ++++ | − | + | Cytotoxic |
| PA84 | clinical | + | − | + | Cytotoxic |
| PA85 | clinical | − | − | + | Cytotoxic |
| PA86 | clinical | − | − | − | - |
| PA87 | clinical | +++ | + | − | Invasion |
| PA88 | clinical | ++ | − | + | Cytotoxic |
| PA89 | clinical | +++ | + | − | Invasion |
| PA90 | clinical | +++ | + | − | Invasion |
| PA91 | clinical | +++ | − | + | Cytotoxic |
| PA92 | clinical | +++ | + | − | Invasion |
| PA005 | oral | − | + | − | Invasion |
| PA006 | oral | − | − | − | - |
| PA009 | oral | + | + | − | Invasion |
| PA010 | oral | − | + | − | Invasion |
| PA011 | oral | − | + | − | Invasion |
| PA022 | oral | + | + | − | Invasion |
| PA023 | oral | + | + | − | Invasion |
| PA024 | oral | + | + | − | Invasion |
| PA025 | oral | +++ | + | − | Invasion |
| Antibiotic (Sensitivity a) | |||
|---|---|---|---|
| Imipenem | Gentamicin | Ceftazidime | |
| LCL12 | 4 μg/mL (S) | 4 μg/mL (S) | 256 μg/mL (R) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, L.-C.; Tsai, Y.-C.; Lin, N.-T. Phage–Antibiotic Synergy Enhances Biofilm Eradication and Survival in a Zebrafish Model of Pseudomonas aeruginosa Infection. Int. J. Mol. Sci. 2025, 26, 5337. https://doi.org/10.3390/ijms26115337
Lin L-C, Tsai Y-C, Lin N-T. Phage–Antibiotic Synergy Enhances Biofilm Eradication and Survival in a Zebrafish Model of Pseudomonas aeruginosa Infection. International Journal of Molecular Sciences. 2025; 26(11):5337. https://doi.org/10.3390/ijms26115337
Chicago/Turabian StyleLin, Ling-Chun, Yu-Chuan Tsai, and Nien-Tsung Lin. 2025. "Phage–Antibiotic Synergy Enhances Biofilm Eradication and Survival in a Zebrafish Model of Pseudomonas aeruginosa Infection" International Journal of Molecular Sciences 26, no. 11: 5337. https://doi.org/10.3390/ijms26115337
APA StyleLin, L.-C., Tsai, Y.-C., & Lin, N.-T. (2025). Phage–Antibiotic Synergy Enhances Biofilm Eradication and Survival in a Zebrafish Model of Pseudomonas aeruginosa Infection. International Journal of Molecular Sciences, 26(11), 5337. https://doi.org/10.3390/ijms26115337





