Construction of SNP Fingerprinting and Genetic Diversity Analysis of Eggplant Based on KASP Technology
Abstract
1. Introduction
2. Results
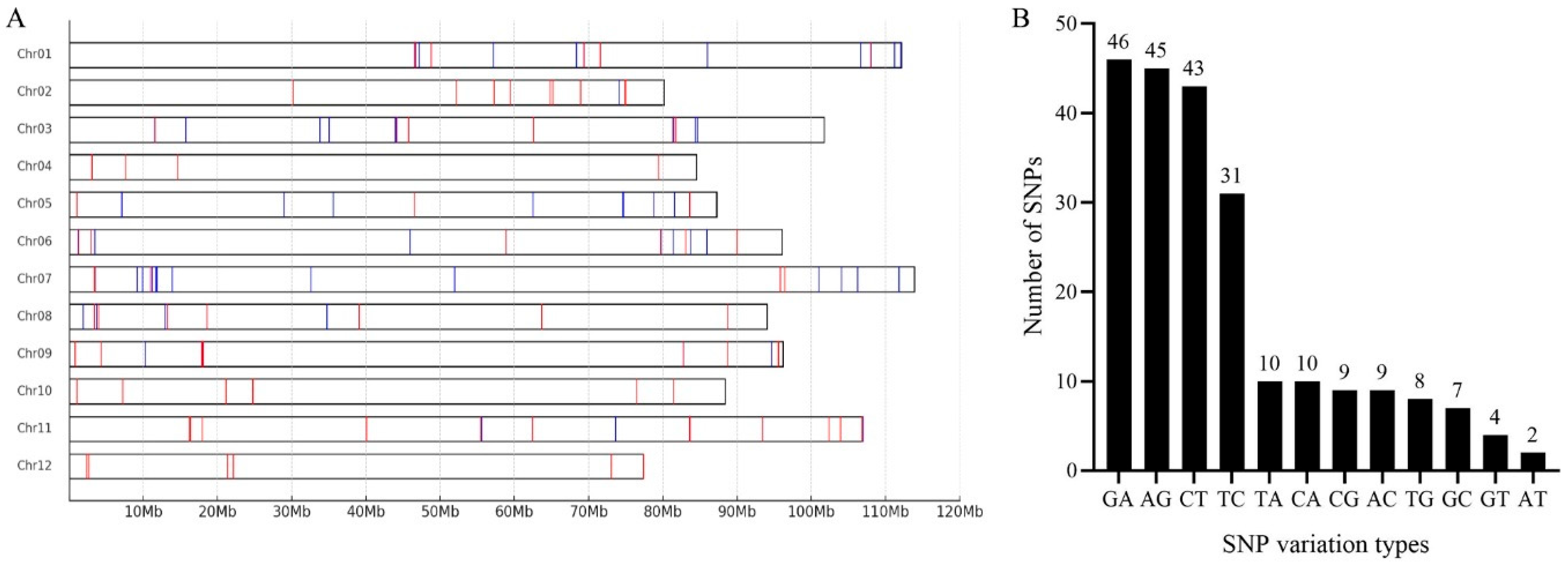
2.1. Identification of High-Quality SNPs
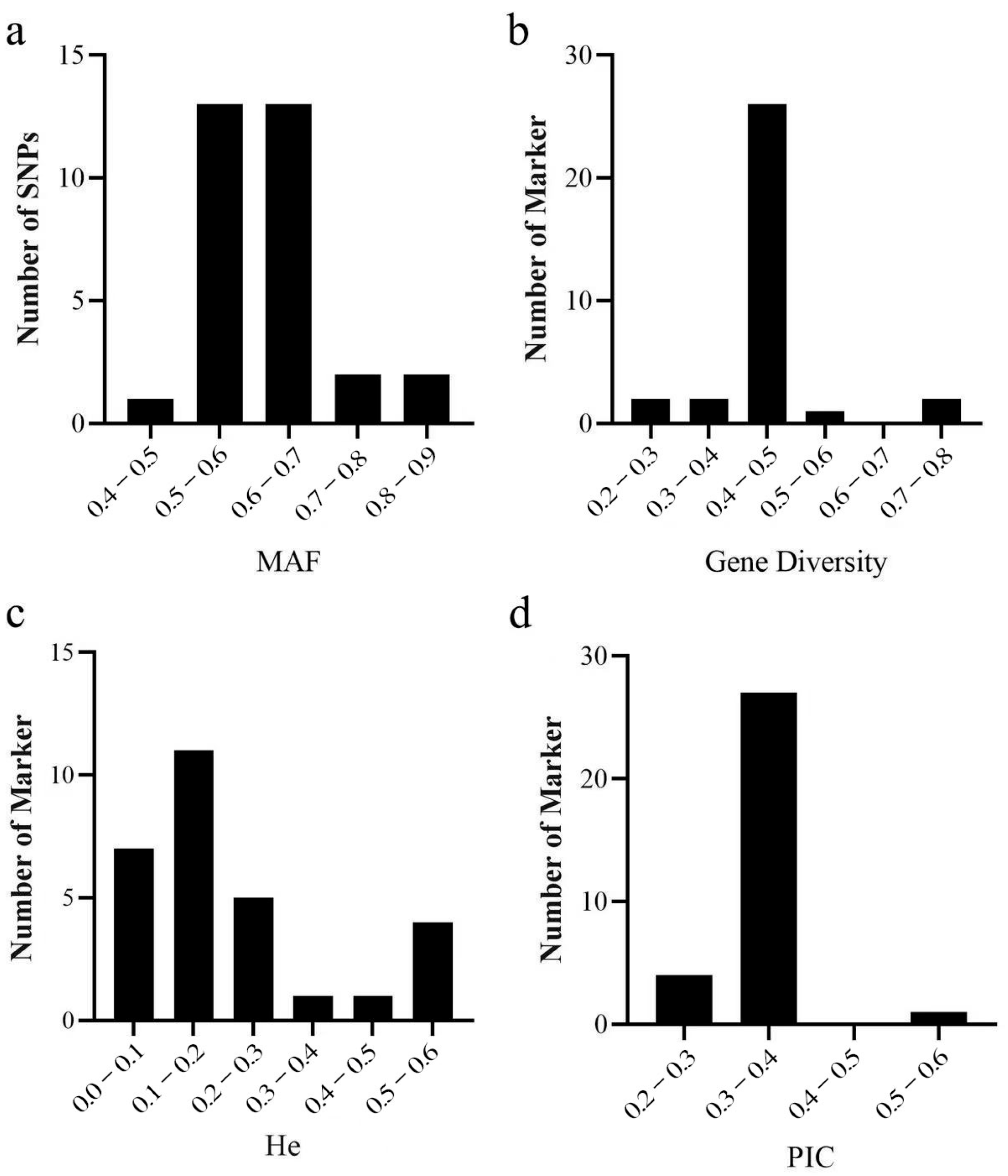
2.2. Development and Genotyping of KASP
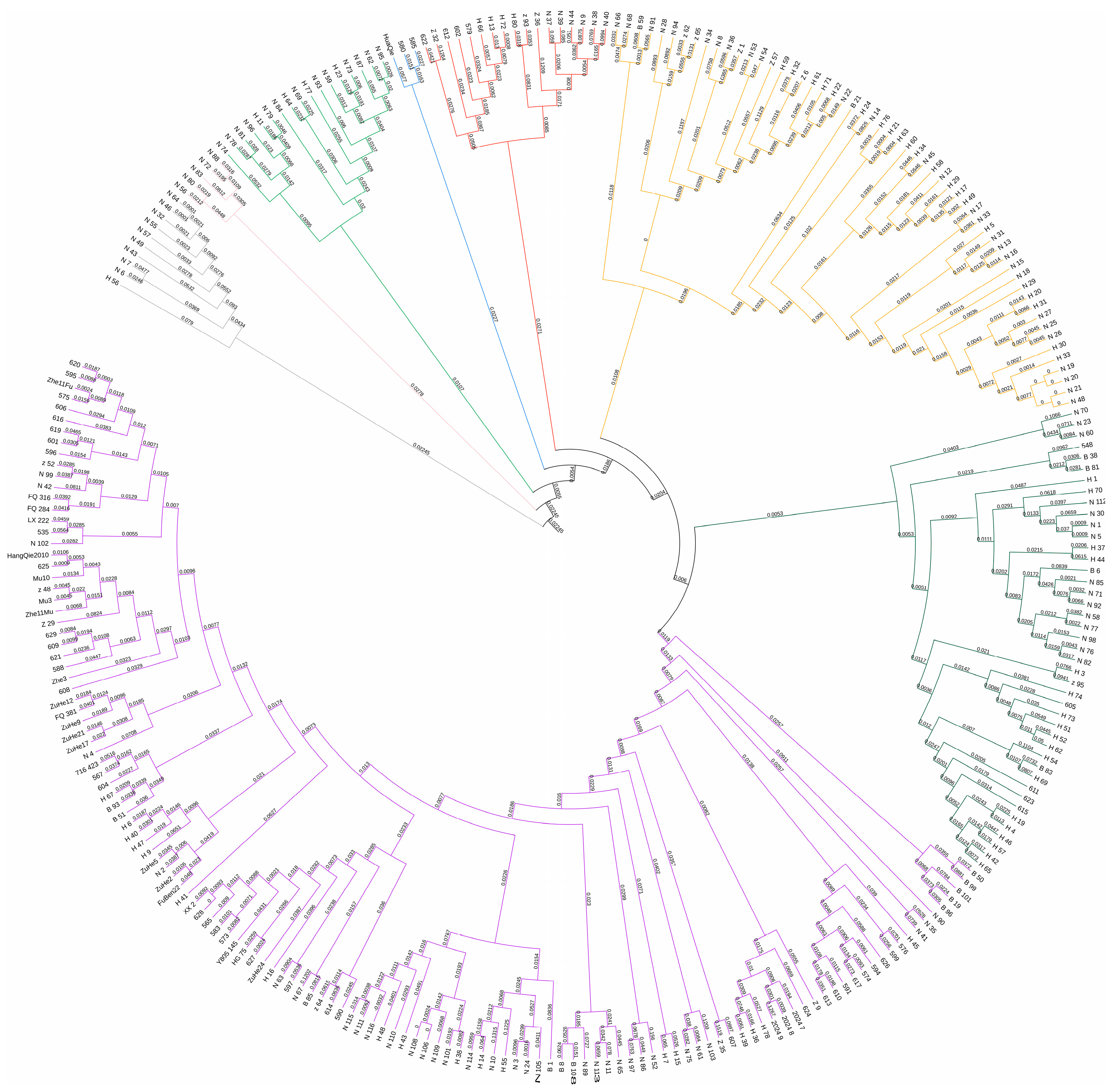
2.3. Analysis of Genetic Diversity
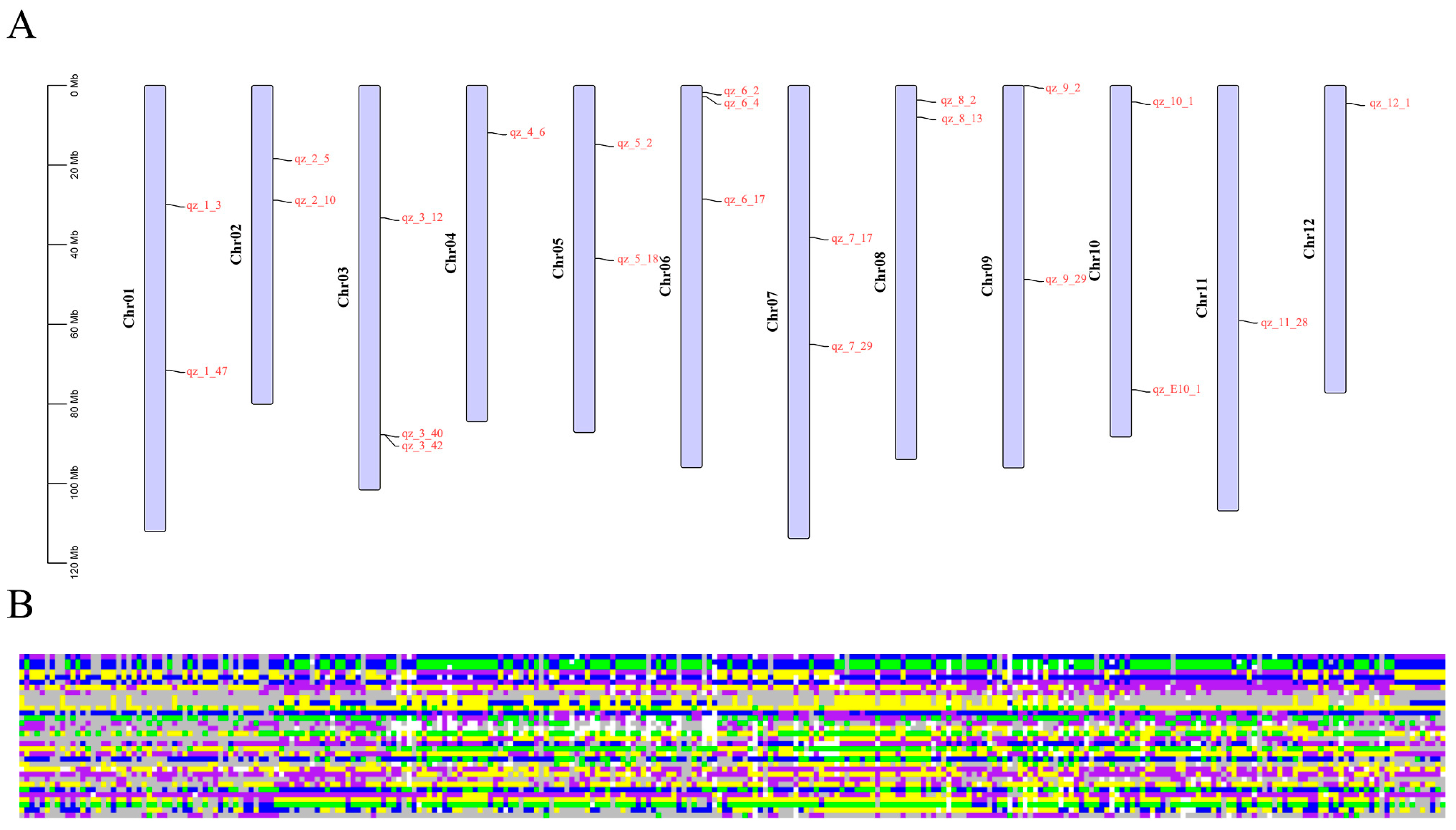
2.4. Construction of SNP Fingerprints
2.5. Construction of Core Germplasm
3. Discussion
4. Materials and Methods
4.1. Plant Materials and DNA Extraction
4.2. SNPs Identification
4.3. KASP Development and Genotyping
4.4. Population Differentiation and Genetic Diversity Indices
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daunay, M.C.; Janick, J. History and Iconography of Eggplant. Chron. Hortic. 2007, 47, 16–22. [Google Scholar]
- Sękara, A.; Cebula, S.; Kunicki, E. Cultivated eggplants-origin, breeding objectives and genetic resources, a review. Folia Hort. 2007, 19, 97–114. [Google Scholar]
- Han, S.W.; Tae, J.; Kim, J.A.; Kim, D.K.; Seo, G.S.; Yun, K.J.; Choi, S.C.; Kim, T.H.; Nah, Y.H.; Lee, Y.M. The aqueous extract of Solanum melongena inhibits PAR2 agonist-induced inflammation. Clin. Chim. Acta. 2003, 328, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, D.J.; Kendall, C.W.; Marchie, A.; Faulkner, D.A.; Wong, J.M.; de Souza, R.; Emam, A.; Parker, T.L.; Vidgen, E.; Trautwein, E.A.; et al. Direct comparison of a dietary portfolio of cholesterol-lowering foods with a statin in hypercholesterolemic participants. Am. J. Clin. Nutr. 2005, 81, 380–387. [Google Scholar] [CrossRef]
- Kashyap, V.; Vinod Kumar, S.; Collonnier, C.; Fusari, F.; Haicour, R.; Rotino, G.L.; Sihachakr, D.; Rajam, M.V. Biotechnology of eggplant. Sci. Hortic. 2003, 97, 1–25. [Google Scholar] [CrossRef]
- Yunnan Institute of Botany. Flora of Yunnan. Volume 2: Seed Plants; Science Press: Beijing, China, 1979; pp. 563–591. [Google Scholar]
- Cao, J.S.; Qin, L. Germplasm Resources of Horticultural Plants; China Agricultural Press: Beijing, China, 2005; pp. 1–4. [Google Scholar]
- Marshall, D.R. Crop genetic resources: Current and emerging issues. In Plant Population Genetics, Breeding and Genetic Resources; Cambridge University Press: Cambridge, UK, 1990; pp. 367–388. [Google Scholar]
- Tirnaz, S.; Batley, J. Epigenetics: Potentials and challenges in crop breeding. Mol. Plant. 2019, 12, 1309–1311. [Google Scholar] [CrossRef]
- Yuan, J.; Cao, H.; Qin, W.; Yang, S.; Zhang, D.; Zhu, L.; Song, H.; Zhang, Q. Genomic and modern biotechnological strategies for enhancing salt tolerance in crops. New Crops 2025, 2, 2949–9526. [Google Scholar] [CrossRef]
- Slavin, J.L.; Lloyd, B. Health Benefits of Fruits and Vegetables. Adv. Nutr. 2012, 3, 506–516. [Google Scholar] [CrossRef]
- Daraboina, R.; Cooper, O.; Amini, M. Segmentation of organic food consumers: A revelation of purchase factors in organic food markets. J. Retail. Consum. Serv. 2024, 78, 103710. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Jeffreys, A.J.; Wilson, V.; Thein, S.L. Hypervariable ‘minisatellite’ regions in human DNA. Nature 1985, 314, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.S.; Sargeant, L.L.; King, T.J.; Mattick, J.S.; Georges, M.; Hetzel, D.J. The conservation of dinucleotide microsatellites among mammalian genomes allows the use of heterologous PCR primer pairs in closely related species. Genomics 1991, 10, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, S.; Kanagarajan, S.; Ponnusamy, S. Efficiency of RAPD and ISSR markers system in accessing genetic variation of rice bean (Vigna umbellata) landraces. Electron. J. Biotechnol. 2008, 11, 32–41. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S. The new genomics: Global views of biology. Science 1996, 274, 536–539. [Google Scholar] [CrossRef]
- Li, G.; Quiros, C.F. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001, 103, 455–461. [Google Scholar] [CrossRef]
- Hu, J.; Vick, B.A. Target region amplification polymorphism: A novel marker technique for plant genotyping. Plant Mol. Biol. Rep. 2003, 21, 289–294. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Start Codon Targeted (SCoT) Polymorphism: A Simple, Novel DNA Marker Technique for Generating Gene-Targeted Markers in Plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Yang, G.; Chen, S.; Chen, L.; Sun, K.; Huang, C.; Zhou, D.; Huang, Y.; Wang, J.; Liu, Y.; Wang, H.; et al. Development of a core SNP arrays based on the KASP method for molecular breeding of rice. Rice 2019, 12, 21. [Google Scholar] [CrossRef]
- Chen, Z.; Tang, D.; Ni, J.; Li, P.; Liu, J. Development of genic KASP SNP markers from RNA-Seq data for map-based cloning and marker-assisted selection in maize. BMC Plant Biol. 2021, 21, 157. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.O.; Zhang, N.; Jin, H.; Si, H. Genetic analysis of potato breeding collection using single-nucleotide polymorphism (SNP) markers. Plants 2023, 12, 1895. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.S.; Zhao, N.; Li, H.; Zhai, H.; He, S.Z.; Liu, Q.C. SSR fingerprinting of 203 sweetpotato (Ipomoea batatas (L.) Lam.) varieties. J. Integr. Agric. 2018, 17, 86–93. [Google Scholar] [CrossRef]
- Ongom, P.O.; Fatokun, C.; Togola, A.; Salvo, S.; Oyebode, O.G.; Ahmad, M.S.; Jockson, I.D.; Bala, G.; Boukar, O. Molecular Fingerprinting and Hybridity Authentication in Cowpea Using Single Nucleotide Polymorphism Based Kompetitive Allele-Specific PCR Assay. Front. Plant Sci. 2021, 12, 734117. [Google Scholar] [CrossRef]
- Ou, T.; Wu, Z.; Tian, C.; Yang, Y.; Gong, W.; Niu, J.; Li, Z. Development of Genome-Wide SSR Markers in Leymus chinensis with Genetic Diversity Analysis and DNA Fingerprints. Int. J. Mol. Sci. 2025, 26, 918. [Google Scholar] [CrossRef]
- Wu, F.; Cai, G.; Xi, P.; Guo, Y.; Xu, M.; Li, A. Genetic Diversity Analysis and Fingerprint Construction for 87 Passionfruit (Passiflora spp.) Germplasm Accessions on the Basis of SSR Fluorescence Markers. Int. J. Mol. Sci. 2024, 25, 10815. [Google Scholar] [CrossRef]
- Shen, Y.; Wang, J.; Shaw, R.K.; Yu, H.; Sheng, X.; Zhao, Z.; Li, S.; Gu, H. Development of GBTS and KASP panels for genetic diversity, population structure, and fingerprinting of a large collection of Broccoli (Brassica oleracea L. var. italica) in China. Front. Plant Sci. 2021, 12, 655254. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Lyu, M.J.; Liu, J.; Wu, J.N.; Wang, Q.; Xie, T.Y.; Li, H.; Chen, R.; Sun, D.; Yang, Y.; et al. Construction of an SNP fingerprinting database and population genetic analysis of 329 cauliflower cultivars. BMC Plant Biol. 2022, 22, 522. [Google Scholar] [CrossRef]
- Bibi, A.C.; Marountas, J.; Katsarou, K.; Kollias, A.; Pavlidis, P.; Goumenaki, E.; Kafetzopoulos, D. Nuclear SSR-based genetic diversity and STRUCTURE analysis of Greek tomato landraces and the Greek Tomato Database (GTD). Plant Genet. Resour. 2024, 22, 107–116. [Google Scholar] [CrossRef]
- Shahzad, R.; Jamil, S.; Rahman, S.U.; Yasmeen, E.; Iqbal, M.Z. Development of a core set of simple sequence repeat markers for DNA fingerprinting of tomato germplasm. Pak. J. Bot. 2024, 56, 989–999. [Google Scholar]
- Park, G.; Choi, Y.; Jung, J.K.; Shim, E.J.; Kang, M.Y.; Sim, S.C.; Chung, S.M.; Lee, G.P. Genetic Diversity Assessment and Cultivar Identification of Cucumber (Cucumis sativus L.) Using the Fluidigm Single Nucleotide Polymorphism Assay. Plants 2021, 10, 395. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Falcón, J.E.; Vilanova, S.; Plazas, M.; Prohens, J. Diversity, relationships, and genetic fingerprinting of the Listada de Gandía eggplant landrace using genomic SSRs and EST-SSRs. Sci. Hortic. 2011, 129, 238–246. [Google Scholar] [CrossRef]
- Vilanova, S.; Manzur, J.P.; Prohens, J. Development and characterization of genomic simple sequence repeat markers in eggplant and their application to the study of diversity and relationships in a collection of different cultivar types and origins. Mol. Breed. 2012, 30, 647–660. [Google Scholar] [CrossRef]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 11769. [Google Scholar] [CrossRef]
- Liu, W.; Qian, Z.; Zhang, J.; Yang, J.; Wu, M.; Barchi, L.; Zhao, H.; Sun, H.; Cui, Y.; Wen, C. Impact of fruit shape selection on genetic structure and diversity uncovered from genome-wide perfect SNPs genotyping in eggplant. Mol. Breed. 2019, 39, 140. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Han, R.; Zhang, F.; Mao, A.; Luo, J.; Dong, B.; Liu, H.; Tang, H.; Zhang, J.; et al. Target SSR-Seq: A novel SSR genotyping technology associate with perfect SSRs in genetic analysis of cucumber varieties. Front. Plant Sci. 2019, 10, 531. [Google Scholar] [CrossRef]
- Gessese, M.; Bariana, H.; Wong, D.; Hayden, M.; Bansal, U. Molecular Mapping of Stripe Rust Resistance Gene Yr81 in a Common Wheat Landrace Aus27430. Plant Dis. 2019, 103, 1166–1171. [Google Scholar] [CrossRef]
- Zhu, Z.; Cao, Q.; Han, D.; Wu, J.; Wu, L.; Tong, J.; Xu, X.; Yan, J.; Zhang, Y.; Xu, K.; et al. Molecular characterization and validation of adult-plant stripe rust resistance gene Yr86 in Chinese wheat cultivar Zhongmai 895. Theor. Appl. Genet. 2023, 136, 142. [Google Scholar] [CrossRef]
- Pang, H.; Ai, J.; Wang, W.; Hu, T.; Hu, H.; Wang, J.; Yan, Y.; Wu, X.; Bao, C.; Wei, Q. Fine mapping of QTL-fl3.1 reveal SmeFL as the candidate gene regulating fruit length in eggplant (Solanum melongena L.). Veget. Res. 2024, 4, e028. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Petr, D.; Auton, A.; Goncalo, A.; Cornelis, A.; Banks, E.; Mark, A. The variant call format and VCF tools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]

| Original Germplasm | Core Germplasm | MR | CE | SH | HE | NE | CV |
|---|---|---|---|---|---|---|---|
| 280 | 56 | 0.36 | 0.36 | 3.41 | 0.61 | 1.00 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Pang, H.; Hu, N.; Hu, H.; Hu, T.; Yan, Y.; Wang, J.; Ai, J.; Bao, C.; Wei, Q. Construction of SNP Fingerprinting and Genetic Diversity Analysis of Eggplant Based on KASP Technology. Int. J. Mol. Sci. 2025, 26, 5312. https://doi.org/10.3390/ijms26115312
Wang W, Pang H, Hu N, Hu H, Hu T, Yan Y, Wang J, Ai J, Bao C, Wei Q. Construction of SNP Fingerprinting and Genetic Diversity Analysis of Eggplant Based on KASP Technology. International Journal of Molecular Sciences. 2025; 26(11):5312. https://doi.org/10.3390/ijms26115312
Chicago/Turabian StyleWang, Wuhong, Hongtao Pang, Na Hu, Haijiao Hu, Tianhua Hu, Yaqin Yan, Jinglei Wang, Jiaqi Ai, Chonglai Bao, and Qingzhen Wei. 2025. "Construction of SNP Fingerprinting and Genetic Diversity Analysis of Eggplant Based on KASP Technology" International Journal of Molecular Sciences 26, no. 11: 5312. https://doi.org/10.3390/ijms26115312
APA StyleWang, W., Pang, H., Hu, N., Hu, H., Hu, T., Yan, Y., Wang, J., Ai, J., Bao, C., & Wei, Q. (2025). Construction of SNP Fingerprinting and Genetic Diversity Analysis of Eggplant Based on KASP Technology. International Journal of Molecular Sciences, 26(11), 5312. https://doi.org/10.3390/ijms26115312






