Acute Lymphoblastic Leukemia and Associated HLA-A, B, DRB1, and DQB1 Molecules: A Moroccan Pediatric Case–Control Study
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
| Characteristics | Acute Lymphoblastic Leukemia Patients n (%) |
|---|---|
| Total patient | 70 |
| Median age, years, range | 9.25 (0.5–18) |
| Type of acute lymphoid leukemia | |
| ALL B | 39 (55.71) |
| ALL T | 23 (32.85) |
| Unclassified | 8 (11.42) |
| Gender | |
| Female | 26 (62.85) |
| Male | 44 (37.14) |
| ABO groups | |
| A+ | 17 (24.28) |
| A− | 0 (0) |
| B+ | 14 (20) |
| B− | 0 (0) |
| AB+ | 3 (4.28) |
| AB− | 0 (0) |
| O+ | 25 (35.71) |
| O- | 3 (4.28) |
| Unclassified | 8 (11.42) |
| Children’s race | |
| White | 61 (87.14) |
| Black | 9 (12.85) |
2.2. Distribution of HLA Loci and Allele Groups in Patients with ALL and Controls
- HLA-A allele groups.
| HLA-A | Frequency of Allele Groups in Patients with ALL 2n = 140; n (%) | Frequency of Allele Groups in Control Group 2n = 272; n (%) | p-Value |
|---|---|---|---|
| A*02 | 26 (18.57%) | 65 (23.89%) | 0.26 |
| A*30 | 7 (5.00%) | 24 (8.82%) | 0.23 |
| A*68 | 15 (10.71%) | 7 (2.57%) | 0.001 |
| A*01 | 12 (8.57%) | 38 (13.97%) | 0.15 |
| A*03 | 17 (12.14%) | 26 (9.55%) | 0.52 |
| A*24 | 9 (6.42%) | 15 (5.51%) | 0.87 |
| A*33 | 5 (3.57%) | 18 (6.61%) | 0.29 |
| A*29 | 5 (3.57%) | 11 (4.04%) | 0.97 |
| A*11 | 8 (5.71%) | 6 (2.20%) | 0.11 |
| A*32 | 7 (5.00%) | 13 (4.77%) | 0.88 |
| A*23 | 13 (9.28%) | 24 (8.82%) | 0.97 |
| A*26 | 4 (1.92%) | 5 (1.83%) | 0.75 |
| A*31 | 2 (2.85%) | 5 (1.83%) | 0.92 |
| A*34 | 2 (2.85%) | 4 (1.47%) | 0.68 |
| A*36 | 1 (0.71%) | 0 (0.00%) | 0.73 |
| A*25 | 1 (0.71%) | 1 (0.36%) | 0.78 |
| A*28 | 2 (2.85%) | 0 (0.00%) | 0.21 |
| A*66 | 2 (2.85%) | 6 (2.20%) | 0.86 |
| A*74 | 1 (0.71%) | 0 (0.00%) | 0.75 |
| A*80 | 1 (0.71%) | 4 (1.47%) | 0.85 |
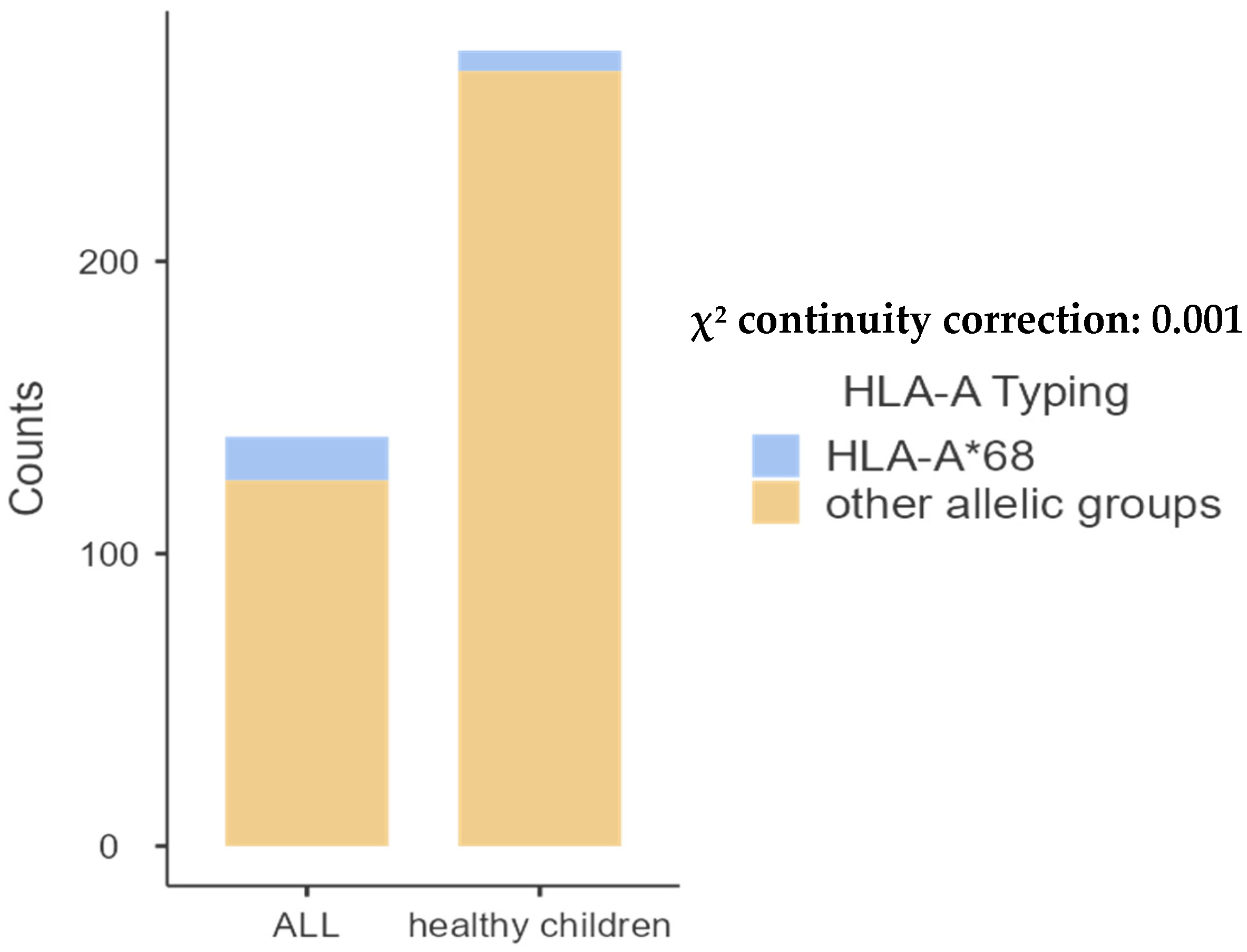
- HLA-B allele groups.
| HLA-B | Frequency of Allele Groups in Patients with ALL 2n = 140; n (%) | Frequency of Allele Groups in Control Group 2n = 272; n (%) | p-Value |
|---|---|---|---|
| B*15 | 3 (2.14%) | 10 (3.67%) | 0.58 |
| B*13 | 0 (0.00%) | 5 (1.83%) | 0.25 |
| B*07 | 11 (7.85%) | 12 (4.41%) | 0.22 |
| B*14 | 11 (7.85%) | 7 (2.57%) | 0.02 |
| B*44 | 15 (10.71%) | 29 (10.66%) | 0.87 |
| B*50 | 12 (8.57%) | 17 (6.25%) | 0.50 |
| B*35 | 4 (2.85%) | 15 (5.51%) | 0.33 |
| B*51 | 12 (8.57%) | 12 (4.41%) | 0.13 |
| B*08 | 7 (5%) | 12 (4.41%) | 0.98 |
| B*18 | 7 (5%) | 10 (3.67%) | 0.70 |
| B*49 | 9 (6.42%) | 16 (5.88%) | <0.001 |
| B*41 | 3 (2.14%) | 11 (4.04%) | 0.47 |
| B*27 | 6 (4.28%) | 9 (3.30%) | 0.82 |
| B*42 | 3 (2.14%) | 6 (2.20%) | 0.75 |
| B*53 | 4 (2.85%) | 11 (4.04%) | 0.74 |
| B*45 | 7 (5%) | 20 (7.35%) | 0.48 |
| B*38 | 5 (3.57%) | 8 (2.94%) | 0.96 |
| B*40 | 0 (0%) | 10 (3.67%) | 0.05 |
| B*78 | 1 (0.71%) | 2 (0.73%) | 0.55 |
| B*57 | 2 (1.42%) | 17 (6.25%) | 0.04 |
| B*52 | 2 (1.42%) | 5 (1.83%) | 0.92 |
| B*17 | 1 (0.71%) | 2 (0.73%) | 0.55 |
| B*39 | 3 (2.14%) | 7 (2.57%) | 0.94 |
| B*58 | 5 (3.57%) | 11 (4.04%) | 0.97 |
| B*55 | 0 (0.0%) | 2 (0.73%) | 0.78 |
| B*37 | 2 (1.42%) | 0 (1.04%) | 0.21 |
| B*72 | 3 (2.14%) | 3 (1.10%) | 0.68 |
| B*63 | 2 (1.42%) | 0 (0.0%) | 0.21 |
| B*62 | 0 (0.0%) | 2 (0.73%) | 0.78 |
| B*55 | 0 (0.0%) | 2 (0.73%) | 0.78 |
| B*25 | 0 (0.0%) | 1 (0.36%) | 0.73 |

- HLA-DRB1 allele groups.
| HLA-DRB1 | Frequency of Allele Groups in Patients with ALL 2n = 110; n (%) | Frequency of Allele Groups in Control Group 2n = 190; n (%) | p-Value |
|---|---|---|---|
| DRB1*03 | 24 (24.00%) | 33 (17.36%) | 0.42 |
| DRB1*13 | 17 (15.45%) | 38 (20%) | 0.40 |
| DRB1*04 | 14 (12.72%) | 29 (15.26%) | 0.66 |
| DRB1*15 | 10 (9.09%) | 28 (14.73%) | 0.21 |
| DRB1*11 | 11 (10%) | 9 (4.73%) | 0.12 |
| DRB1*07 | 17 (15.45%) | 33 (17.36%) | 0.78 |
| DRB1*01 | 12 (10.90%) | 2 (1.05%) | 0.0002 |
| DRB1*08 | 2 (1.81%) | 8 (4.21%) | 0.43 |
| DRB1*10 | 1 (0.90%) | 1 (0.52%) | 0.73 |
| DRB1*14 | 2 (1.81%) | 4 (2.10%) | 0.79 |
| DRB1*09 | 0 (0.00%) | 4 (2.10%) | 0.31 |
| DRB1*16 | 0 (0.00%) | 1 (0.52%) | 0.78 |
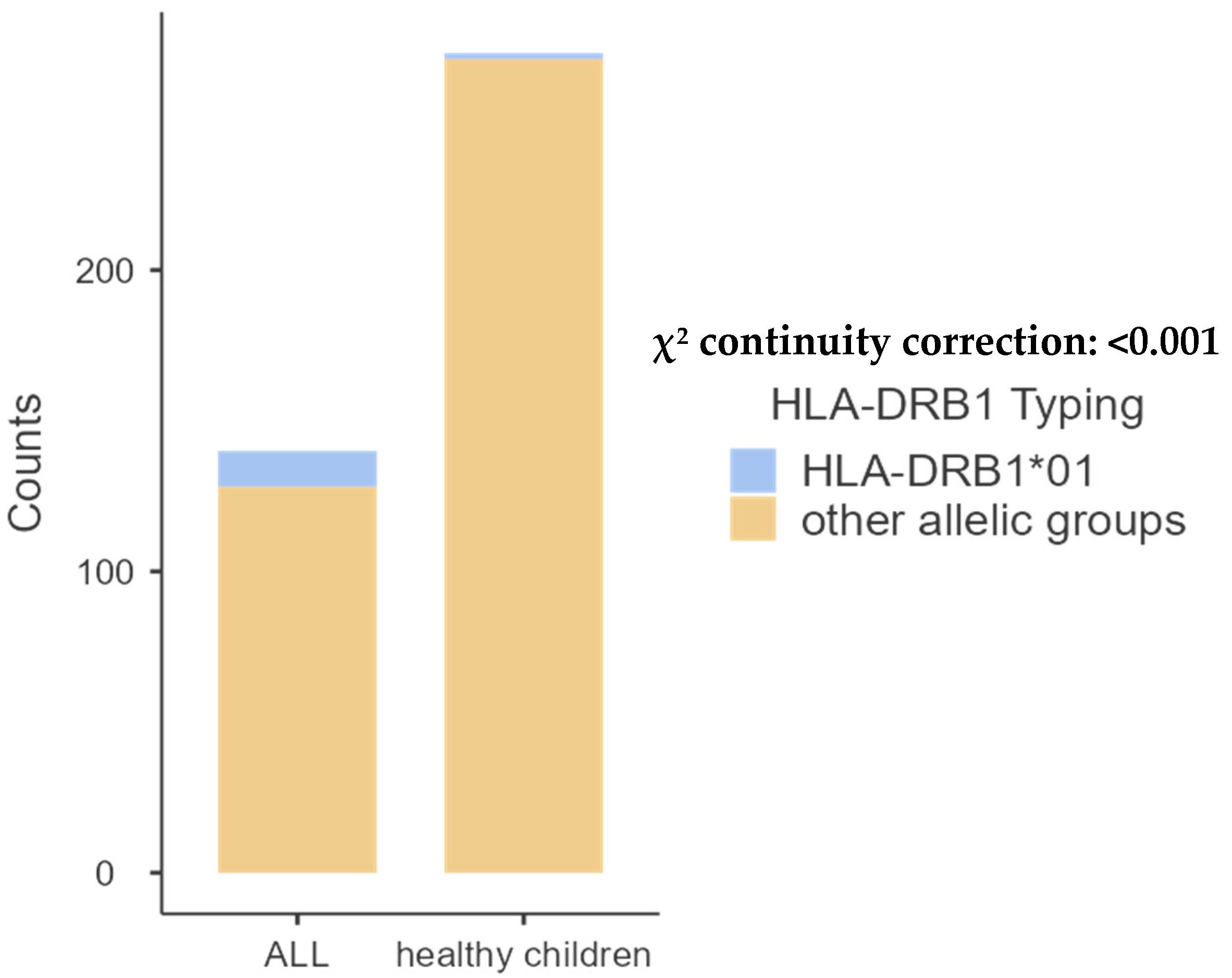
- HLA-DQB1 allele groups.
| HLA-DQB1 | Frequency of Allele Groups in Patients with ALL 2n = 110; n (%) | Frequency of Allele Groups in Control Group 2n = 190; n (%) | p-Value |
|---|---|---|---|
| DQB1*02 | 38 (34.54%) | 69 (36.31%) | 0.85 |
| DQB1*03 | 25 (22.72%) | 46 (24.21%) | 0.88 |
| DQB1*04 | 5 (4.54%) | 15 (7.89%) | 0.37 |
| DQB1*05 | 16 (14.54%) | 7 (3.68%) | 0.001 |
| DQB1*06 | 26 (23.63%) | 53(27.89%) | 0.50 |
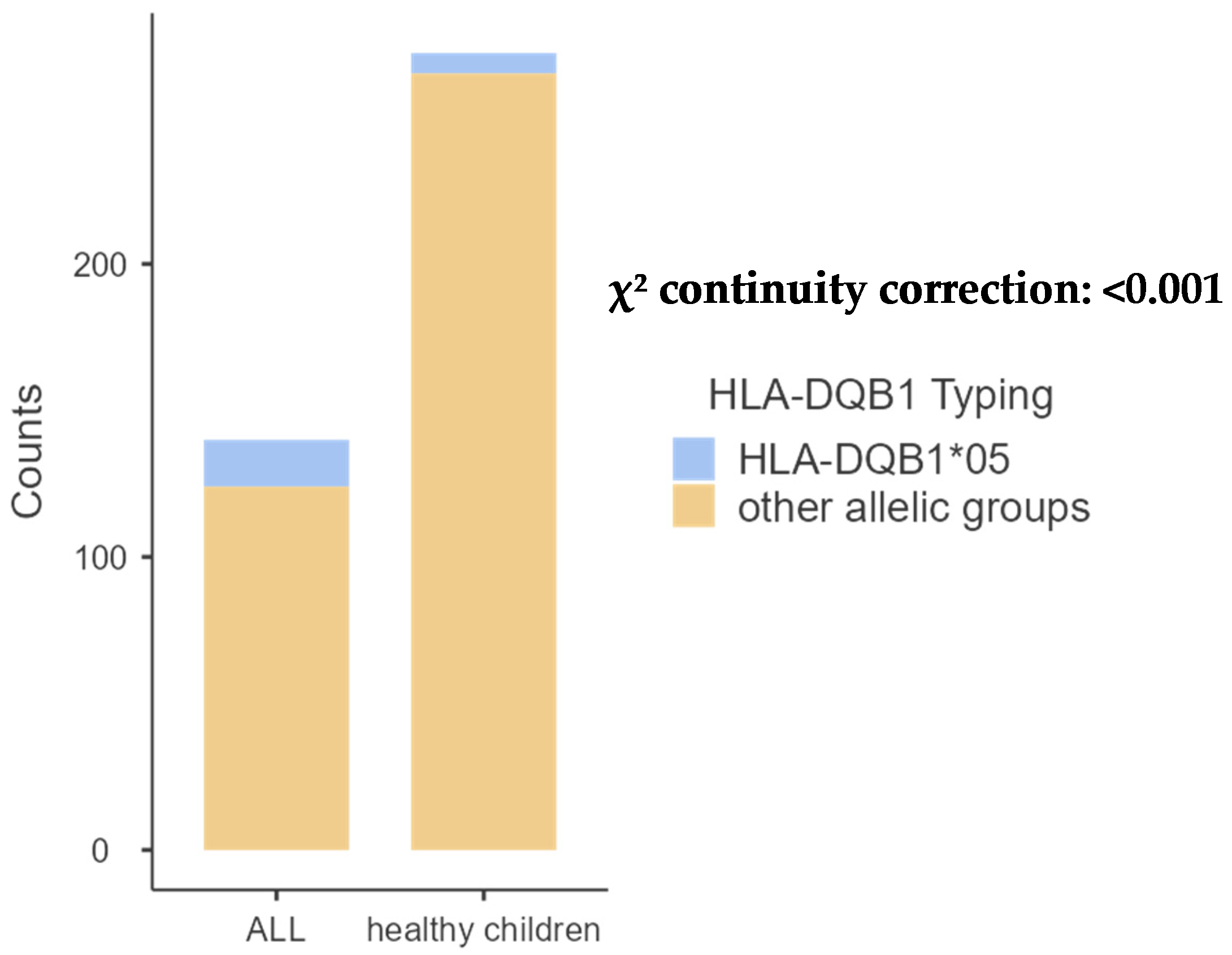
2.3. Distribution of HLA Loci and Allele Groups in Patients with BCP-ALL and Controls
- HLA-A allele groups.
| HLA-A | Frequency of Allele Groups in Patients with BCP-ALL 2n = 78; n (%) | Frequency of Allele Groups in Control Group 2n = 272; n (%) | p-Value |
|---|---|---|---|
| A*02 | 13 (16.66%) | 65 (23.89%) | 0.23 |
| A*30 | 4 (5.12%) | 24 (8.82%) | 0.41 |
| A*68 | 3 (3.84%) | 7 (2.57%) | 0.83 |
| A*01 | 6 (7.69%) | 38 (13.97%) | 0.20 |
| A*03 | 11 (14.10%) | 26 (9.55%) | 0.34 |
| A*24 | 6 (7.69%) | 15 (5.51%) | 0.65 |
| A*33 | 4 (5.12%) | 18 (6.61%) | 0.83 |
| A*29 | 3 (3.84%) | 11 (4.04%) | 0.80 |
| A*11 | 5 (6.41%) | 6 (2.20%) | 0.21 |
| A*32 | 6 (7.69%) | 13 (4.77%) | 0.13 |
| A*23 | 9 (11.53%) | 24 (8.82%) | 0.61 |
| A*26 | 3 (3.84%) | 5 (1.83%) | 0.51 |
| A*34 | 1 (1.28%) | 4 (1.47%) | 0.67 |
| A*25 | 1 (1.28%) | 1 (0.36%) | 0.92 |
| A*28 | 2 (2.56%) | 0 (0.00%) | 0.50 |
| A*66 | 1 (1.28%) | 6 (2.20%) | 0.95 |
- HLA-B allele groups.
| HLA-B | Frequency of Allele Groups in Patients with BCP-ALL 2n = 78; n (%) | Frequency of Allele Groups in Control Group 2n = 272; n (%) | p-Value |
|---|---|---|---|
| B*15 | 1 (1.28%) | 10 (3.67%) | 0.50 |
| B*13 | 0 (0.00%) | 5 (1.83%) | 0.50 |
| B*07 | 7 (8.97%) | 12 (4.41%) | 0.18 |
| B*14 | 5 (6.41%) | 7 (2.57%) | 0.88 |
| B*44 | 10 (12.82%) | 29 (10.66%) | 0.74 |
| B*50 | 7 (8.97%) | 17 (6.25%) | 0.55 |
| B*35 | 3 (3.84%) | 15 (5.51%) | 0.76 |
| B*51 | 9 (11.53%) | 12 (4.41%) | 0.03 |
| B*08 | 5 (6.41%) | 12 (4.41%) | 0.67 |
| B*18 | 3 (3.84%) | 10 (3.67%) | 0.78 |
| B*49 | 4 (5.12%) | 16 (5.88%) | 0.98 |
| B*41 | 2 (2.56%) | 11 (4.04%) | 0.78 |
| B*27 | 2 (2.56%) | 9 (3.30%) | 0.97 |
| B*42 | 1 (1.82%) | 6 (2.20%) | 0.95 |
| B*53 | 2 (2.56%) | 11 (4.04%) | 0.78 |
| B*45 | 5 (6.41%) | 20 (7.35%) | 0.97 |
| B*38 | 3 (3.84%) | 8 (2.94%) | 0.97 |
| B*40 | 0 (0.00%) | 10 (3.67%) | 0.18 |
| B*78 | 1 (1.28%) | 2 (0.73%) | 0.81 |
| B*57 | 1 (1.28%) | 17 (6.25%) | 0.14 |
| B*52 | 2 (2.56%) | 5 (1.83%) | 0.95 |
| B*17 | 0 (0.00%) | 2 (0.73%) | 0.92 |
| B*39 | 0 (0.00%) | 7 (2.57%) | 0.33 |
| B*58 | 1 (1.28%) | 11 (4.04%) | 0.40 |
| B*37 | 1 (1.28%) | 0 (0.00%) | 0.50 |
| B*72 | 2 (2.56%) | 3 (1.10%) | 0.67 |
| B*63 | 1 (1.28%) | 0 (0.0%) | 0.50 |
| B*62 | 0 (0.00%) | 2 (0.73%) | 0.92 |
| B*55 | 0 (0.00%) | 2 (0.73%) | 0.92 |
| B*25 | 0 (0.00%) | 1 (0.36%) | 0.50 |
- HLA-DRB1 allele groups.
| HLA-DRB1 | Frequency of Allele Groups in Patients with BCP-ALL 2n = 72; n (%) | Frequency of Allele Groups in Control Group 2n = 190; n (%) | p-Value |
|---|---|---|---|
| DRB1*03 | 17 (23.61%) | 33 (17.36%) | 0.33 |
| DRB1*13 | 11 (15.27%) | 38 (20%) | 0.48 |
| DRB1*04 | 10 (13.88%) | 29 (15.26%) | 0.93 |
| DRB1*15 | 7 (9.72%) | 28 (14.73%) | 0.38 |
| DRB1*11 | 6 (8.33%) | 9 (4.73%) | 0.41 |
| DRB1*07 | 9 (12.50%) | 33 (17.36%) | 0.44 |
| DRB1*01 | 10 (13.88%) | 2 (1.05%) | 0.00004 |
| DRB1*08 | 1 (1.38%) | 8 (4.21%) | 0.45 |
| DRB1*14 | 1 (1.38%) | 4 (2.10%) | 0.89 |
| DRB1*09 | 0 (0.00%) | 4 (2.10%) | 0.49 |
| DRB1*16 | 0 (0.00%) | 1 (0.52%) | 0.61 |
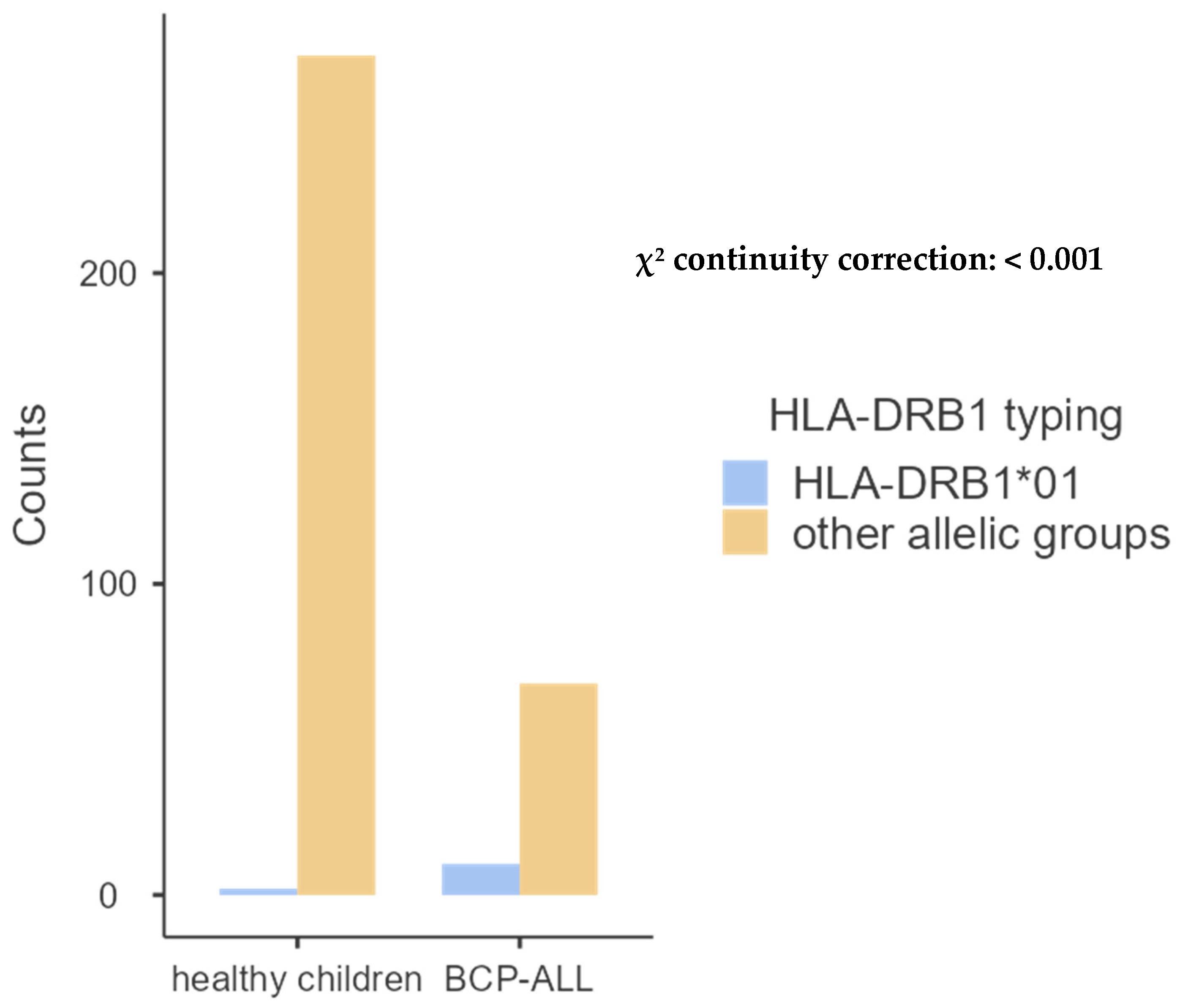
- HLA-DQB1 allele groups.
| HLA-DQB1 | Frequency of Allele Groups in Patients with ALL B 2n = 72; n (%) | Frequency of Allele Groups in Control Group 2n = 190; n (%) | p-Value |
|---|---|---|---|
| DQB1*02 | 24 (33.33%) | 69 (36.31%) | 0.75 |
| DQB1*03 | 17 (23.61%) | 46 (24.21%) | 0.95 |
| DQB1*04 | 2 (2.77%) | 15 (7.89%) | 0.22 |
| DQB1*05 | 11 (15.27%) | 7 (3.68%) | 0.002 |
| DQB1*06 | 18 (25.00%) | 53 (27.89%) | 0.75 |

2.4. Distribution of HLA Loci and Allele Groups in Patients with TCP-ALL and Controls
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Ethical Considerations
4.3. Sample Collection and Processing
4.4. DNA Extraction
4.5. HLA Typing
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALL | Acute lymphoid leukemia |
| SSOs | Sequence-specific oligonucleotides |
| SSPs | Sequence-specific primers |
| PCR | Polymerase chain reaction |
| HLA | Human leukocyte antigen |
| BCP-ALL | B cell precursor acute lymphoblastic leukemia |
| TCP-ALL | T-cell precursor acute lymphoblastic leukemia |
| MHC | Major histocompatibility complex |
| IARC | International Agency for Research on cancer |
References
- Andriescu, E.C.; Coughlin, C.C.; Cheng, C.E.; Prajapati, V.H.; Huang, J.T.; Schmidt, B.A.; Degar, B.A.; Aplenc, R.; Pillai, V.; Yan, A.C.; et al. Pediatric leukemia cutis: A case series. Pediatr. Dermatol. 2019, 36, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Milan, T.; Canaj, H.; Villeneuve, C.; Ghosh, A.; Barabé, F.; Cellot, S.; Wilhelm, B.T. Pediatric leukemia: Moving toward more accurate models. Exp. Hematol. 2019, 74, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hosking, F.J.; Leslie, S.; Dilthey, A.; Moutsianas, L.; Wang, Y.; Dobbins, S.E.; Papaemmanuil, E.; Sheridan, E.; Kinsey, S.E.; Lightfoot, T.; et al. MHC variation and risk of childhood B-cell precursor acute lymphoblastic leukemia. Blood 2011, 117, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- Ibrahimova, A.; Pommert, L.; Breese, E.H. Acute Leukemia in Infants. Curr. Oncol. Rep. 2021, 23, 27. [Google Scholar] [CrossRef]
- Cancer Today. Available online: https://gco.iarc.who.int/today/ (accessed on 14 August 2024).
- Mishra, V.C.; Raina, V.; Sharma, G. HLA association with leukemia: A review of the literature. Gene Rep. 2020, 21, 100939. [Google Scholar] [CrossRef]
- El Karaaoui, A.; Tamim, H.; El Achkar, H.; Fermanian, P.; Abbas, F.; Keleshian, S.; Muwakkit, S.; Mahfouz, R. Association of Human Leukocyte Antigens (HLA) profile and acute lymphoblastic leukemia in Lebanese pediatric patients: A first report from Lebanon. Hum. Gene 2022, 33, 201072. [Google Scholar] [CrossRef]
- Klitz, W.; Gragert, L.; Trachtenberg, E. Spectrum of HLA associations: The case of medically refractory pediatric acute lymphoblastic leukemia. Immunogenetics 2012, 64, 409–419. [Google Scholar] [CrossRef]
- Fernández-Torres, J.; Flores-Jiménez, D.; Arroyo-Pérez, A.; Granados, J.; López-Reyes, A. HLA-B*40 Allele Plays a Role in the Development of Acute Leukemia in Mexican Population: A Case-Control Study. BioMed Res. Int. 2013, 2013, 705862. [Google Scholar] [CrossRef]
- Urayama, K.Y.; Thompson, P.D.; Taylor, G.M.; Trachtenberg, E.A.; Chokkalingam, A.P. Genetic Variation in the Extended Major Histocompatibility Complex and Susceptibility to Childhood Acute Lymphoblastic Leukemia: A Review of the Evidence. Front. Oncol. 2013, 3, 300. Available online: https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2013.00300/full (accessed on 14 August 2024). [CrossRef]
- Ozdilli, K.; Oguz, F.S.; Anak, S.; Kekik, C.; Carin, M.; Gedikoglu, G. The frequency of HLA class I and II alleles in Turkish childhood acute leukaemia patients. J. Int. Med. Res. 2010, 38, 1835–1844. [Google Scholar] [CrossRef]
- Solanki, H.; Tiwari, A.K.; Raina, V.; Sharma, G. Association study of HLA class I and class II alleles with childhood acute lymphoblastic leukemia in Indian patients. Gene Rep. 2021, 23, 101086. [Google Scholar] [CrossRef]
- Constantinescu, I.; Boșcaiu, V.; Cianga, P.; Dinu, A.A.; Gai, E.; Melinte, M.; Moise, A. The frequency of HLA alleles in the Romanian population. Immunogenetics 2016, 68, 167–178. [Google Scholar] [CrossRef]
- Risk Factors for Childhood Leukemia|American Cancer Society. Available online: https://www.cancer.org/cancer/types/leukemia-in-children/causes-risks-prevention/risk-factors.html (accessed on 14 August 2024).
- Buffler, P.A.; Kwan, M.L.; Reynolds, P.; Urayama, K.Y. Environmental and Genetic Risk Factors for Childhood Leukemia: Appraising the Evidence. Cancer Investig. 2005, 23, 60–75. [Google Scholar] [CrossRef]
- Kabbaj, M.; Oudghiri, M.; Naya, A.; Naamane, H.; El Turk, J.; Bennani, S.; Hassar, M. HLA-A, -B, -DRB1 alleles and haplotypes frequencies in Moroccan patients with leukemia. Ann. Biol. Clin. 2010, 68, 291–296. [Google Scholar] [CrossRef]
- Orouji, E.; Tavakkol Afshari, J.; Badiee, Z.; Shirdel, A.; Alipour, A. Association between HLA-DQB1 gene and patients with acute lymphoblastic leukemia (ALL). Int. J. Hematol. 2012, 95, 551–555. Available online: http://link.springer.com/10.1007/s12185-012-1051-8 (accessed on 25 January 2025). [CrossRef]
- Kohansal Vajari, M.; Ehsan, M.; Ghiasi, S.; Fooladi, S.; Karami, N.; Hassanshahi, G.; Fatemi, A. Human Leukocyte Antigen Alleles (HLA-A, HLA-B, and HLA-DRB1) are associated with Acute Lymphoblastic Leukemia (ALL): A Case-Control Study in a Sample of Iranian Population. Asian Pac. J. Cancer Prev. 2024, 25, 1507–1513. [Google Scholar] [CrossRef]
- Güleç, R.D.; Arslan, F.D. Frequencies of HLA Alleles in Patients with Acute Lymphoblastic and Myeloid Leukemia. Med. Sci. Discov. 2023, 10, 539–545. [Google Scholar] [CrossRef]
- Patıroğlu, T.; Akar, H.H. The Frequency of HLA-A, HLA-B, and HLA-DRB1 Alleles in Patients with Acute Lymphoblastic Leukemia in the Turkish Population: A Case-Control Study. Turk. J. Haematol. 2016, 33, 339–345. [Google Scholar] [CrossRef]
- Hassan, N.; Idris, S.Z.; Chang, K.M.; Osman, R.; Ibrahim, H.M.; Dhaliwal, J.S.; Abdullah, M. High Variability in HLA-DRB1*03, a Predisposing Allele in Acute Lymphoblastic Leukemia. Iran. J. Blood Cancer 2024, 16, 24–33. [Google Scholar] [CrossRef]
- El Ansary, M.M.; Mohammed, L.A.; Hassan, T.H.; Baraka, A.; Ahmed, A.A. Human leukocyte antigen-DRB1 polymorphism in childhood acute lymphoblastic leukemia. Mol. Clin. Oncol. 2015, 3, 425–429. [Google Scholar] [CrossRef][Green Version]
- Nouar, N.H.; Yafour, N.; Youcef, B.Y.; Bouhass, R.; Chekkal, M.; Brahimi, M.; Bekadja, M.; Sahraoui, T. HLA-B*58 and HLA-B*27 Play a Role in the Development of Acute Leukemia: A Case Control Study. Asian Pac. J. Cancer Prev. 2024, 25, 169–173. Available online: https://journal.waocp.org/article_90969.html (accessed on 26 April 2025). [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laaziri, K.; Zyad, A.; Laaziaf, E.M.; Houdzi, J.E.; Elhanafi, F.; Brahim, I.; Lakhouaja, N.; Hazime, R.; Ammara, M.; Naya, A.; et al. Acute Lymphoblastic Leukemia and Associated HLA-A, B, DRB1, and DQB1 Molecules: A Moroccan Pediatric Case–Control Study. Int. J. Mol. Sci. 2025, 26, 5295. https://doi.org/10.3390/ijms26115295
Laaziri K, Zyad A, Laaziaf EM, Houdzi JE, Elhanafi F, Brahim I, Lakhouaja N, Hazime R, Ammara M, Naya A, et al. Acute Lymphoblastic Leukemia and Associated HLA-A, B, DRB1, and DQB1 Molecules: A Moroccan Pediatric Case–Control Study. International Journal of Molecular Sciences. 2025; 26(11):5295. https://doi.org/10.3390/ijms26115295
Chicago/Turabian StyleLaaziri, Khalid, Abdelmajid Zyad, El Mehdi Laaziaf, Jamila El Houdzi, Fatimaezzahra Elhanafi, Ikram Brahim, Nadia Lakhouaja, Raja Hazime, Mounia Ammara, Abdellah Naya, and et al. 2025. "Acute Lymphoblastic Leukemia and Associated HLA-A, B, DRB1, and DQB1 Molecules: A Moroccan Pediatric Case–Control Study" International Journal of Molecular Sciences 26, no. 11: 5295. https://doi.org/10.3390/ijms26115295
APA StyleLaaziri, K., Zyad, A., Laaziaf, E. M., Houdzi, J. E., Elhanafi, F., Brahim, I., Lakhouaja, N., Hazime, R., Ammara, M., Naya, A., & Admou, B. (2025). Acute Lymphoblastic Leukemia and Associated HLA-A, B, DRB1, and DQB1 Molecules: A Moroccan Pediatric Case–Control Study. International Journal of Molecular Sciences, 26(11), 5295. https://doi.org/10.3390/ijms26115295







