Fine Mapping and Screening of Candidate Gene for Yellow-to-Green Mutation in Snap Bean (Phaseolus vulgaris L.) Leaf Color
Abstract
1. Introduction
2. Results
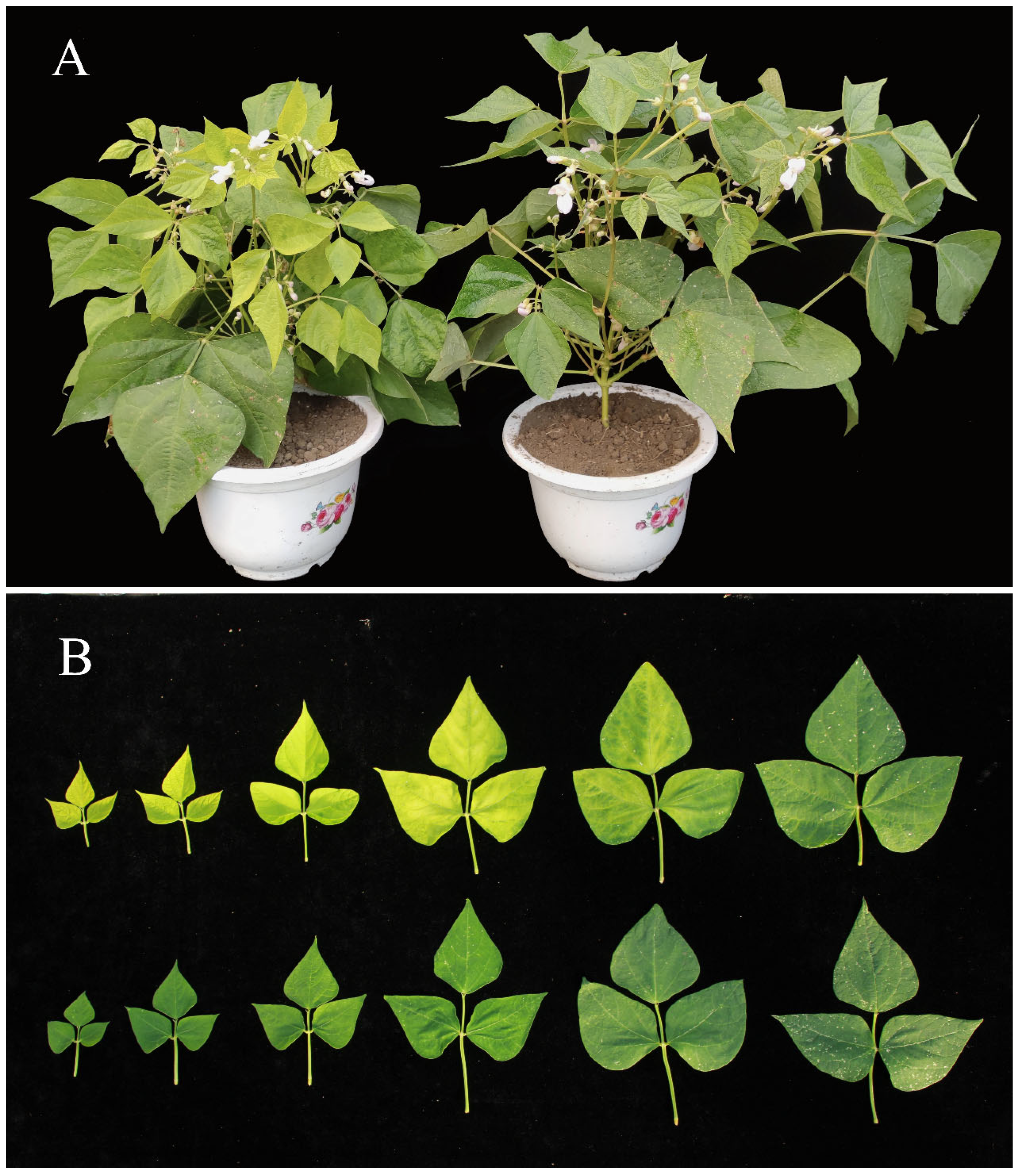
2.1. Morphological Characteristics of ytg-1 Mutant
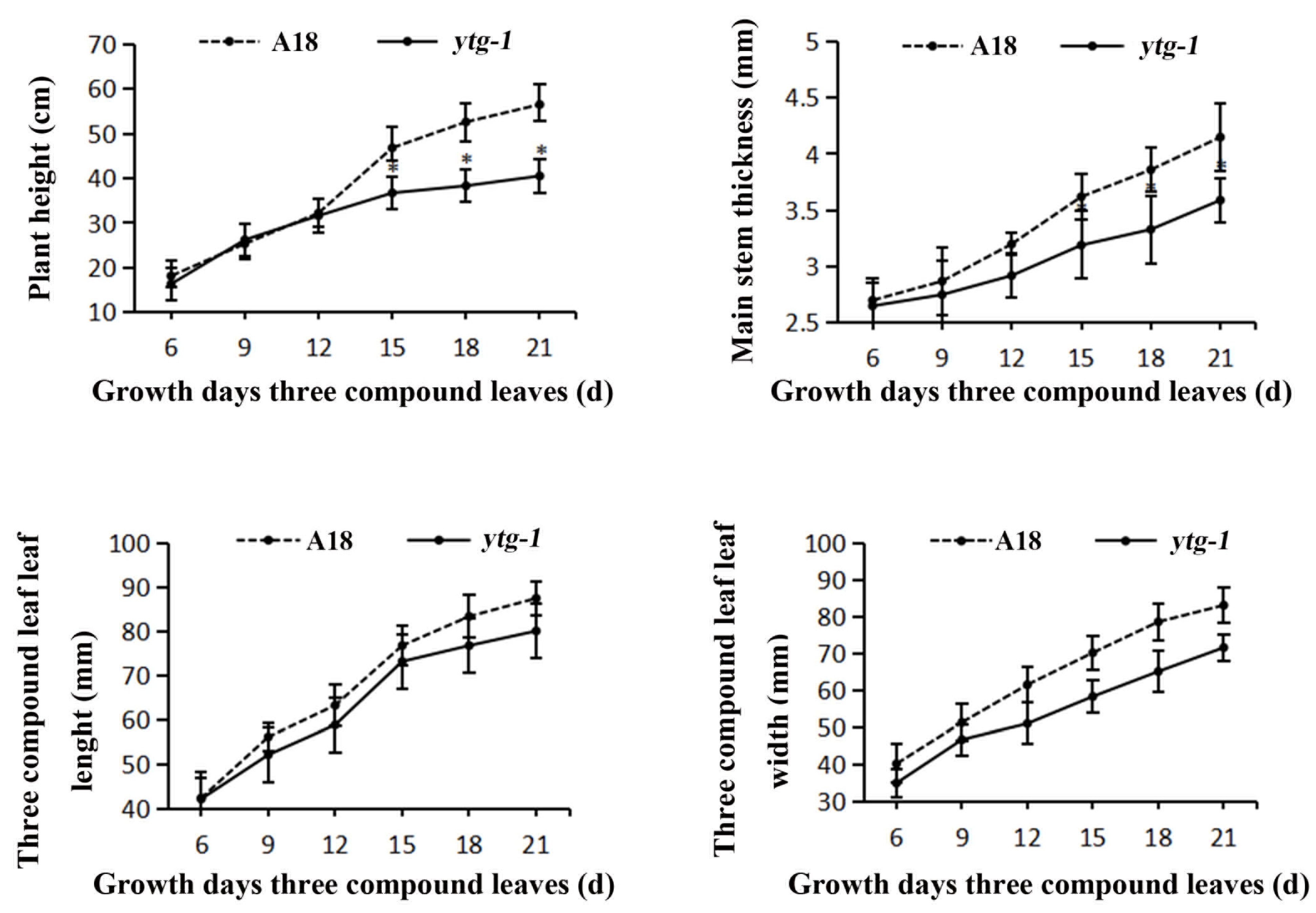
2.2. Changes in Agronomic Traits of ytg-1 Mutant
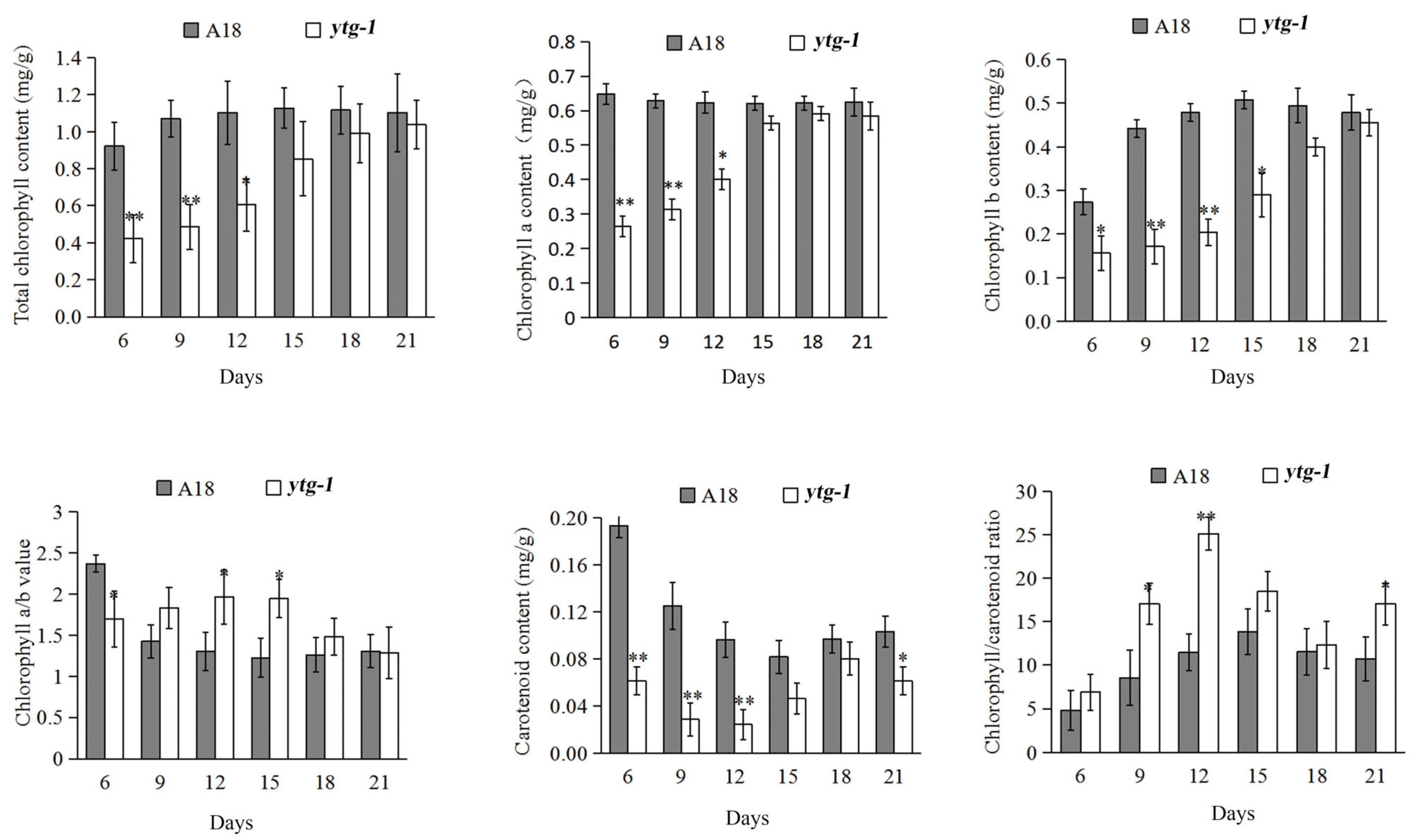
2.3. Analysis of Photosynthetic Pigment Content
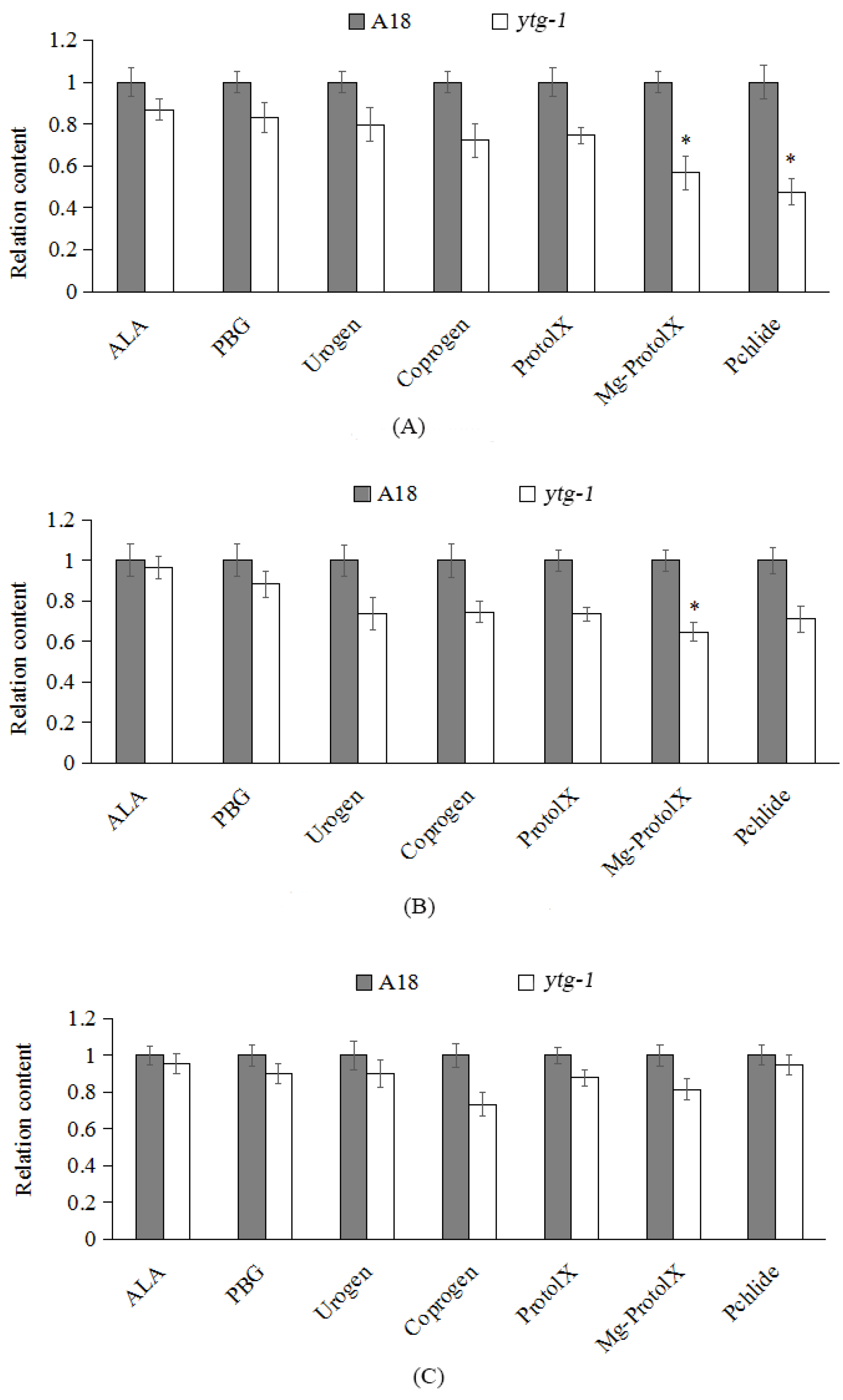
2.4. Analysis of the Content of Intermediate Metabolites in Chlorophyll Synthesis in ytg-1
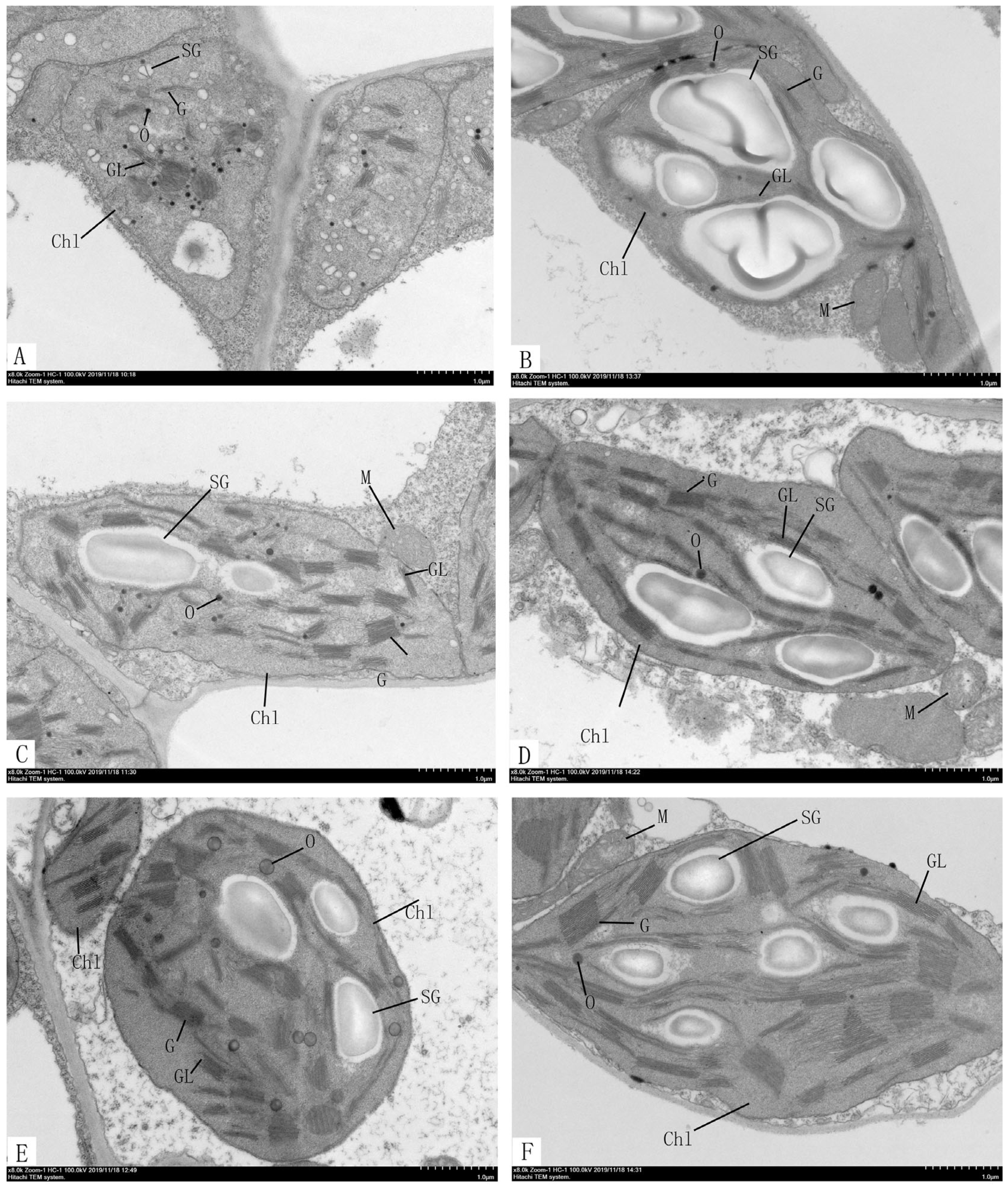
2.5. Ultrastructural Analysis of Chloroplast in ytg-1
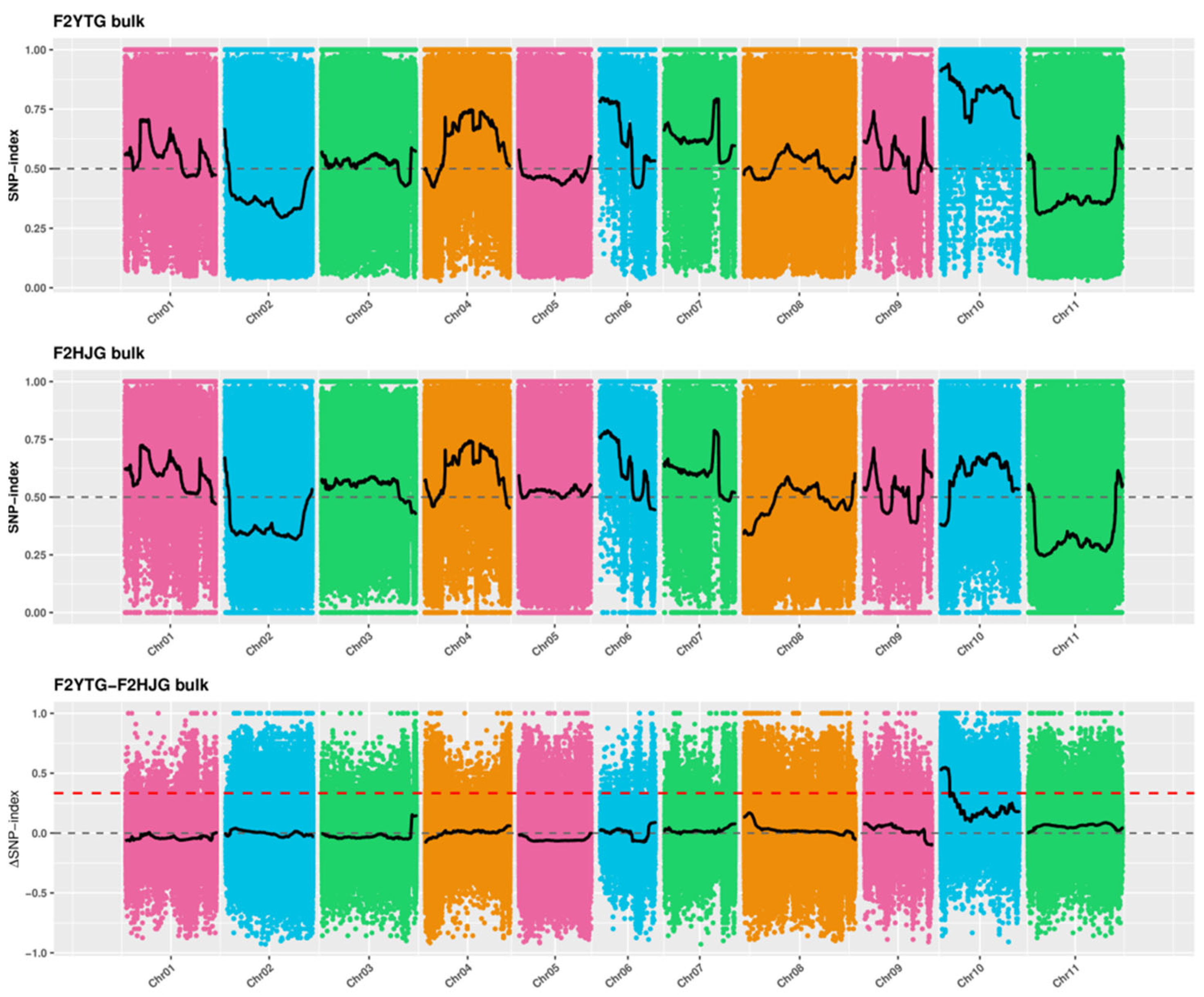
2.6. Genetic Analysis of the Phenotype of Leaf Color Variation in Mutant ytg-1
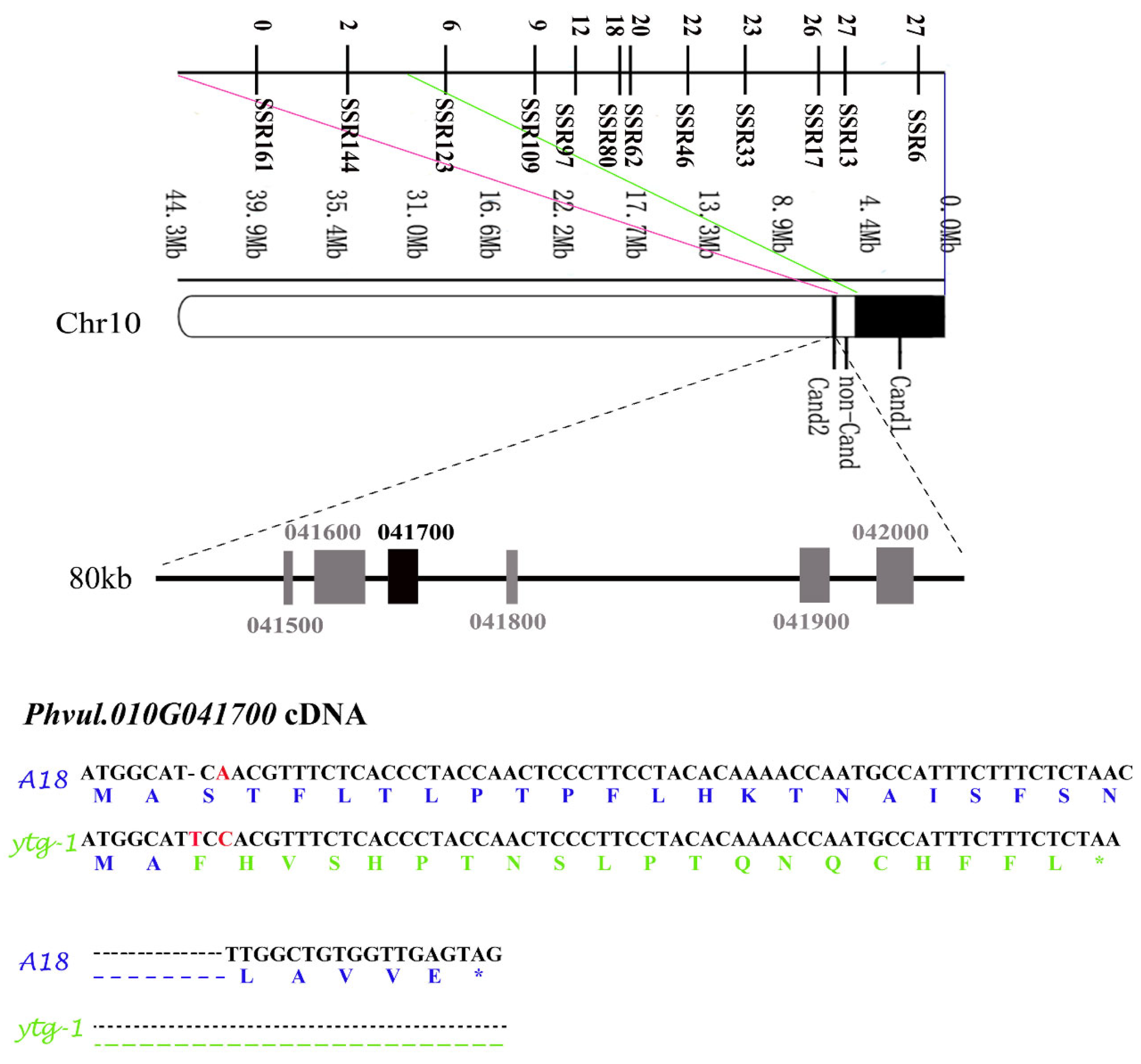
2.7. Fine Mapping of pvytg-1
2.8. Candidate Gene Screening and Sequence Analysis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Determination of Agronomic Trait Parameters
4.3. Determination of Photosynthetic Pigment Content
4.4. Determination of the Intermediate Metabolites in Chlorophyll Biosynthesis
4.5. Observation of the Ultrastructure of Chloroplasts
4.6. Genetic Analysis of Leaf Yellow-to-Green Mutant Characteristics
4.7. Bulked-Segregant Analysis Coupled with Whole-Genome Sequencing (BSA-Seq) Analysis
4.8. Fine Mapping of pvytg-1 Mutated Gene
4.9. Gene Cloning and Sequence Comparison
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suzuki, J.Y.; Bollivar, D.W.; Bauer, C.E. Genetic analysis of chlorophyll biosynthesis. Annu. Rev. Genet. 1997, 31, 61–89. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.-J.; Zhang, H.-Q.; Wang, Y.; He, F.; Liu, J.-L.; Xiao, X.; Shu, Z.-F.; Li, W.; Wang, G.-H. Mapped Clone and Functional Analysis of Leaf-Color Gene Ygl7 in a Rice Hybrid (Oryza sativa L. ssp. indica). PLoS ONE 2014, 9, e99564. [Google Scholar] [CrossRef]
- Hong, M.J.; Kim, J.-B.; Yoon, Y.H.; Kim, S.H.; Ahn, J.-W.; Jeong, I.Y.; Kang, S.-Y.; Seo, Y.W.; Kim, D.S. The effects of chronic gamma irradiation on oxidative stress response and the expression of anthocyanin biosynthesis-related genes in wheat (Triticum aestivum L.). Int. J. Radiat. Biol. 2014, 90, 1218–1228. [Google Scholar] [CrossRef]
- Vaculík, M.; Pavlovič, A.; Lux, A. Silicon alleviates cadmium toxicity by enhanced photosynthetic rate and modified bundle sheath’s cell chloroplasts ultrastructure in maize. Ecotoxicol. Environ. Saf. 2015, 120, 66–73. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Xie, J.; Ding, W.; Li, Y.; Yue, Y.; Wang, L. Biochemical and Comparative Transcriptome Analyses Reveal Key Genes Involved in Major Metabolic Regulation Related to Colored Leaf Formation in Osmanthus fragrans ‘Yinbi Shuanghui’ during Development. Biomolecules 2020, 10, 549. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Yoo, J.-H.; Yoo, S.-C.; Cho, S.-H.; Koh, H.-J.; Seo, H.S.; Paek, N.-C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef]
- Zhu, X.; Guo, S.; Wang, Z.; Du, Q.; Xing, Y.; Zhang, T.; Shen, W.; Sang, X.; Ling, Y.; He, G. Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa). BMC Plant Biol. 2016, 16, 134. [Google Scholar] [CrossRef]
- Liu, N.-T.; Jane, W.-N.; Tsay, H.-S.; Wu, H.; Chang, W.-C.; Lin, C.-S. Chloroplast genome aberration in micropropagation-derived albino Bambusa edulis mutants, ab1 and ab2. Plant Cell, Tissue Organ Cult. Plant Cell Tissue Organ Cult. (PCTOC) 2007, 88, 147–156. [Google Scholar] [CrossRef]
- Caredda, S.; Doncoeur, C.; Devaux, P.; Sangwan, R.S.; Clément, C. Plastid differentiation during androgenesis in albino and non-albino producing cultivars of barley (Hordeum vulgare L.). Sex. Plant Reprod. 2000, 13, 95–104. [Google Scholar] [CrossRef]
- Liu, C.; Zhu, H.; Xing, Y.; Tan, J.; Chen, X.; Zhang, J.; Peng, H.; Xie, Q.; Zhang, Z. Albino Leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J. Exp. Bot. 2016, 67, 5339–5347. [Google Scholar] [CrossRef]
- Wu, W.; Elsheery, N.; Wei, Q.; Zhang, L.; Huang, J. Defective Etioplasts Observed in Variegation Mutants May Reveal the Light-Independent Regulation of White/Yellow Sectors of Arabidopsis Leaves. J. Integr. Plant Biol. 2011, 53, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Parks, B.M.; Quail, P.H. Phytochrome-Deficient hy1 and hy2 Long Hypocotyl Mutants of Arabidopsis Are Defective in Phytochrome Chromophore Biosynthesis. Plant Cell 1991, 3, 1177–1186. [Google Scholar] [CrossRef]
- Archer, E.K.; Bonnett, H.T. Characterization of a Virescent Chloroplast Mutant of Tobacco. Plant Physiol. 1987, 83, 920–925. [Google Scholar] [CrossRef]
- Sheng, Z.; Lv, Y.; Li, W.; Luo, R.; Wei, X.; Xie, L.; Jiao, G.; Shao, G.; Wang, J.; Tang, S.; et al. Yellow-Leaf 1 encodes a magnesium-protoporphyrin IX monomethyl ester cyclase, involved in chlorophyll biosynthesis in rice (Oryza sativa L.). PLoS ONE 2017, 12, e0177989. [Google Scholar] [CrossRef]
- Liu, C.; Li, Y.; Liu, D.; Yan, Z.; Feng, G.; Yang, X. Blocked chlorophyll synthesis leads to the production of golden snap bean pods. Mol. Genet. Genom. 2020, 295, 1325–1337. [Google Scholar] [CrossRef]
- Liu, L.; Jia, L.; Xu, L.; Deng, L.; Zhao, P.; Zeng, W. Mutation of iPGAM using the CRISPR/Cas9 system affects pollen vitality and chlorophyll synthesis in Nicotiana tabacum. Plant Biotechnol. Rep. 2021, 15, 217–227. [Google Scholar] [CrossRef]
- Trick, M.; Adamski, N.M.; Mugford, S.G.; Jiang, C.-C.; Febrer, M.; Uauy, C. Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol. 2012, 12, 14. [Google Scholar] [CrossRef]
- Ramirez-Gonzalez, R.H.; Segovia, V.; Bird, N.; Fenwick, P.; Holdgate, S.; Berry, S.; Jack, P.; Caccamo, M.; Uauy, C. RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnol. J. 2014, 13, 613–624. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Z.; Zhang, Y.; Li, C.; Feng, H. Identification and fine mapping of a stay-green gene (Brnye1) in pakchoi (Brassica campestris L. ssp. chinensis). Theor. Appl. Genet. 2018, 131, 673–684. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, Z.; Shan, X.; Li, C.; Tang, X.; Chi, M.; Feng, H. Physiological properties and chlorophyll biosynthesis in a Pak-choi (Brassica rapa L. ssp chinensis) yellow leaf mutant, pylm. Acta Physiol. Plant. 2017, 39, 22. [Google Scholar] [CrossRef]
- Nguyen, B.; Ma, R.; Tang, W.K.; Shi, D.; Tolia, N.H. Crystal structure of P. falciparum Cpn60 bound to ATP reveals an open dynamic conformation before substrate binding. Sci. Rep. 2021, 11, 5930–5936. [Google Scholar] [CrossRef] [PubMed]
- Kupper, M.; Gupta, S.K.; Feldhaar, H.; Gross, R. Versatile roles of the chaperonin GroEL in microorganism-insect interactions. FEMS Microbiol. Lett. 2014, 353, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Rustad, T.; Mahajan, G.; Kumar, A.; Rao, K.V.; Banerjee, S.; Sherman, D.R.; Mande, S.C. Towards understanding the biological function of the unusual chaperonin Cpn60.1 (GroEL1) of Mycobacterium tuberculosis. Tuberculosis 2016, 97, 137–146. [Google Scholar] [CrossRef]
- Koumoto, Y.; Shimada, T.; Kondo, M.; Hara-Nishimura, I.; Nishimura, M. Chloroplasts Have a Novel Cpn10 in Addition to Cpn20 as Co-chaperonins in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 29688–29694. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Dei, M. Benzyladenine-induced stimulation of 5-aminolevulinic acid accumulation under various light intensities in levulinic acid-treated cotyledons of etiolated cucumber. Physiol. Plant. 1985, 64, 153–160. [Google Scholar] [CrossRef]
- Bogorad, L. Porphyrin synthesis. Methods Enzymol. 1962, 5, 885–895. [Google Scholar] [CrossRef]
- Plöscher, M.; Granvogl, B.; Reisinger, V.; Eichacker, L.A. Identification of the N-termini of NADPH: Protochlorophyllide oxidoreductase A and B from barley etioplasts (Hordeum vulgare L.). FEBS J. 2003, 276, 1074–1081. [Google Scholar] [CrossRef]
| Strain | Pod Length (cm) | Pod Width (cm) | Single Plant Yield (g) | Number of Pods Per Plant (number) | 100 Grain Weight (g) |
|---|---|---|---|---|---|
| ytg-1 | 14.87 ± 0.06 ** | 1.01 ± 0.04 | 166.67 ± 5.77 | 37.00 ± 2.65 * | 38.38 ± 0.22 ** |
| A18 | 16.77 ± 0.15 | 1.05 ± 0.01 | 150.00 ± 10.00 | 31.33 ± 1.15 | 37.11 ± 0.11 |
| Generations | Total | Green Leaves | Yellow to Green Leaves | Segregation Ration | χ2 |
|---|---|---|---|---|---|
| P1 (ytg-1) | 50 | 0 | 50 | ||
| P2 (A18) | 50 | 50 | 0 | ||
| F1 (P1 × P2) | 32 | 32 | 0 | ||
| F1 (P2 × P1) | 28 | 28 | 0 | ||
| BC1 (F1 × P1) | 33 | 18 | 15 | 1.2:1 | 1.2 |
| BC2 (F1 × P2) | 29 | 29 | 0 | ||
| F2 | 1233 | 916 | 317 | 2.89:1 | 0.956 |
| Gene | Name | Location | Function Notes |
|---|---|---|---|
| Phvul.010G041500 | - | 6303589–6304203 | - |
| Phvul.010G041600 | - | 6307176–6312920 | - |
| Phvul.010G041700 | CPN10-1 | 6314835–6318150 | 10 kDa chaperonin 1, chloroplastic |
| Phvul.010G041800 | ATL4 | 6329479–6330555 | E3 ubiquitin-protein ligase ATL4 |
| Phvul.010G041900 | - | 6358186–6361597 | - |
| Phvul.010G042000 | At5g59540 | 6365314–6369368 | 1-aminocyclopropane-1-carboxylate oxidase homolog 12 |
| Markers | Upstream Primer (5′-3′) | Downstream Primer (5′-3′) | Location |
|---|---|---|---|
| SSR6 | GCAGTTGCAGTCATTGTATAG | GGAAAGAAATTGGGAAAGACAG | 433,325–433,425 |
| SSR13 | CCTCATTGACTTGCTTCATT | CAACTCCTTGTTTGACCAA | 1,113,712–1,113,812 |
| SSR17 | CTGCTTACATCTTTGTCCTTC | CTAGTGTTGACCATTTGAGTG | 1,364,003–1,365,003 |
| SSR33 | TAGTTGGTGGTCGTCCTAT | TGGTCATTCTTAGTGGTTGT | 1,950,571–1,950,871 |
| SSR46 | GATGGAATGTCAGTGAGGTA | CAGTGTTGTAGAGGTTGGA | 2,402,344–2,402,644 |
| SSR62 | TTGGAACACAACCACTCAT | ACTCCTTTAGAAACCTCTCTC | 2,833,661–2,833,961 |
| SSR80 | CTCTCCACTCTTCTTCTTCTT | GAGGAACTGCGATAACTAATG | 2,912,191–2,912,318 |
| SSR97 | CCTACCCATTCTTGAATAACC | CTTGCTCAGCTCACTCTC | 3,054,694–30,554,714 |
| SSR109 | TCCACTGCTACTGTGTCTA | CTCCAATCCATCATCATCATC | 3,497,326–3,497,626 |
| SSR123 | ACATCGTGACGGAGAACA | AGTAGGAAGCAATGCCATC | 4,571,601–4,571,619 |
| SSR144 | TCTTCTTCCTCTCACTTCTC | CTCCGCATAAGCAGACAA | 5,315,083–5,315,102 |
| SSR161 | CACGATGGTTGGAGTTAGAA | AGTGGCTATCTATATGTGGAAGG | 6,092,486–6,092,505 |
| Gene ID | F-Primer (5′-3′) | R-Primer (5′-3′) |
|---|---|---|
| pvytg-1 | AGCAACTGAGTAGATAAACCTAA | AGTTGGTCTTATTAACACCATTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Liu, D.; Feng, G.; Zhang, T.; Qin, X.; Sun, Z.; Yan, Z.; Yang, X. Fine Mapping and Screening of Candidate Gene for Yellow-to-Green Mutation in Snap Bean (Phaseolus vulgaris L.) Leaf Color. Int. J. Mol. Sci. 2025, 26, 5293. https://doi.org/10.3390/ijms26115293
Liu C, Liu D, Feng G, Zhang T, Qin X, Sun Z, Yan Z, Yang X. Fine Mapping and Screening of Candidate Gene for Yellow-to-Green Mutation in Snap Bean (Phaseolus vulgaris L.) Leaf Color. International Journal of Molecular Sciences. 2025; 26(11):5293. https://doi.org/10.3390/ijms26115293
Chicago/Turabian StyleLiu, Chang, Dajun Liu, Guojun Feng, Taifeng Zhang, Xiangkai Qin, Zhuang Sun, Zhishan Yan, and Xiaoxu Yang. 2025. "Fine Mapping and Screening of Candidate Gene for Yellow-to-Green Mutation in Snap Bean (Phaseolus vulgaris L.) Leaf Color" International Journal of Molecular Sciences 26, no. 11: 5293. https://doi.org/10.3390/ijms26115293
APA StyleLiu, C., Liu, D., Feng, G., Zhang, T., Qin, X., Sun, Z., Yan, Z., & Yang, X. (2025). Fine Mapping and Screening of Candidate Gene for Yellow-to-Green Mutation in Snap Bean (Phaseolus vulgaris L.) Leaf Color. International Journal of Molecular Sciences, 26(11), 5293. https://doi.org/10.3390/ijms26115293




