Molecular Mass and Isoelectric Point Analysis of Cytokinin Sequences in the Wheat Genome
Abstract
1. Introduction
2. Results
2.1. The Number of Chromosomes for Cytokinin Riboside 5′-Monophosphate Phosphoribohydrolase Genes Through NCBI and Ensembl Plants Databases
2.2. The Relative Synonymous Codon Usage (RSCU)
2.3. The Values of Molecular Weight and Theoretical Isoelectric Point
3. Discussion
4. Materials and Methods
4.1. The Wheat Genome Characterization
4.2. Principal Component Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2016, 67, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Yamburenko, M.V.; Kieber, J.J.; Schaller, G.E. Dynamic patterns of expression for genes regulating cytokinin metabolism and signaling during rice inflorescence development. PLoS ONE 2017, 12, e0176060. [Google Scholar] [CrossRef] [PubMed]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmülling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef] [PubMed]
- Pavlů, J.; Novák, J.; Koukalová, V.; Luklová, M.; Brzobohatý, B.; Černý, M. Cytokinin at the crossroads of abiotic stress signalling pathways. Int. J. Mol. Sci. 2018, 19, 2450. [Google Scholar] [CrossRef]
- Prasad, R. Cytokinin and Its Key Role to Enrich the Plant Nutrients and Growth Under Adverse Conditions-An Update. Frontiers. An Update. Front. Genet. 2022, 13, 883924. [Google Scholar] [CrossRef]
- Wang, W.; Hao, Q.; Tian, F.; Li, Q.; Wang, W. Cytokinin-Regulated Sucrose Metabolism in Stay-Green Wheat Phenotype. PLoS ONE 2016, 11, e0161351. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grewal, S.; Yang, C.Y.; Scholefield, D.; Ashling, S.; Ghosh, S.; Swarbreck, D.; Collins, J.; Yao, E.; Sen, T.Z.; Wilson, M.; et al. Chromosome-scale genome assembly of bread wheat’s wild relative Triticum timopheevii. Sci. Data 2024, 11, 420. [Google Scholar] [CrossRef]
- Dwivedi, S.K.; Kumar, S.A.; Mondal, S.U.; Malviya, N.U.; Dubey, R.A. Physiological basis of cytokinin induced drought tolerance in wheat (Triticum aestivum L.). J. AgriSearch 2014, 1, 139–144. [Google Scholar]
- Criado, M.; Caputo, C.; Roberts, I.; Castro, M.; Barneix, A. Cytokinin-induced changes of nitrogen remobilization and chloroplast ultrastructure in wheat (Triticum aestivum). J. Plant Physiol. 2009, 166, 1775–1785. [Google Scholar] [CrossRef]
- Bariah, I.; Keidar-Friedman, D.; Kashkush, K. Identification and characterization of large scale genomic rearrangements during wheat evolution. PLoS ONE 2020, 15, e0231323. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Khan, A.; Hashem, A.; Abd_Allah, E.; Al-Harrasi, A. The molecular mass and isoelectric point of plant proteomes. BMC Genom. 2019, 20, 631. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, B.; Ogonowska, H.; Szala, K.; Bajguz, A.; Orczyk, W.; Nadolska-Orczyk, A. Silencing of TaCKX1 Mediates Expression of Other TaCKX Genes to Increase Yield Parameters in Wheat. Int. J. Mol. Sci. 2020, 21, 4809. [Google Scholar] [CrossRef] [PubMed]
- Mameaux, S.; Cockram, J.; Thiel, T.; Steuernagel, B.; Stein, N.; Taudien, S.; Jack, P.; Werner, P.; Gray, J.C.; Greenland, A.J.; et al. Molecular, phylogenetic and comparative genomic analysis of the cytokinin oxidase/dehydrogenase gene family in the Poaceae. Plant Biotechnol. J. 2012, 10, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.K.; Zhao, Q.; Wang, Y.; Zhao, J.J.; Qiao, L.; Wu, B.B.; Yan, S.X.; Zheng, J.; Zheng, X.W. Comparative Analysis of Genomic and Transcriptome Sequences Reveals Divergent Patterns of Codon Bias in Wheat and Its Ancestor Species. Front. Genet. 2021, 12, 732432. [Google Scholar] [CrossRef]
- Zhang, J.; Vankova, R.; Malbeck, J.; Dobrev, P.; Xu, Y.; Chong, K.; Neff, M. AtSOFL1 and AtSOFL2 Act Redundantly as Positive Modulators of the Endogenous Content of Specific Cytokinins in Arabidopsis. PLoS ONE 2009, 4, e8236. [Google Scholar] [CrossRef]
- Youden, W.J.; Denny, F.E. Factors influencing the pH equilibrium known as the isoelectric point of plant tissue. Am. J. Bot. Bot. Soc. Am. 1926, 13, 743–753. [Google Scholar] [CrossRef]
- Al-Khayri, J.M.; Alshegaihi, R.M.; Mahgoub, E.I.; Mansour, E.; Atallah, O.O.; Sattar, M.N.; Al-Mssallem, M.Q.; Alessa, F.M.; Aldaej, M.I.; Hassanin, A.A. Association of High and Low Molecular Weight Glutenin Subunits with Gluten Strength in Tetraploid Durum Wheat (Triticum turgidum spp. Durum L.). Plants 2023, 12, 1416. [Google Scholar] [CrossRef]
- Gun, L.; Yumiao, R.; Haixian, P.; Liang, Z. Comprehensive Analysis and Comparison on the Codon Usage Pattern of Whole Mycobacterium tuberculosis Coding Genome from Different Area. Biomed. Res. Int. 2018, 2018, 3574976. [Google Scholar] [CrossRef]
- Ahlinder, J.; Hall, D.; Suontama, M.; Sillanpää, M. Principal component analysis revisited: Fast multitrait genetic evaluations with smooth convergence. G3 Genes|Genomes|Genet. 2024, 14, jkae228. [Google Scholar] [CrossRef]
- Yang, Q.; Xin, C.; Xiao, Q.; Lin, Y.; Li, L.; Zhao, J. Codon usage bias in chloroplast genes implicate adaptive evolution of four ginger species. Front. Plant Sci. 2023, 14, 1304264. [Google Scholar] [CrossRef]
- Khalid, A.; Hameed, A.; Tahir, M.F. Wheat quality. A review on chemical composition, nutritional attributes, grain anatomy, types, classification, and function of seed storage proteins in bread making quality. Front. Nutr. 2023, 10, 1053196. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.H.; Smith, D.K.; Rabadan, R.; Peiris, M.; Poon, L.L. Codon usage bias and the evolution of influenza a viruses. Codon usage biases of influenza virus. BMC Evol. Biol. 2010, 10, 253. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.; Appel, R.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Yin, H.; Liu, W.; Hu, X.; Jia, J.; Liu, M.; Wei, J.; Cheng, Y.; Gong, X.; Li, Q.; Yan, W.; et al. A wheat phytohormone atlas spanning major tissues across the entire life cycle provides novel insights into cytokinin and jasmonic acid interplay. Mol. Plant 2025, 18, 366–382. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Q.R.; Nguyen, D.H.; Wingerd, M.A.; Church, G.M.; Steffen, M.A. Molecular weight assessment of proteins in total proteome profiles using 1D-PAGE and LC/MS/MS. Proteome Sci. 2005, 3, 6. [Google Scholar] [CrossRef]
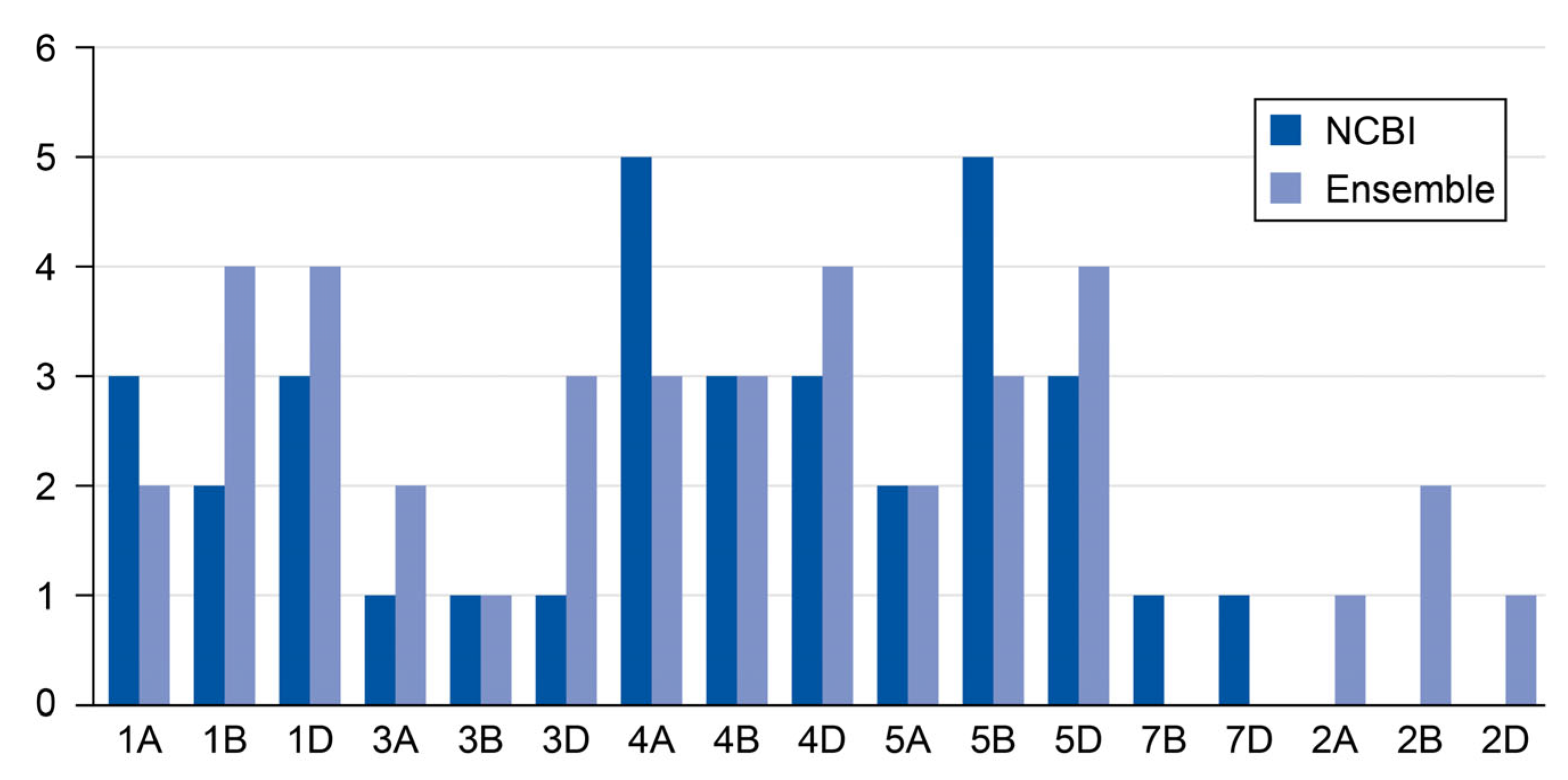
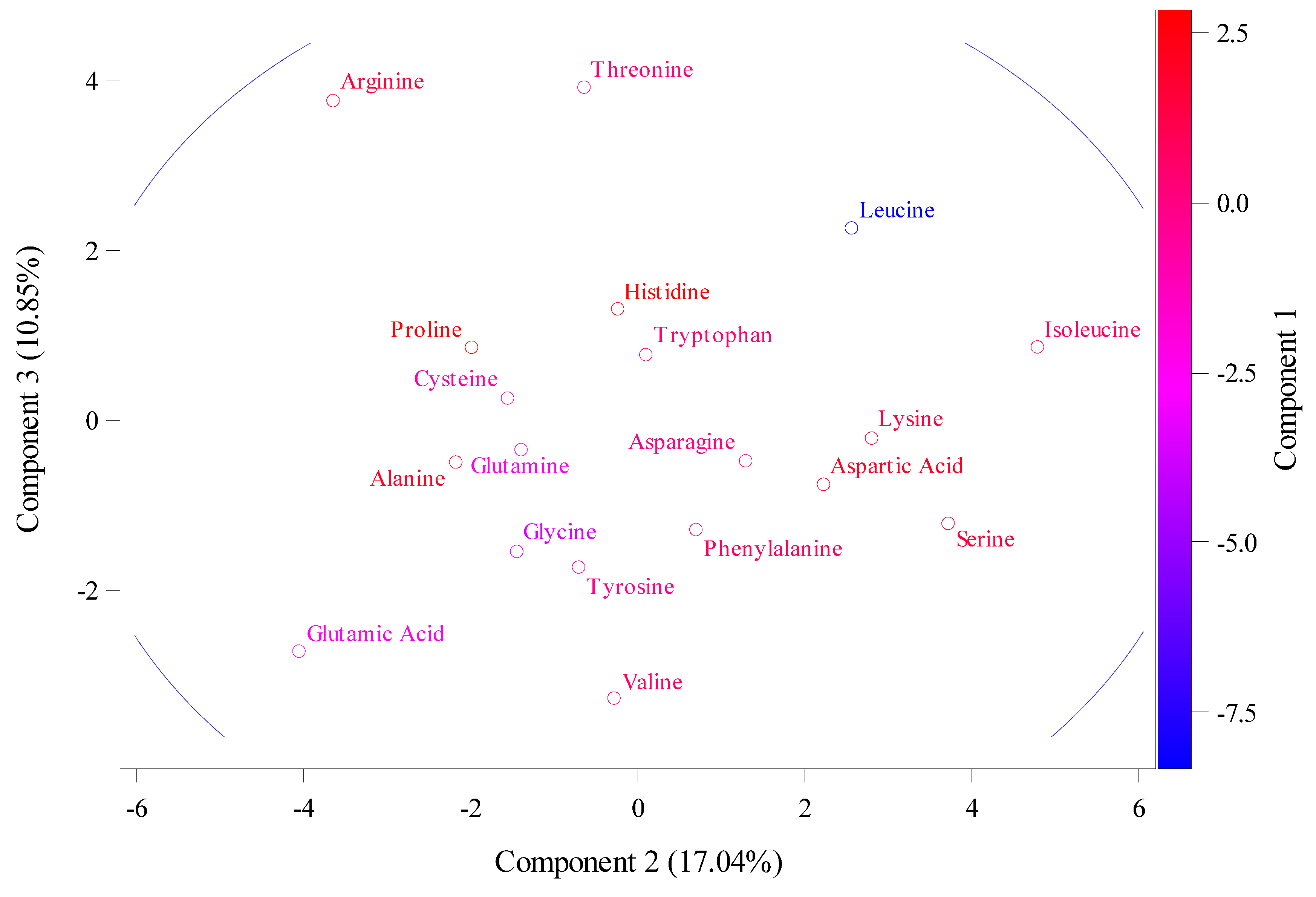
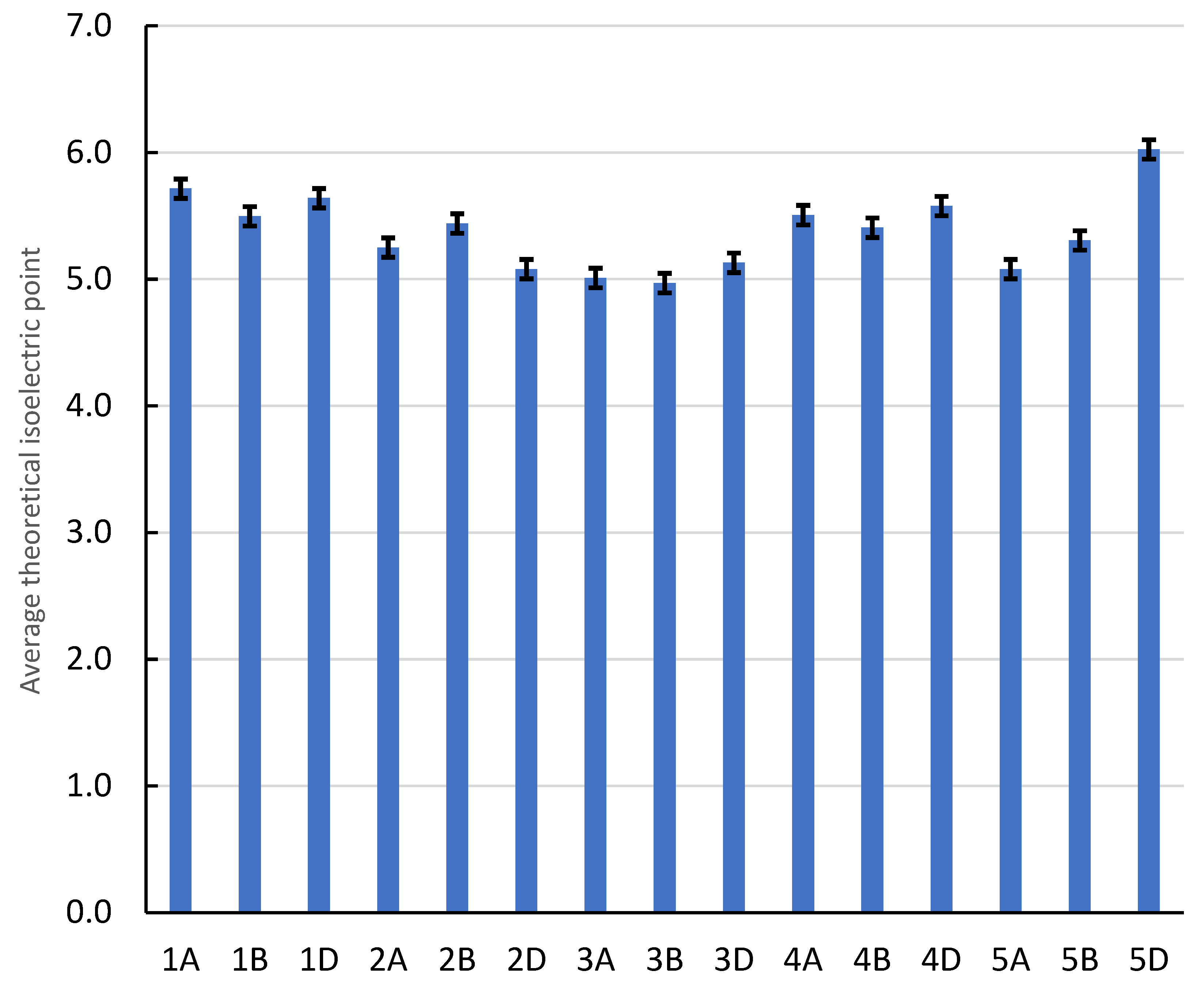

| Location | Bp | Molecular Weight (g/mol) | Theoretical Isoelectric Point (pI) | Instability Index (II) | Aliphatic Index (AI) | Hydrophobicity (HY) |
|---|---|---|---|---|---|---|
| 4A | 218 | 22,991.1 | 5.38 | 38.33 | 93.53 | −0.105 |
| 2A | 243 | 26,618.74 | 5.25 | 40.31 | 91.44 | −0.096 |
| 4B | 205 | 22,513.91 | 5.78 | 41.69 | 94.63 | 0.008 |
| 3A | 234 | 25,265.85 | 5.21 | 34.3 | 95 | −0.017 |
| 1D | 215 | 23,152.61 | 5.77 | 54.16 | 92.98 | 0.035 |
| 3D | 241 | 25,737.38 | 5.21 | 33.91 | 93.49 | 0.01 |
| 1B | 268 | 28,401.58 | 5.06 | 38.27 | 96.12 | 0.071 |
| 1D | 239 | 26,037.71 | 5.66 | 43.3 | 84.1 | −0.38 |
| 1A | 208 | 22,818.19 | 6.13 | 35.3 | 97.5 | −0.045 |
| 2B | 280 | 30,712.62 | 5.71 | 39.74 | 95.71 | −0.03 |
| 2B | 246 | 26,897.94 | 5.17 | 38.94 | 93.09 | −0.118 |
| 5D | 235 | 24,482.96 | 6.22 | 42.68 | 88.04 | 0.008 |
| 3A | 229 | 24,220.69 | 4.81 | 29.84 | 88.69 | 0.04 |
| 4D | 245 | 25,850.59 | 6.6 | 38.16 | 89.59 | −0.049 |
| 5D | 248 | 26,288.03 | 6.23 | 50.36 | 88.91 | −0.108 |
| 5D | 247 | 26,070.71 | 6.17 | 41.8 | 86.52 | −0.082 |
| 5A | 246 | 26,030.62 | 5.23 | 38.94 | 88.82 | −0.063 |
| 1B | 208 | 22,819.17 | 5.98 | 36.23 | 97.5 | −0.045 |
| 5B | 241 | 25,631.11 | 5.15 | 35.92 | 92.24 | −0.012 |
| 1D | 273 | 28,817.09 | 5.15 | 36.73 | 39.99 | 0.067 |
| 2D | 246 | 26,898.99 | 5.08 | 43.64 | 94.31 | −0.085 |
| 1A | 215 | 23,217.63 | 5.3 | 50 | 92.05 | 0.02 |
| 5B | 241 | 25,631.11 | 5.15 | 35.92 | 92.24 | −0.012 |
| 4A | 206 | 22,487.78 | 5.15 | 37.95 | 95.58 | 0.052 |
| 4D | 225 | 23,599.69 | 5.26 | 36.46 | 93.24 | −0.106 |
| 1B | 215 | 23,221.66 | 5.45 | 48 | 91.12 | −0.003 |
| 5B | 246 | 26,101.79 | 5.62 | 41.79 | 91.99 | −0.002 |
| 4D | 250 | 26,176.7 | 5.28 | 36.82 | 87.8 | −0.004 |
| 3D | 226 | 24,032.55 | 4.97 | 28.66 | 88.98 | 0.023 |
| 5A | 245 | 26,310.95 | 4.93 | 39.99 | 95.51 | −0.001 |
| 1D | 208 | 22,819.17 | 5.98 | 36.23 | 97.5 | −0.045 |
| 3B | 226 | 24,032.55 | 4.97 | 27.99 | 88.98 | 0.023 |
| 4A | 248 | 26,111.76 | 5.99 | 48.45 | 88.06 | −0.105 |
| 5D | 249 | 26,428.14 | 5.48 | 43.07 | 90.48 | −0.032 |
| 3D | 239 | 25,637.3 | 5.21 | 34.82 | 94.64 | 0.014 |
| 4B | 205 | 22,282.61 | 5.29 | 34.03 | 97.51 | 0.107 |
| 4D | 205 | 22,315.99 | 5.17 | 36.73 | 96.05 | 0.08 |
| 4B | 226 | 23,572.58 | 5.15 | 37.49 | 91.15 | −0.125 |
| Mean | 233.947 | 25,058.883 | 5.455 | 39.130 | 90.923 | −0.029 |
| StD | 19.385 | 1982.593 | 0.439 | 5.748 | 9.140 | 0.084 |
| NCBI | Ensembl | |||
|---|---|---|---|---|
| Gene Symbol ID | Chro. | Gene Symbol ID | Chro. | |
| 1 | LOC123045576 | 1A | TraesCS4A02G277200 | 4A |
| 2 | LOC123045860 | 1A | TraesCS2A02G380600 | 2A |
| 3 | LOC123061754 | 3A | TraesCS4B02G250400 | 4B |
| 4 | LOC123065219 | 1A | TraesCS3A02G211100 | 3A |
| 5 | LOC123070388 | 3B | TraesCS1D02G367400 | 1D |
| 6 | LOC123078846 | 3D | TraesCS3D02G213900 | 3D |
| 7 | LOC123080662 | 1B | TraesCS1B02G471300 | 1B |
| 8 | LOC123082392 | 4A | TraesCS1D02G003500 | 1D |
| 9 | LOC123083722 | 4A | TraesCS1A02G156100 | 1A |
| 10 | LOC123087480 | 4A | TraesCS2B02G397600 | 2B |
| 11 | LOC123087481 | 4A | TraesCS5D02G568400 | 5D |
| 12 | LOC123088143 | 4A | TraesCS3A02G251500 | 3A |
| 13 | LOC123093512 | 4B | TraesCS4A02G317900 | 4A |
| 14 | LOC123094293 | 4B | TraesCS5D02G568500 | 5D |
| 15 | LOC123095076 | 4B | TraesCS5B02G561400 | 5B |
| 16 | LOC123097019 | 4D | TraesCS5A02G347400 | 5A |
| 17 | LOC123098787 | 4D | TraesCS1B02G173200 | 1B |
| 18 | LOC123099424 | 4D | TraesCS5B02G348600 | 5B |
| 19 | LOC123107636 | 5A | TraesCS1D02G444500 | 1D |
| 21 | LOC123107637 | 5A | TraesCS2D02G376900 | 2D |
| 22 | LOC123112988 | 5B | TraesCS1A02G362500 | 1A |
| 23 | LOC123112989 | 5B | TraesCS5B02G561300 | 5B |
| 24 | LOC123114455 | 5B | TraesCS4A02G413200 | 4A |
| 25 | LOC123116784 | 5B | TraesCS4D02G033800 | 4D |
| 26 | LOC123117873 | 5B | TraesCS1B02G379700 | 1B |
| 27 | LOC123121452 | 1B | TraesCS5B02G348400 | 5B |
| 28 | LOC123123922 | 5D | TraesCS5B02G348300 | 5B |
| 29 | LOC123125354 | 5D | TraesCS3D02G251900 | 3D |
| 30 | LOC123126254 | 5D | TraesCS5A02G347500 | 5A |
| 31 | LOC123157422 | 7B | TraesCS1D02G154700 | 1D |
| 32 | LOC123160435 | 1D | TraesCS3B02G281000 | 3B |
| 33 | LOC123161849 | 1D | TraesCS4A02G318100 | 4A |
| 34 | LOC123165646 | 7D | TraesCS5D02G353600 | 5D |
| 35 | LOC123181086 | 1D | TraesCS3B02G241600 | 3B |
| 36 | TraesCS4B02G313800 | 4B | ||
| 37 | TraesCS4D02G310800 | 4D | ||
| 38 | TraesCS4B02G035700 | 4B | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almutairi, M.M.; Alzahrani, S.M. Molecular Mass and Isoelectric Point Analysis of Cytokinin Sequences in the Wheat Genome. Int. J. Mol. Sci. 2025, 26, 5270. https://doi.org/10.3390/ijms26115270
Almutairi MM, Alzahrani SM. Molecular Mass and Isoelectric Point Analysis of Cytokinin Sequences in the Wheat Genome. International Journal of Molecular Sciences. 2025; 26(11):5270. https://doi.org/10.3390/ijms26115270
Chicago/Turabian StyleAlmutairi, Meshal M., and Saad M. Alzahrani. 2025. "Molecular Mass and Isoelectric Point Analysis of Cytokinin Sequences in the Wheat Genome" International Journal of Molecular Sciences 26, no. 11: 5270. https://doi.org/10.3390/ijms26115270
APA StyleAlmutairi, M. M., & Alzahrani, S. M. (2025). Molecular Mass and Isoelectric Point Analysis of Cytokinin Sequences in the Wheat Genome. International Journal of Molecular Sciences, 26(11), 5270. https://doi.org/10.3390/ijms26115270







