Characterization of HLA-A/HLA-B/HLA-C/HLA-DRB1 Haplotypes in Romanian Stem Cell Donors Through High-Resolution Next-Generation Sequencing
Abstract
1. Introduction
2. Results
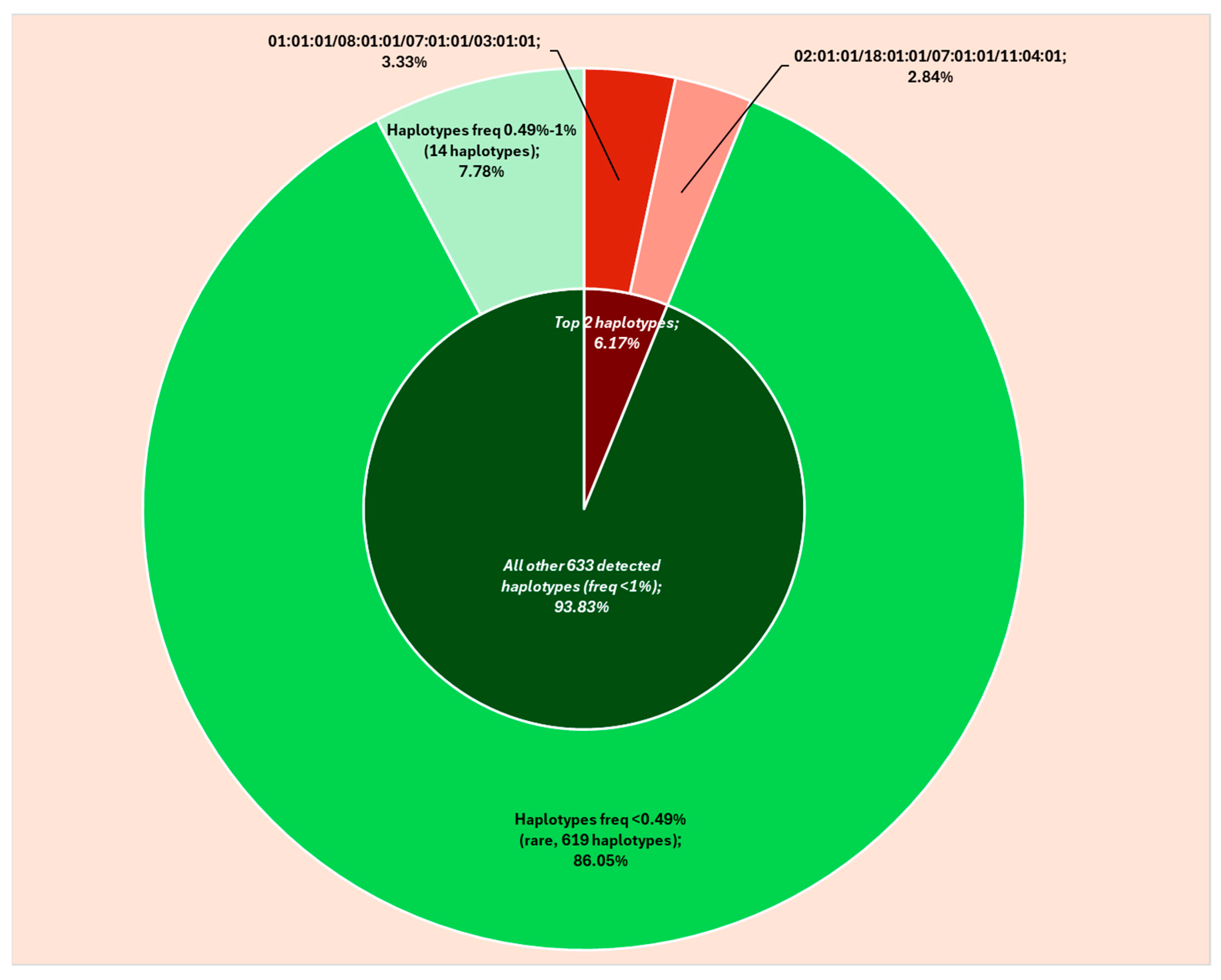
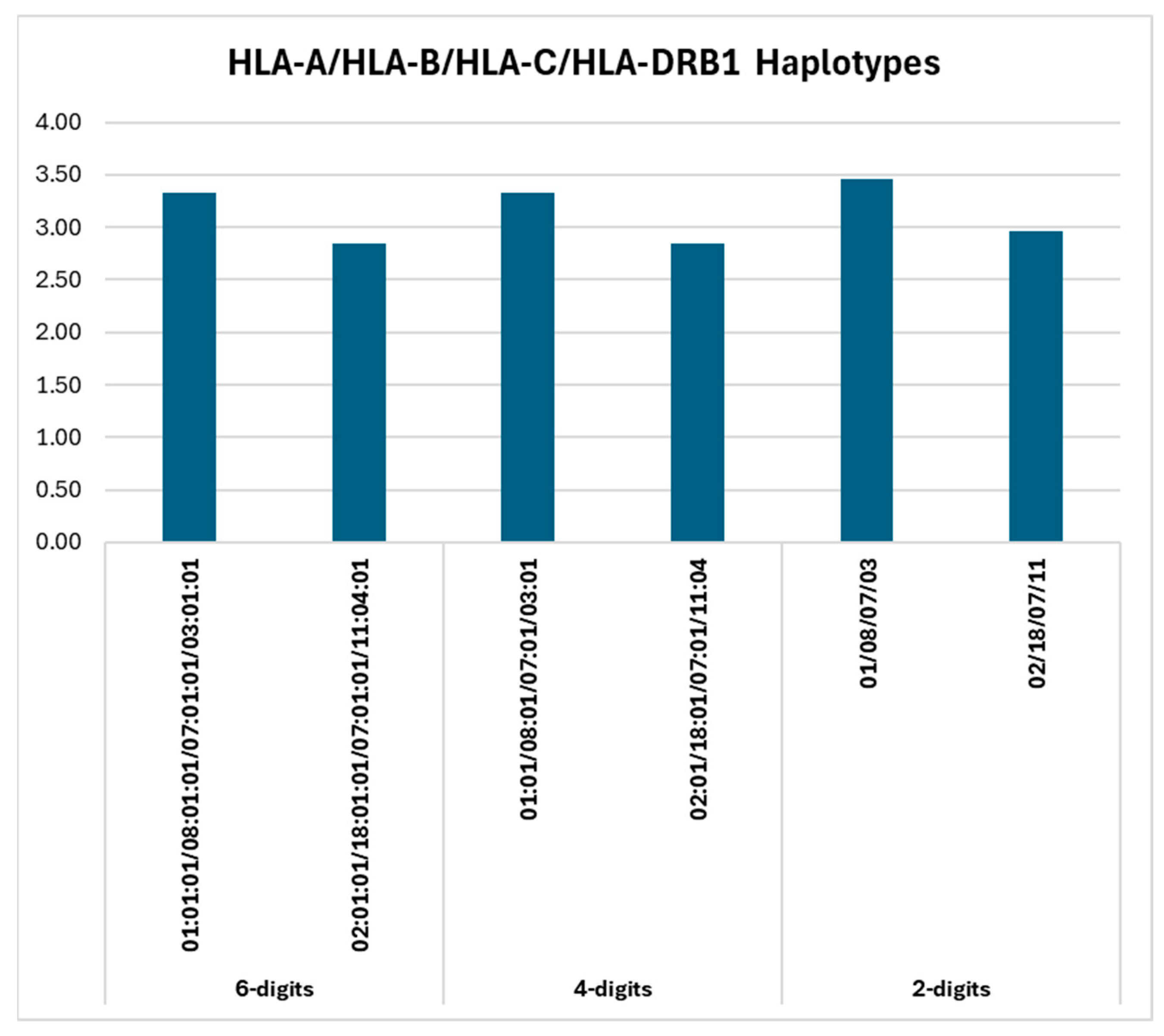
2.1. HLA Haplotypes: 6-Digits Resolution
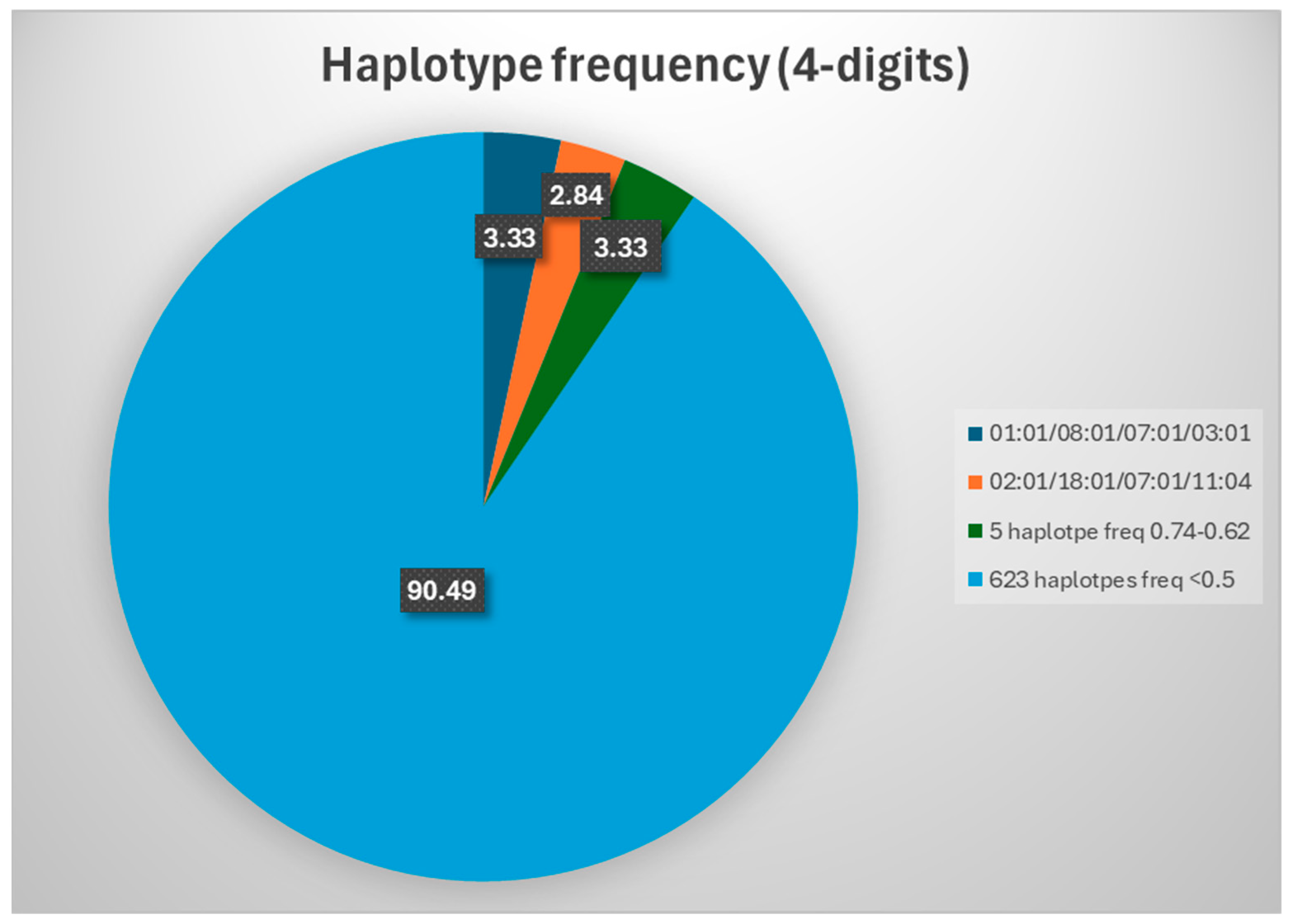
2.2. HLA Haplotypes: 4-Digits Resolution
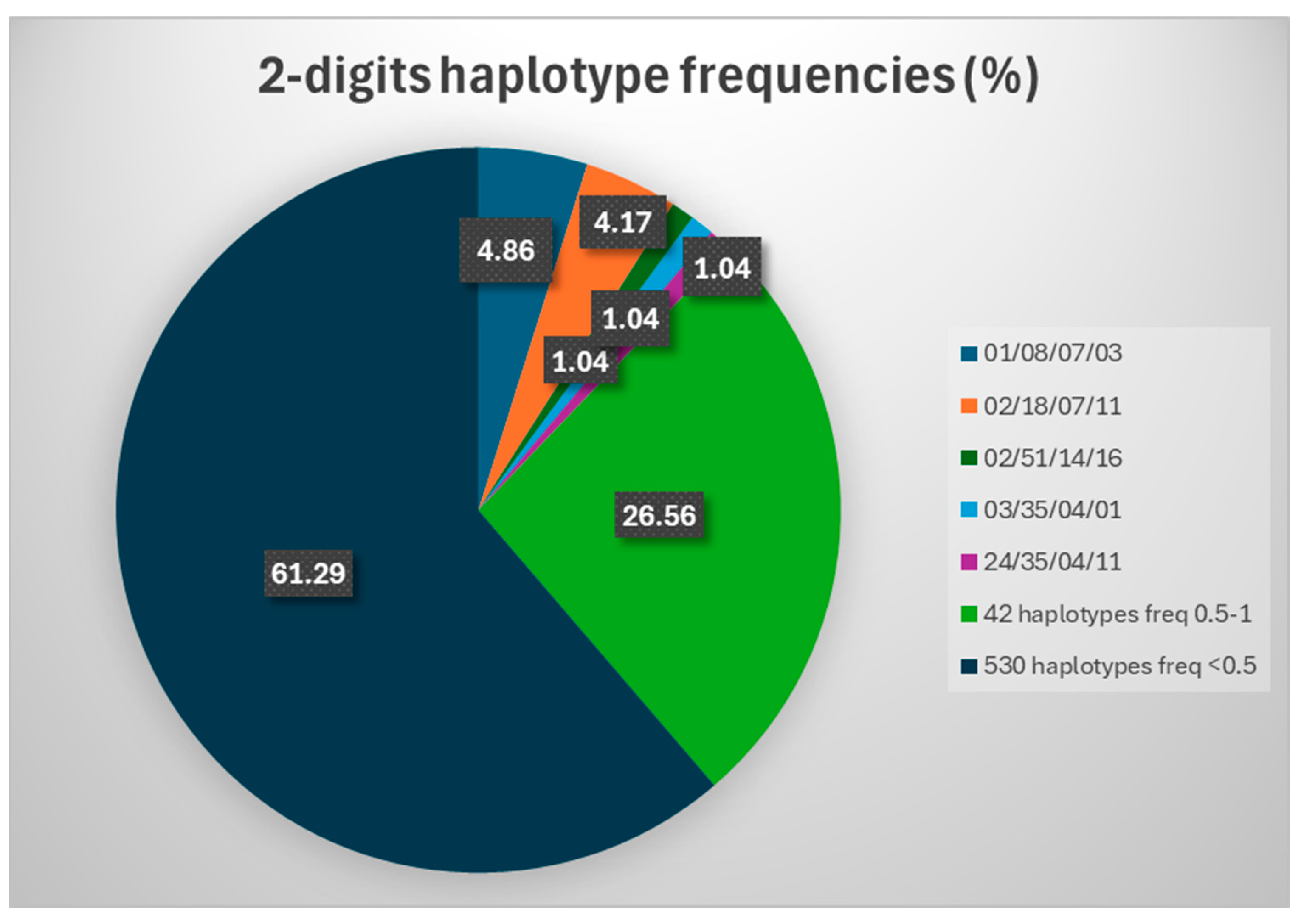
2.3. HLA Haplotypes: 2-Digit Resolution
3. Discussions
4. Material and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kirijas, M.; Genadieva Stavrik, S.; Senev, A.; Efinska Mladenovska, O.; Petlichkovski, A. HLA-A, -B, -C and -DRB1 allele and haplotype frequencies in the Macedonian population based on a family study. Hum. Immunol. 2018, 79, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Mack, S.J.; Tu, B.; Lazaro, A.; Yang, R.; Lancaster, A.K.; Cao, K.; Ng, J.; Hurley, C.K. HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies distinguish Eastern European Americans from the general European American population. Tissue Antigens 2008, 73, 17–32. [Google Scholar] [CrossRef]
- Sanchez-Mazas, A.; Buhler, S.; Nunes, J.M. A new HLA map of Europe: Regional genetic variation and its implication for peopling history, disease-association studies and tissue transplantation. Hum. Hered. 2013, 76, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz-Villena, A.; Martinez-Laso, J.; Moscoso, J.; Livshits, G.; Zamora, J.; Gomez-Casado, E.; Silvera-Redondo, C.; Melvin, K.; Crawford, M.H. HLA genes in the Chuvashian population from European Russia: Admixture of Central European and Mediterranean populations. Hum. Biol. 2003, 75, 375–392. [Google Scholar] [CrossRef][Green Version]
- Tokić, S.; Žižkova, V.; Štefanić, M.; Glavaš-Obrovac, L.; Marczi, S.; Samardžija, M.; Sikorova, K.; Petrek, M. HLA-A, -B, -C, -DRB1, -DQA1, and -DQB1 allele and haplotype frequencies defined by next generation sequencing in a population of East Croatia blood donors. Sci. Rep. 2020, 10, 5513. [Google Scholar] [CrossRef]
- Grubic, Z. HLA allele and haplotype diversity in the Croatian population: State of the art. HLA 2018, 92 (Suppl. S2), 51–56. [Google Scholar] [CrossRef] [PubMed]
- Suslova, T.A.; Burmistrova, A.L.; Chernova, M.S.; Khromova, E.B.; Lupar, E.I.; Timofeeva, S.V.; Devald, I.V.; Vavilov, M.N.; Darke, C. HLA gene and haplotype frequencies in Russians, Bashkirs and Tatars, living in the Chelyabinsk Region (Russian South Urals). Int. J. Immunogenet. 2012, 39, 394–408. [Google Scholar] [CrossRef]
- Sulcebe, G.; Sanchez-Mazas, A.; Tiercy, J.M.; Shyti, E.; Mone, I.; Ylli, Z.; Kardhashi, V. HLA allele and haplotype frequencies in the Albanian population and their relationship with the other European populations. Int. J. Immunogenet. 2009, 36, 337–343. [Google Scholar] [CrossRef]
- Nowak, J.; Mika-Witkowska, R.; Polak, M.; Zajko, M.; Rogatko-Koroś, M.; Graczyk-Pol, E.; Lange, A. Allele and extended haplotype polymorphism of HLA-A, -C, -B, -DRB1 and -DQB1 loci in Polish population and genetic affinities to other populations. Tissue Antigens 2008, 71, 193–205. [Google Scholar] [CrossRef]
- Fernandez Vina, M.A.; Hollenbach, J.A.; Lyke, K.E.; Sztein, M.B.; Maiers, M.; Klitz, W.; Cano, P.; Mack, S.; Single, R.; Brautbar, C.; et al. Tracking human migrations by the analysis of the distribution of HLA alleles, lineages and haplotypes in closed and open populations. Philosophical transactions of the Royal Society of London. Ser. B Biol. Sci. 2012, 367, 820–829. [Google Scholar] [CrossRef]
- Grubic, Z.; Burek Kamenaric, M.; Mikulic, M.; Stingl Jankovic, K.; Maskalan, M.; Zunec, R. HLA-A, HLA-B and HLA-DRB1 allele and haplotype diversity among volunteer bone marrow donors from Croatia. Int. J. Immunogenet. 2014, 41, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, I.; Boșcaiu, V.; Cianga, P.; Dinu, A.A.; Gai, E.; Melinte, M.; Moise, A. The frequency of HLA alleles in the Romanian population. Immunogenetics 2016, 68, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Arrieta-Bolaños, E.; Hernández-Zaragoza, D.I.; Barquera, R. An HLA map of the world: A comparison of HLA frequencies in 200 worldwide populations reveals diverse patterns for class I and class II. Front. Genet. 2023, 14, 866407. [Google Scholar] [CrossRef]
- Matevosyan, L.; Chattopadhyay, S.; Madelian, V.; Avagyan, S.; Nazaretyan, M.; Hyussian, A.; Vardapetyan, E.; Arutunyan, R.; Jordan, F. HLA-A, HLA-B, and HLA-DRB1 allele distribution in a large Armenian population sample. Tissue Antigens 2011, 78, 21–30. [Google Scholar] [CrossRef]
- Arnaiz-Villena, A.; Gomez-Casado, E.; Martinez-Laso, J. Population genetic relationships between Mediterranean populations determined by HLA allele distribution and a historic perspective. Tissue Antigens 2002, 60, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Drenovska, K.; Ivanova, M.; Vassileva, S.; Shahid, M.A.; Naumova, E. Association of specific HLA alleles and haplotypes with pemphigus vulgaris in the Bulgarian population. Front. Immunol. 2022, 13, 901386. [Google Scholar] [CrossRef]
- Hajjej, A.; Abdrakhmanova, S.; Turganbekova, A.; Almawi, W.Y. HLA allele and haplotype frequencies in Kazakhstani Russians and their relationship with other populations. HLA 2023, 101, 249–261. [Google Scholar] [CrossRef]
- Vică, M.L.; Matei, H.V.; Bondor, C.I.; Nicula, G.Z.; Siserman, C.V.; Loga, L.; Dican, L. HLA Polymorphisms and Haplotype Diversity in Transylvania, Romania. J. Immunol. Res. 2019, 2019, 1342762. [Google Scholar] [CrossRef]
- Cano, P.; Testi, M.; Andreani, M.; Khoriaty, E.; Bou Monsef, J.; Galluccio, T.; Troiano, M.; Fernandez-Vina, M.; Inati, A. HLA population genetics: A Lebanese population. Tissue Antigens 2012, 80, 341–355. [Google Scholar] [CrossRef]
- Creary, L.E.; Gangavarapu, S.; Mallempati, K.C.; Montero-Martín, G.; Caillier, S.J.; Santaniello, A.; Hollenbach, J.A.; Oksenberg, J.R.; Fernández-Viña, M.A. Next-generation sequencing reveals new information about HLA allele and haplotype diversity in a large European American population. Hum. Immunol. 2019, 80, 807–822. [Google Scholar] [CrossRef]
- Obradovic, J.; Todosijevic, J.; Jurisic, V. Application of the conventional and novel methods in testing EGFR variants for NSCLC patients in the last 10 years through different regions: A systematic review. Mol. Biol. Rep. 2021, 48, 3593–3604. [Google Scholar] [CrossRef] [PubMed]
- Caragea, A.M.; Ursu, R.-I.; Maruntelu, I.; Tizu, M.; Constantinescu, A.-E.; Tălăngescu, A.; Constantinescu, I. High Resolution HLA-A, HLA-B, and HLA-C Allele Frequencies in Romanian Hematopoietic Stem Cell Donors. Int. J. Mol. Sci. 2024, 25, 8837. [Google Scholar] [CrossRef] [PubMed]
- Caragea, M.A.; Ursu, I.R.; Visan, D.L.; Maruntelu, I.; Iordache, P.; Constantinescu, A.; Tizu, M.; Tălăngescu, A.; Constantinescu, I. High-Resolution HLA-DRB1 Allele Frequencies in a Romanian Cohort of Stem Cell Donors. Balkan J. Med. Genet. 2024, 27, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mazas, A.; Nunes, J.M. PGAE HLA Consortium of the 18th International HLA and Immunogenetics Workshop. The most frequent HLA alleles around the world: A fundamental synopsis. Best Pract. Res. Clin. Haematol. 2024, 37, 101559. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Soormally, A.R.; Hayhurst, J.D.; Marsh, S.G.E. The IPD-IMGT/HLA Database–New developments in reporting HLA variation. Hum. Immunol. 2016, 77, 233–237. [Google Scholar] [CrossRef]
- Gonzalez-Galarza, F.F.; McCabe, A.; Santos, E.J.M.D.; Jones, J.; Takeshita, L.; Ortega-Rivera, N.D.; Cid-Pavon, G.M.D.; Ramsbottom, K.; Ghattaoraya, G.; Alfirevic, A.; et al. Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef]
- Allele Frequency Database. Available online: http://www.allelefrequencies.net (accessed on 2 January 2025).
- Gonzalez-Galarza, F.F.; Christmas, S.; Middleton, D.; Jones, A.R. Allele frequency net: A database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011, 39, D913–D919. [Google Scholar] [CrossRef]
| HLA-A Allele | AF (Allele Frequency) | HLA-B Allele | AF | HLA-C Allele | AF | HLA-DRB1 Alleles | AF |
|---|---|---|---|---|---|---|---|
| A*02:01:01 | 26.11% | B*18:01:01 | 11.25% | C*07:01:01 | 17.36% | DRB1*16:01:01 | 12.59% |
| A*01:01:01 | 12.50% | B*51:01:01 | 10.83% | C*04:01:01 | 13.47% | DRB1*11:04:01 | 12.10% |
| A*24:02:01 | 11.67% | B*08:01:01 | 7.78% | C*12:03:01 | 10.69% | DRB1*03:01:01 | 11.98% |
| A*03:01:01 | 9.72% | B*35:01:01 | 5% | C*06:02:01 | 9.44% | DRB1*07:01:01 | 9.26% |
| A*11:01:01, A*32:01:01 | 6.25% | B*35:03:01 | 4.58% | C*02:02:02 | 8.75% | DRB1*01:01:01 | 7.28% |
| HLA-A/HLA-B/HLA-C/HLA-DRB1 Haplotypes | No. | Haplotype Frequency (HF) (%) |
|---|---|---|
| 01:01:01/08:01:01/07:01:01/03:01:01 | 27 | 3.33 |
| 02:01:01/18:01:01/07:01:01/11:04:01 | 23 | 2.84 |
| 02:01:01/51:01:01/14:02:01/16:01:01 | 6 | 0.74 |
| 24:02:01/35:02:01/04:01:01/11:04:01 | 6 | 0.74 |
| 02:01:01/27:02:01/02:02:02/16:01:01 | 5 | 0.62 |
| 03:01:01/35:01:01/04:01:01/01:01:01 | 5 | 0.62 |
| 26:01:01/38:01:01/12:03:01/13:01:01 | 5 | 0.62 |
| 01:01:01/13:02:01/06:02:01/07:01:01 | 4 | 0.49 |
| 01:01:01/52:01:01/12:02:02/15:02:01 | 4 | 0.49 |
| 01:01:01/57:01:01/06:02:01/07:01:01 | 4 | 0.49 |
| 02:01:01/07:02:01/07:02:01/15:01:01 | 4 | 0.49 |
| 02:01:01/13:02:01/06:02:01/07:01:01 | 4 | 0.49 |
| 02:01:01/44:27:01/07:04:01/16:01:01 | 4 | 0.49 |
| 23:01:01/44:03:01/04:01:01/07:01:01 | 4 | 0.49 |
| 24:02:01/13:02:01/06:02:01/07:01:01 | 4 | 0.49 |
| 25:01:01/18:01:01/12:03:01/15:01:01 | 4 | 0.49 |
| HLA-A/HLA-B/HLA-C/HLA-DRB1 Haplotypes | No. | Haplotype Frequency (%) |
|---|---|---|
| 01:01/08:01/07:01/03:01 | 27 | 3.33 |
| 02:01/18:01/07:01/11:04 | 23 | 2.84 |
| 02:01/51:01/14:02/16:01 | 6 | 0.74 |
| 24:02/35:02/04:01/11:04 | 6 | 0.74 |
| 02:01/27:02/02:02/16:01 | 5 | 0.62 |
| 03:01/35:01/04:01/01:01 | 5 | 0.62 |
| 26:01/38:01/12:03/13:01 | 5 | 0.62 |
| 01:01/13:02/06:02/07:01 | 4 | 0.49 |
| 01:01/52:01/12:02/15:02 | 4 | 0.49 |
| 01:01/57:01/06:02/07:01 | 4 | 0.49 |
| 02:01/07:02/07:02/15:01 | 4 | 0.49 |
| 02:01/13:02/06:02/07:01 | 4 | 0.49 |
| 02:01/44:27/07:04/16:01 | 4 | 0.49 |
| 11:01/55:01/03:03/16:01 | 4 | 0.49 |
| 23:01/44:03/04:01/07:01 | 4 | 0.49 |
| 24:02/13:02/06:02/07:01 | 4 | 0.49 |
| 25:01/18:01/12:03/15:01 | 4 | 0.49 |
| HLA-A/HLA-B/HLA-C/HLA-DRB1 Haplotypes | No. | Haplotype Frequency (%) |
|---|---|---|
| 01/08/07/03 | 28 | 3.46 |
| 02/18/07/11 | 24 | 2.96 |
| 02/51/14/16 | 6 | 0.74 |
| 03/35/04/01 | 6 | 0.74 |
| 24/35/04/11 | 6 | 0.74 |
| 02/08/07/03 | 5 | 0.62 |
| 02/13/06/07 | 5 | 0.62 |
| 02/27/02/16 | 5 | 0.62 |
| 02/35/04/11 | 5 | 0.62 |
| 02/35/04/14 | 5 | 0.62 |
| 02/44/07/16 | 5 | 0.62 |
| 26/38/12/13 | 5 | 0.62 |
| 32/35/04/11 | 5 | 0.62 |
| 01/13/06/07 | 4 | 0.49 |
| 01/52/12/15 | 4 | 0.49 |
| 01/57/06/07 | 4 | 0.49 |
| 02/07/07/15 | 4 | 0.49 |
| 02/52/12/15 | 4 | 0.49 |
| 11/35/04/11 | 4 | 0.49 |
| 11/55/03/16 | 4 | 0.49 |
| 23/44/04/07 | 4 | 0.49 |
| 24/13/06/07 | 4 | 0.49 |
| 24/35/04/16 | 4 | 0.49 |
| 25/18/12/15 | 4 | 0.49 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caragea, A.M.; Ursu, R.-I.; Bohîlțea, L.C.; Iordache, P.; Constantinescu, A.-E.; Constantinescu, I. Characterization of HLA-A/HLA-B/HLA-C/HLA-DRB1 Haplotypes in Romanian Stem Cell Donors Through High-Resolution Next-Generation Sequencing. Int. J. Mol. Sci. 2025, 26, 5250. https://doi.org/10.3390/ijms26115250
Caragea AM, Ursu R-I, Bohîlțea LC, Iordache P, Constantinescu A-E, Constantinescu I. Characterization of HLA-A/HLA-B/HLA-C/HLA-DRB1 Haplotypes in Romanian Stem Cell Donors Through High-Resolution Next-Generation Sequencing. International Journal of Molecular Sciences. 2025; 26(11):5250. https://doi.org/10.3390/ijms26115250
Chicago/Turabian StyleCaragea, Andreea Mirela, Radu-Ioan Ursu, Laurențiu Camil Bohîlțea, Paul Iordache, Alexandra-Elena Constantinescu, and Ileana Constantinescu. 2025. "Characterization of HLA-A/HLA-B/HLA-C/HLA-DRB1 Haplotypes in Romanian Stem Cell Donors Through High-Resolution Next-Generation Sequencing" International Journal of Molecular Sciences 26, no. 11: 5250. https://doi.org/10.3390/ijms26115250
APA StyleCaragea, A. M., Ursu, R.-I., Bohîlțea, L. C., Iordache, P., Constantinescu, A.-E., & Constantinescu, I. (2025). Characterization of HLA-A/HLA-B/HLA-C/HLA-DRB1 Haplotypes in Romanian Stem Cell Donors Through High-Resolution Next-Generation Sequencing. International Journal of Molecular Sciences, 26(11), 5250. https://doi.org/10.3390/ijms26115250






