Transcriptomic Responses of Blue Bat Star Patiria pectinifera to Sediment Burial
Abstract
1. Introduction
2. Results
2.1. Survival and Physicochemical Variations at Different Depths of Sediment Burial
2.2. Transcriptomic Sequence Assembly and Functional Annotations
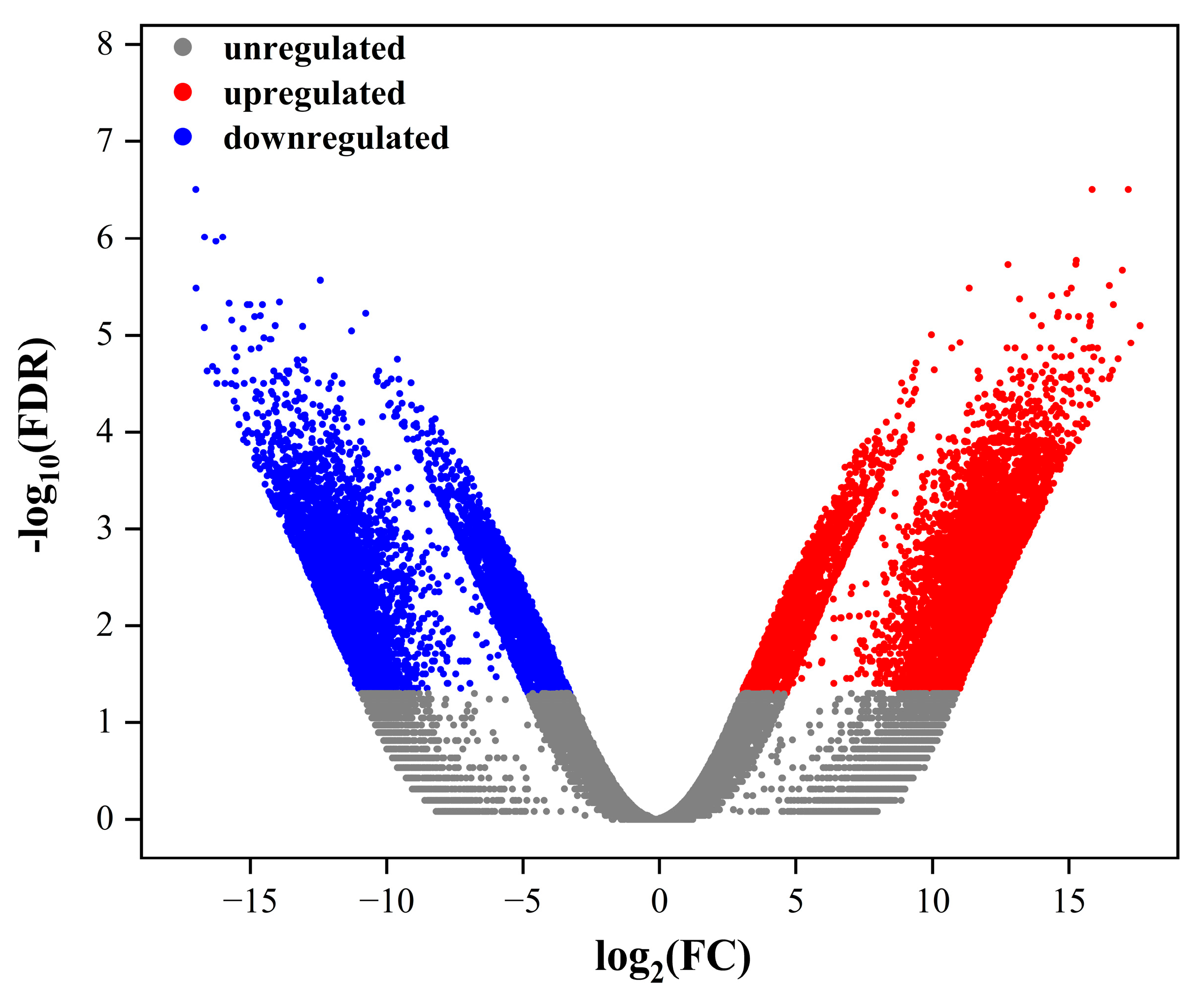
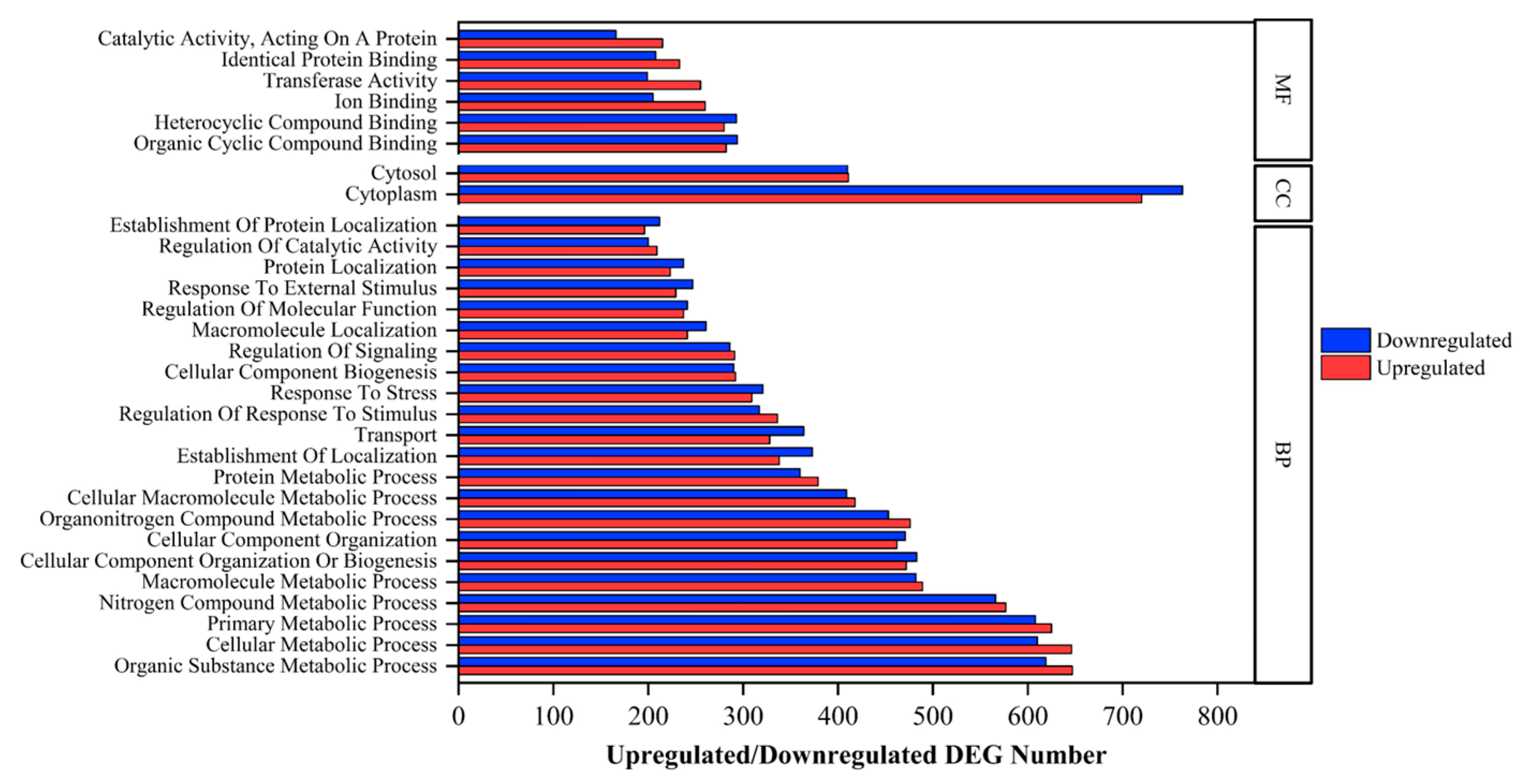
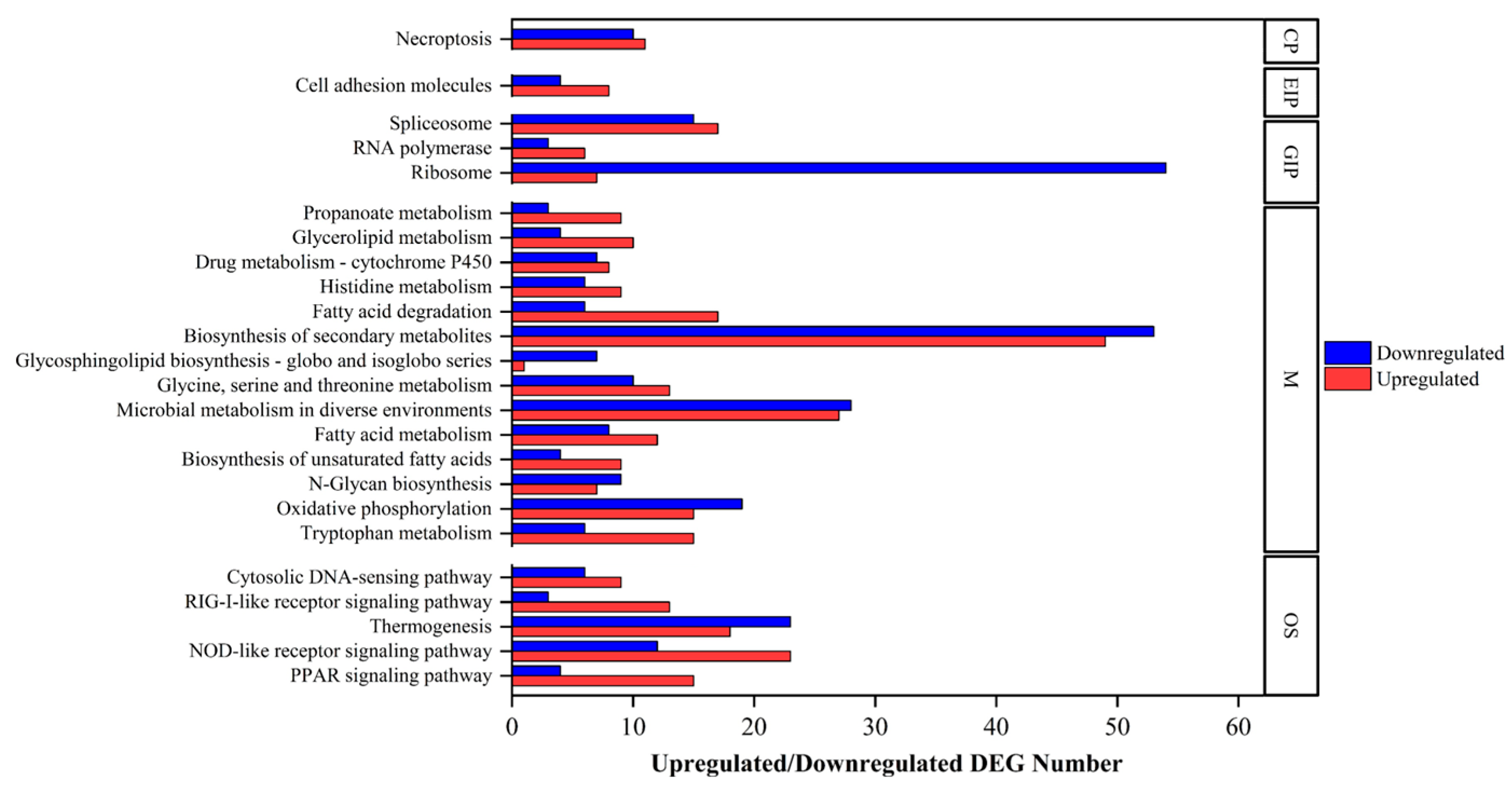
2.3. Annotations of DEGs
2.4. Comparison of DEGs with Other Echinoderms
3. Discussion
3.1. Hypoxic Stress as a Major Physicochemical Determinant in the Sediment Burial
3.2. Metabolic Responses to Hypoxic Stress
3.3. Immune Responses to Hypoxic Stress
3.4. Signaling Pathway Responses to Hypoxic Stress
4. Materials and Methods
4.1. Preparison of Deep-Sea Sediments and Rearing of Patiria pectinifera
4.2. Sediment Burial
4.3. RNA Extraction, Library Preparation and Illumina Hiseq Sequencing
4.4. De Novo Assembly and Functional Annotations
4.5. Differential Expression Analysis and Functional Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BP | Biological process |
| CC | Cellular component |
| CDSs | Coding sequences |
| COG | Clusters of Orthologous Genes |
| CP | Cellular processes |
| DEGs | Differentially expressed genes |
| DO | Dissolved oxygen |
| EIP | Environmental information processing |
| FDR | False discovery rate |
| GIP | Genetic information processing |
| GO | Gene Ontology |
| ISA | International Seabed Authority |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| logFC | Log fold change |
| M | Metabolism |
| MF | Molecular function |
| OS | Organismal systems |
| PPAR | Peroxisome proliferator-activated receptor |
| RPKM | Reads per kilobase of exon per million mapped reads |
| RIG-I-like | Retinoic acid-inducible gene-I-like |
References
- Petersen, S.; Krätschell, A.; Augustin, N.; Jamieson, J.; Hein, J.R.; Hannington, M.D. News from the seabed–Geological characteristics and resource potential of deep-sea mineral resources. Mar. Policy 2016, 70, 175–187. [Google Scholar] [CrossRef]
- Huang, L. Pedogenic ferromanganese nodules and their impacts on nutrient cycles and heavy metal sequestration. Earth-Sci. Rev. 2022, 232, 104147. [Google Scholar] [CrossRef]
- Maciąg, Ł.; Zawadzki, D.; Kozub-Budzyń, G.A.; Piestrzyński, A.; Kotliński, R.A.; Wróbel, R.J. Mineralogy of cobalt-rich ferromanganese crusts from the Perth Abyssal Plain (E Indian Ocean). Minerals 2019, 9, 84. [Google Scholar] [CrossRef]
- Boschen, R.E.; Rowden, A.A.; Clark, M.R.; Gardner, J.P. Mining of deep-sea seafloor massive sulfides: A review of the deposits, their benthic communities, impacts from mining, regulatory frameworks and management strategies. Ocean Coast. Manag. 2013, 84, 54–67. [Google Scholar] [CrossRef]
- Ma, W.; Schott, D.; van Rhee, C. Numerical calculations of environmental impacts for deep sea mining activities. Sci. Total Environ. 2019, 652, 996–1012. [Google Scholar] [CrossRef]
- Clark, M.R.; Durden, J.M.; Christiansen, S. Environmental Impact Assessments for deep-sea mining: Can we improve their future effectiveness? Mar. Policy 2020, 114, 103363. [Google Scholar] [CrossRef]
- Ardito, G.; Rovere, M. Racing the clock: Recent developments and open environmental regulatory issues at the International Seabed Authority on the eve of deep-sea mining. Mar. Policy 2022, 140, 105074. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, X.; Luan, L.; Sha, F.; Liu, X. Technology and equipment of deep-sea mining: State of the art and perspectives. Earth Energy Sci. 2024, 1, 65–84. [Google Scholar] [CrossRef]
- Xu, L.; Li, X.; Liu, Y.; Dou, P.; Hong, Z.; Han, C. A Review of Plume Research in the Collection Process of Deep-Sea Polymetallic Nodules. Water 2024, 16, 3379. [Google Scholar] [CrossRef]
- Carreiro-Silva, M.; Martins, I.; Riou, V.; Raimundo, J.; Caetano, M.; Bettencourt, R.; Rakka, M.; Cerqueira, T.; Godinho, A.; Morato, T.; et al. Mechanical and toxicological effects of deep-sea mining sediment plumes on a habitat-forming cold-water octocoral. Front. Mar. Sci. 2022, 9, 915650. [Google Scholar] [CrossRef]
- Sha, F.; Xi, M.; Wen, Z.; Chen, X.; Zuo, Y.; Xu, J.; Zhang, M.; Niu, H. A review on plumes generation and evolution mechanism during deep-sea polymetallic nodules mining. Ocean Eng. 2024, 298, 117188. [Google Scholar] [CrossRef]
- Jin, S.; Chen, X.; Liu, X.; Zhang, X.; Xie, A.; Yan, J. Research on benthic plume sedimentation characteristics and temperature effect in deep-sea nodule mining operation. Mar. Geol. 2023, 464, 107141. [Google Scholar] [CrossRef]
- Christiansen, B.; Denda, A.; Christiansen, S. Potential effects of deep seabed mining on pelagic and benthopelagic biota. Mar. Policy 2020, 114, 103442. [Google Scholar] [CrossRef]
- Li, X.; Li, S.; Huang, X.; Chen, Y.; Cheng, J.; Zhan, A. Protein-mediated bioadhesion in marine organisms: A review. Mar. Environ. Res. 2021, 170, 105409. [Google Scholar] [CrossRef]
- Leonard, C.; Evans, J.; Knittweis, L.; Aguilar, R.; Alvarez, H.; Borg, J.A.; Garcia, S.; Schembri, P.J. Diversity, distribution, and habitat associations of deep-water echinoderms in the Central Mediterranean. Mar. Biodivers. 2020, 50, 69. [Google Scholar] [CrossRef]
- Parameswaran, U.V.; Jaleel, K.A.; Sanjeevan, V.; Gopal, A.; Vijayan, A.K.; Gupta, G.; Sudhakar, M. Diversity and distribution of echinoderms in the South Eastern Arabian Sea shelf under the influence of seasonal hypoxia. Prog. Oceanogr. 2018, 165, 189–204. [Google Scholar] [CrossRef]
- Lebrato, M.; Iglesias-Rodríguez, D.; Feely, R.A.; Greeley, D.; Jones, D.O.; Suarez-Bosche, N.; Lampitt, R.S.; Cartes, J.E.; Green, D.R.; Alker, B. Global contribution of echinoderms to the marine carbon cycle: CaCO3 budget and benthic compartments. Ecol. Monogr. 2010, 80, 441–467. [Google Scholar] [CrossRef]
- Jones, D.O.; Kaiser, S.; Sweetman, A.K.; Smith, C.R.; Menot, L.; Vink, A.; Trueblood, D.; Greinert, J.; Billett, D.S.; Arbizu, P.M.; et al. Biological responses to disturbance from simulated deep-sea polymetallic nodule mining. PLoS ONE 2017, 12, e0171750. [Google Scholar] [CrossRef]
- Gollner, S.; Kaiser, S.; Menzel, L.; Jones, D.O.; Brown, A.; Mestre, N.C.; Van Oevelen, D.; Menot, L.; Colaço, A.; Canals, M.; et al. Resilience of benthic deep-sea fauna to mining activities. Mar. Environ. Res. 2017, 129, 76–101. [Google Scholar] [CrossRef]
- Levin, L.A.; Amon, D.J.; Lily, H. Challenges to the sustainability of deep-seabed mining. Nat. Sustain. 2020, 3, 784–794. [Google Scholar] [CrossRef]
- Hutchison, Z.L.; Green, D.H.; Burrows, M.T.; Jackson, A.C.; Wilson, B.; Last, K.S. Survival strategies and molecular responses of two marine mussels to gradual burial by sediment. J. Exp. Mar. Biol. Ecol. 2020, 527, 151364. [Google Scholar] [CrossRef]
- Li, X.; Chen, L.; Zhou, Z.; Li, B.; Liu, X. The behavioral and antioxidant response of the bivalve Gomphina veneriformis to sediment burial effect. Acta Oceanol. Sin. 2021, 40, 75–82. [Google Scholar] [CrossRef]
- Hendrick, V.J.; Hutchison, Z.L.; Last, K.S. Sediment burial intolerance of marine macroinvertebrates. PLoS ONE 2016, 11, e0149114. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, Z.L.; Hendrick, V.J.; Burrows, M.T.; Wilson, B.; Last, K.S. Buried alive: The behavioural response of the mussels, Modiolus modiolus and Mytilus edulis to sudden burial by sediment. PLoS ONE 2016, 11, e0151471. [Google Scholar] [CrossRef]
- Bluhm, H. Monitoring megabenthic communities in abyssal manganese nodule sites of the East Pacific Ocean in association with commercial deep-sea mining. Aquat. Conserv. Mar. Freshw. Ecosyst. 1994, 4, 187–201. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Meng, F. Transcriptomic response study of brittle star Ophiothrix exigua to sediment burial. Hydrobiologia 2023, 850, 3645–3654. [Google Scholar] [CrossRef]
- Dan-Sohkawa, M.; Yamanaka, H.; Watanabe, K. Reconstruction of bipinnaria larvae from dissociated embryonic cells of the starfish, Asterina pectinifera. J. Embryol. Exp. Morphol. 1986, 94, 47–60. [Google Scholar] [CrossRef]
- Bolam, S.G. Burial survival of benthic macrofauna following deposition of simulated dredged material. Enviro. Monit. Assess. 2011, 181, 13–27. [Google Scholar] [CrossRef]
- Hinchey, E.K.; Schaffner, L.C.; Hoar, C.C.; Vogt, B.W.; Batte, L.P. Responses of estuarine benthic invertebrates to sediment burial: The importance of mobility and adaptation. Hydrobiologia 2006, 556, 85–98. [Google Scholar] [CrossRef]
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef]
- Guilhermic, C.; Nardelli, M.P.; Mouret, A.; Le Moigne, D.; Howa, H. Short-term response of benthic foraminifera to fine-sediment depositional events simulated in microcosm. Biogeosciences 2023, 20, 3329–3351. [Google Scholar] [CrossRef]
- Lee, Y.; Byeon, E.; Kim, D.-H.; Maszczyk, P.; Wang, M.; Wu, R.S.S.; Jeung, H.-D.; Hwang, U.-K.; Lee, J.-S. Hypoxia in aquatic invertebrates: Occurrence and phenotypic and molecular responses. Aqua. Toxicol. 2023, 263, 106685. [Google Scholar] [CrossRef]
- Sun, J.L.; Zhao, L.L.; Wu, H.; Liu, Q.; Liao, L.; Luo, J.; Lian, W.Q.; Cui, C.; Jin, L.; Ma, J.D. Acute hypoxia changes the mode of glucose and lipid utilization in the liver of the largemouth bass (Micropterus salmoides). Sci. Total Environ. 2020, 713, 135157. [Google Scholar] [CrossRef]
- Zheng, C.; Zhao, Q.; Li, E.; Zhao, D.; Sun, S. Role of hypoxia in the behaviour, physiology, immunity and response mechanisms of crustaceans: A review. Rev. Aquac. 2022, 14, 676–687. [Google Scholar] [CrossRef]
- Genz, J.; Jyde, M.; Svendsen, J.C.; Steffensen, J.F.; Ramløv, H. Excess post-hypoxic oxygen consumption is independent from lactate accumulation in two cyprinid fishes. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2013, 165, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shin, P.; Cheung, S. Comparisons of the metabolic responses of two subtidal nassariid gastropods to hypoxia and re-oxygenation. Mar. Pollut. Bull. 2014, 82, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, X.; Yan, T.; Song, J.; Yu, R.; Zhou, M. Detrimental impact of hypoxia on the mortality, growth, reproduction, and enzyme activities of planktonic mysid Neomysis awatschensis. Aqua. Ecol. 2021, 55, 849–859. [Google Scholar] [CrossRef]
- Hu, M.; Wu, F.; Yuan, M.; Li, Q.; Gu, Y.; Wang, Y.; Liu, Q. Antioxidant responses of triangle sail mussel Hyriopsis cumingii exposed to harmful algae Microcystis aeruginosa and hypoxia. Chemosphere 2015, 139, 541–549. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, W.; Qian, X.; Ji, J.; Ning, X.; Zhu, F.; Yin, S.; Zhang, K. Effects of hypoxia and reoxygenation on apoptosis, oxidative stress, immune response and gut microbiota of Chinese mitten crab, Eriocheir sinensis. Aqua. Toxicol. 2023, 260, 106556. [Google Scholar] [CrossRef]
- Xie, Z.; Wei, S.; Dong, H.; Chen, H.; Zhang, Q.; Liu, W.; Peng, J.; Sokolova, I.M.; Hu, M.; Wang, Y. Hemocyte responses of the oyster Crassostrea hongkongensis exposed to diel-cycling hypoxia and salinity change. Front. Mar. Sci. 2021, 8, 749623. [Google Scholar] [CrossRef]
- Silvestre, F. Signaling pathways of oxidative stress in aquatic organisms exposed to xenobiotics. J. Exp. Zool. A Ecol. Integr. Physiol. 2020, 333, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Wang, H.; Jiang, K.; Yan, X. Transcriptome analysis reveals differential immune related genes expression in Ruditapes philippinarum under hypoxia stress: Potential HIF and NF-κB crosstalk in immune responses in clam. BMC Genom. 2020, 21, 318. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, H.; Zhao, S.; Yu, S.; Xie, S.; Hua, S.; Yan, B.; Xing, C.; Gao, H. Hypoxia stress affects the physiological responses, apoptosis and innate immunity of Kuruma shrimp, Marsupenaeus japonicus. Fish Shellfish Immunol. 2022, 122, 206–214. [Google Scholar] [CrossRef]
- Gorr, T.A.; Gassmann, M.; Wappner, P. Sensing and responding to hypoxia via HIF in model invertebrates. J. Insect Physiol. 2006, 52, 349–364. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, D.-H.; Lee, J.-S.; Lee, M.-C.; Kim, H.S.; Maszczyk, P.; Sakakura, Y.; Yang, Z.; Hagiwara, A.; Park, H.G.; et al. Oxidative stress-mediated deleterious effects of hypoxia in the brackish water flea Diaphanosoma celebensis. Mar. Pollut. Bull. 2024, 205, 116633. [Google Scholar] [CrossRef]
- Sun, Y.; Li, J.; Li, Y.; Wu, Z. Molecular characterization and transcriptional regulation between PPAR and CPT1 in freshwater bivalve Hyriopsis cumingii. Int. J. Biol. Macromol. 2024, 280, 135647. [Google Scholar] [CrossRef]
- Ran, Z.; Kong, F.; Liao, K.; Xu, J.; Liu, X.; Shi, P.; Zhang, M.; Wu, K.; Yan, X. Identification and expression of PPAR in Sinonovacula constricta and their potential regulatory effects on Δ6 fad transcription. J. Ocean Univ. China 2021, 20, 1557–1566. [Google Scholar] [CrossRef]
- D’Aniello, E.; Amodeo, P.; Vitale, R.M. Marine natural and nature-inspired compounds targeting peroxisome proliferator activated receptors (PPARs). Mar. Drugs 2023, 21, 89. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Zhang, L.; Du, Y.; Xu, F.; Li, L.; Zhang, G. Characterization of the mollusc RIG-I/MAVS pathway reveals an archaic antiviral signalling framework in invertebrates. Sci. Rep. 2017, 7, 8217. [Google Scholar] [CrossRef]
- Liang, B.; Su, J. Advances in aquatic animal RIG-I-like receptors. Fish Shellfish Immunol. Rep. 2021, 2, 100012. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, F.; Li, J.; Tong, Y.; Zhang, Y.; Yu, Z. The first invertebrate RIG-I-like receptor (RLR) homolog gene in the pacific oyster Crassostrea gigas. Fish Shellfish Immunol. 2014, 40, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
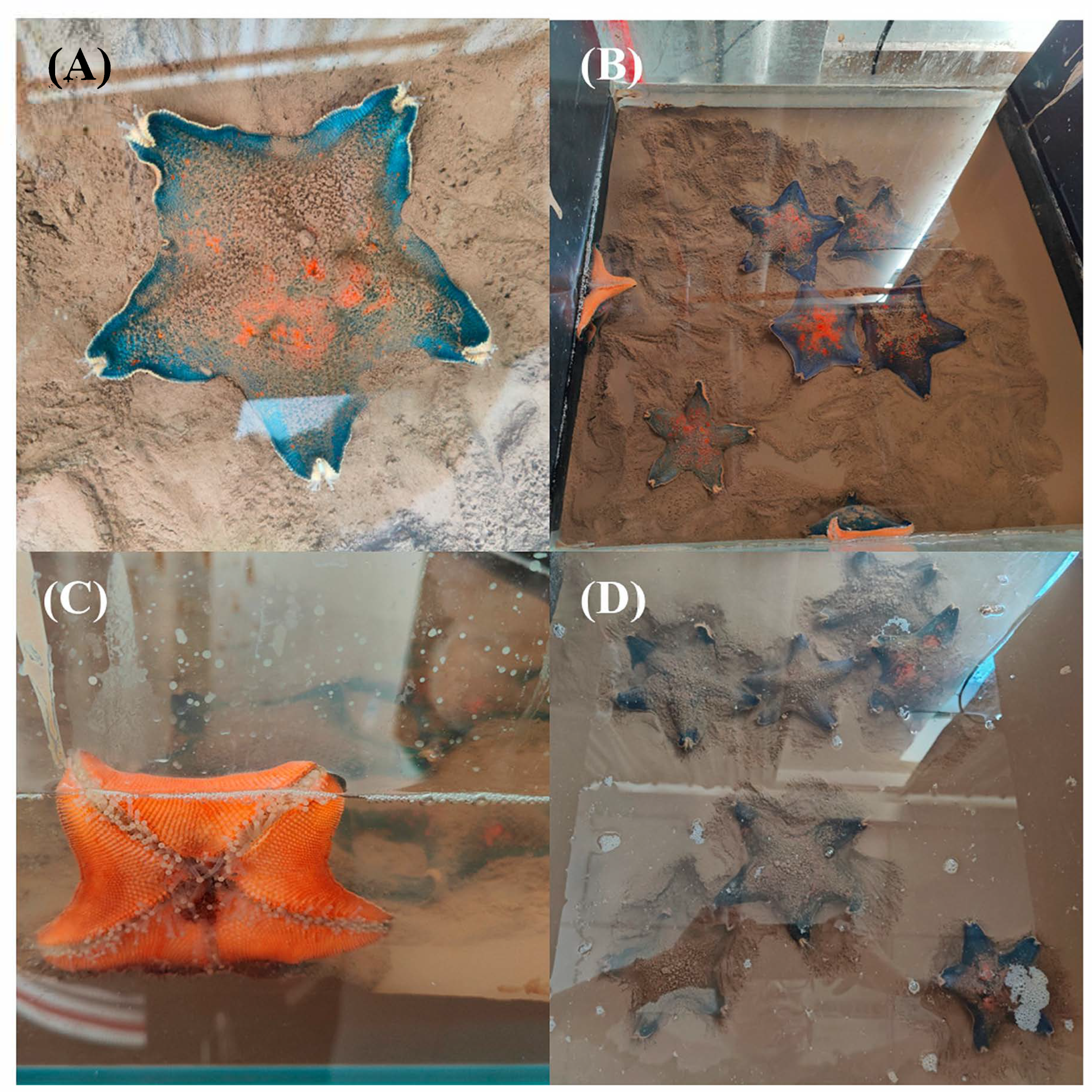
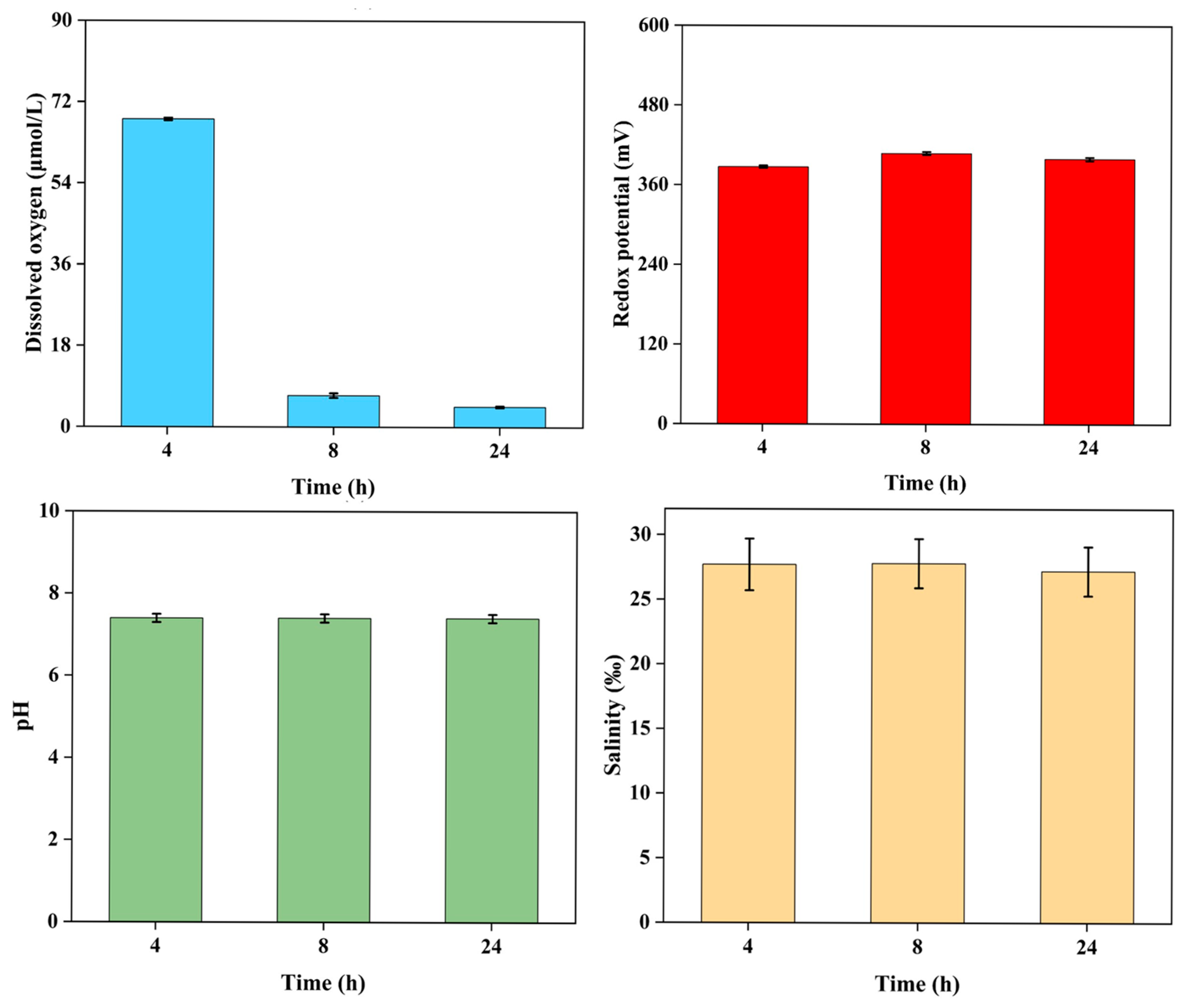
| Sediment Grain Size (μm) | Burial Depth (cm) | Survival Rates (%) |
|---|---|---|
| <63 | 2 | 95.2 |
| 5 | 81.0 | |
| 63 to 250 | 2 | 100.0 |
| 5 | 100.0 |
| Sample | Raw Reads | Raw Bases(bp) | Clean Reads | Clean Bases(bp) | Valid Bases(%) | GC Content(%) |
|---|---|---|---|---|---|---|
| Experimental group 1 | 4,7078,864 | 7,061,829,600 | 44,362,144 | 6,577,106,998 | 93.1 | 45.1 |
| Experimental group 2 | 48,983,870 | 7,347,580,500 | 46,163,352 | 6,844,046,669 | 93.1 | 45.1 |
| Experimental group 3 | 44,980,718 | 6,747,107,700 | 42,781,734 | 6,357,259,233 | 94.2 | 45.2 |
| Control group 1 | 43,523,738 | 6,528,560,700 | 40,288,662 | 5,897,539,644 | 90.3 | 46.0 |
| Control group 2 | 43,174,760 | 6,476,214,000 | 39,964,578 | 5,850,154,113 | 90.3 | 46.0 |
| Control group 3 | 44,645,024 | 6,696,753,600 | 41,962,408 | 6,177,159,182 | 92.2 | 46.3 |
| Class | Species | Accession Number | Size (Mbp) | GC Content (%) | Transcripts | CDSs |
|---|---|---|---|---|---|---|
| Asteroidea | Acanthaster planci | GCF_001949145.1 | 383.8 | 41.5 | 36,221 | 33,201 |
| Asterias amurensis | GCF_032118995.1 | 491.5 | 39.0 | 30,608 | 26,569 | |
| Asterias rubens | GCF_902459465.1 | 417.6 | 39.0 | 28,238 | 24,049 | |
| Patiria miniata | GCF_015706575.1 | 608.3 | 40.5 | 40,283 | 35,403 | |
| Crinoidea | Anneissia japonica | GCF_011630105.1 | 589.6 | 34.5 | 38,110 | 32,774 |
| Antedon mediterranea | GCF_964355755.1 | 354.5 | 33.5 | 33,480 | 25,416 | |
| Echinoidea | Diadema antillarum | GCF_040938485.1 | 1800.0 | 38.5 | 39,579 | 36,155 |
| Lytechinus pictus | GCF_037042905.1 | 811.7 | 36.5 | 37,286 | 29,875 | |
| Lytechinus variegatus | GCF_018143015.1 | 869.6 | 36.5 | 38,196 | 33,669 | |
| Strongylocentrotus purpuratus | GCF_000002235.5 | 921.8 | 37.5 | 45,168 | 38,439 | |
| Holothuroidea | Apostichopus japonicus | GCF_037975245.1 | 671.6 | 37.5 | 57,784 | 45,056 |
| Ophiuroidea | Amphiura filiformis | GCF_039555335.1 | 1600.0 | 36.5 | 46,206 | 39,055 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, H.; Wan, L.; Wang, C.; Sun, C.; Wang, X.; Xu, L. Transcriptomic Responses of Blue Bat Star Patiria pectinifera to Sediment Burial. Int. J. Mol. Sci. 2025, 26, 5208. https://doi.org/10.3390/ijms26115208
Dong H, Wan L, Wang C, Sun C, Wang X, Xu L. Transcriptomic Responses of Blue Bat Star Patiria pectinifera to Sediment Burial. International Journal of Molecular Sciences. 2025; 26(11):5208. https://doi.org/10.3390/ijms26115208
Chicago/Turabian StyleDong, Han, Linli Wan, Chunsheng Wang, Cong Sun, Xiaogu Wang, and Lin Xu. 2025. "Transcriptomic Responses of Blue Bat Star Patiria pectinifera to Sediment Burial" International Journal of Molecular Sciences 26, no. 11: 5208. https://doi.org/10.3390/ijms26115208
APA StyleDong, H., Wan, L., Wang, C., Sun, C., Wang, X., & Xu, L. (2025). Transcriptomic Responses of Blue Bat Star Patiria pectinifera to Sediment Burial. International Journal of Molecular Sciences, 26(11), 5208. https://doi.org/10.3390/ijms26115208








