Transcriptome Analysis of DAMP-Induced Root Growth Regulation and Defense in Foxtail Millet
Abstract
1. Introduction
2. Results
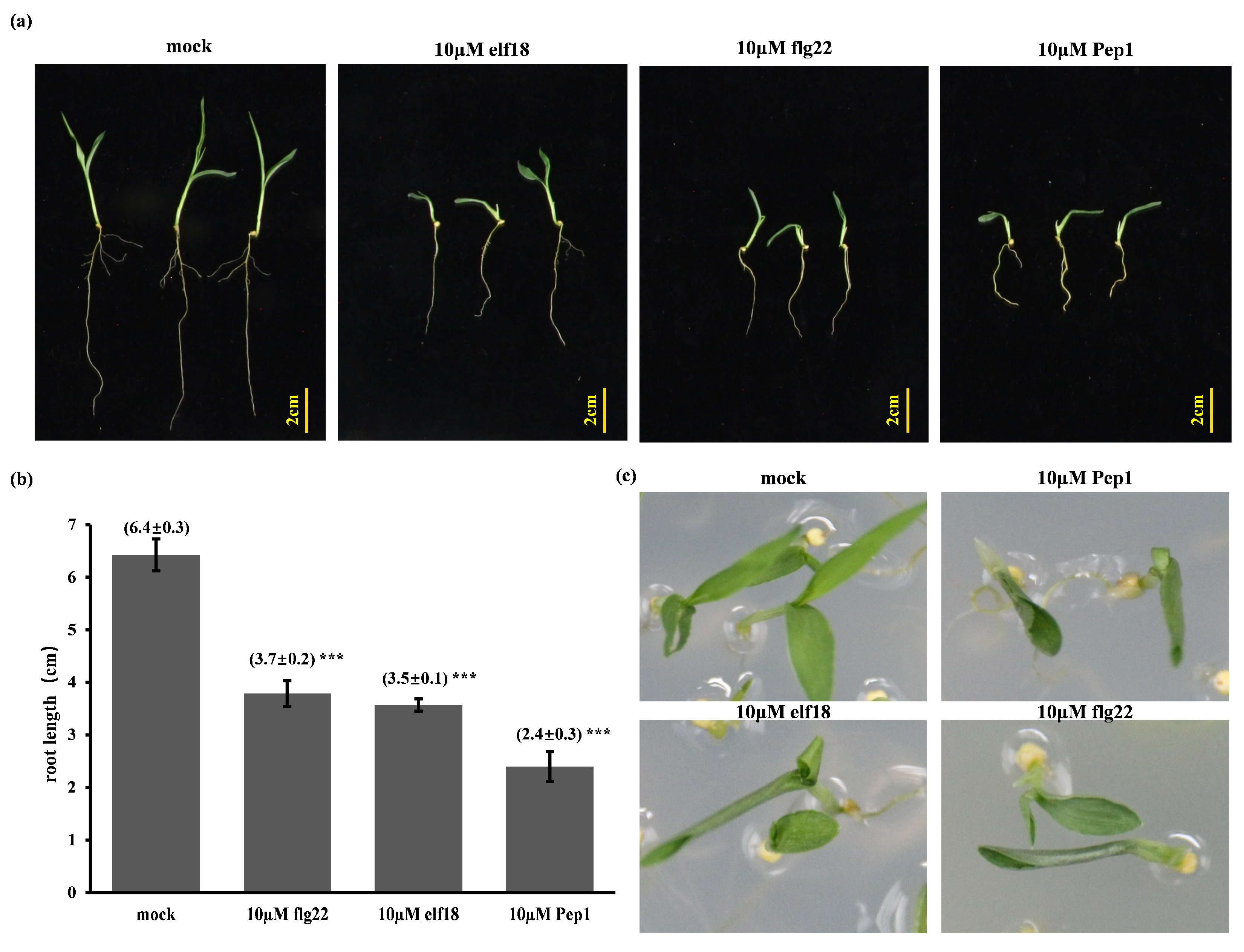
2.1. Elicitor-Induced Immune Responses and Root Growth Inhibition
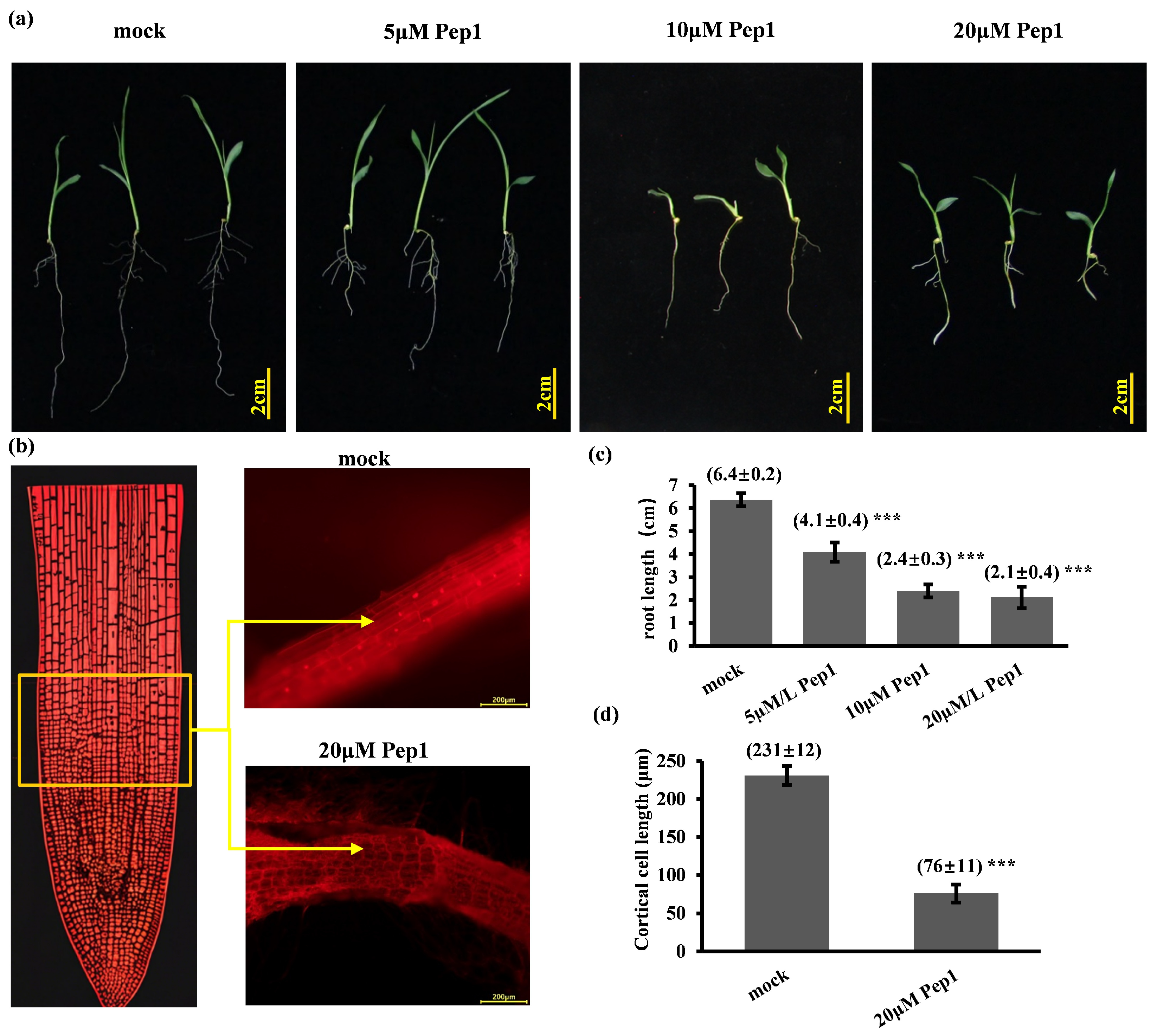
2.2. Synthetic Pep1 Inhibits Foxtail Millet Primary Root Growth
2.3. Transcriptome Analysis of Foxtail Millet in Response to Pep1 Treatment
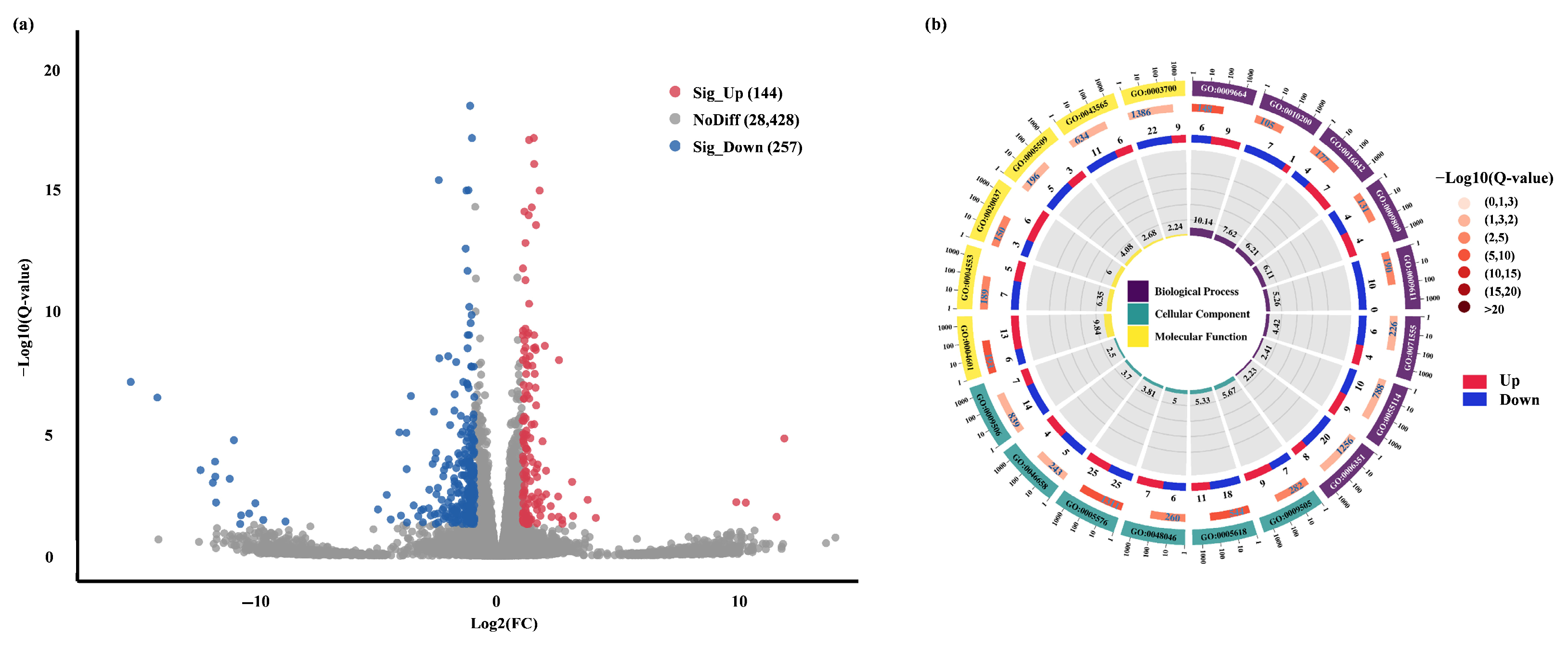
2.3.1. Transcriptome Sequencing Data Quality Assessment
2.3.2. Pep1-Mediated Different Gene Expression and GO Enrichment in Foxtail Millet Roots
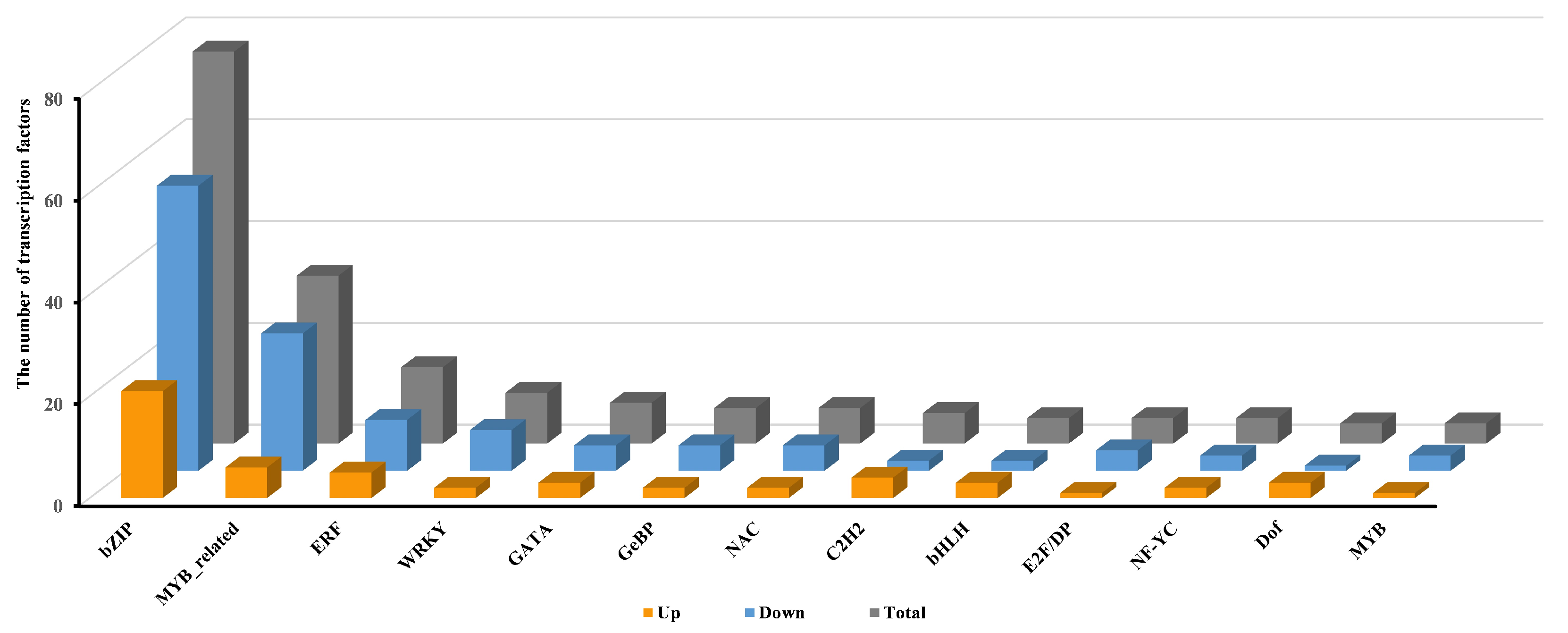
2.3.3. Transcriptional Regulation of Pep1 Response by Transcription Factors in Foxtail Millets
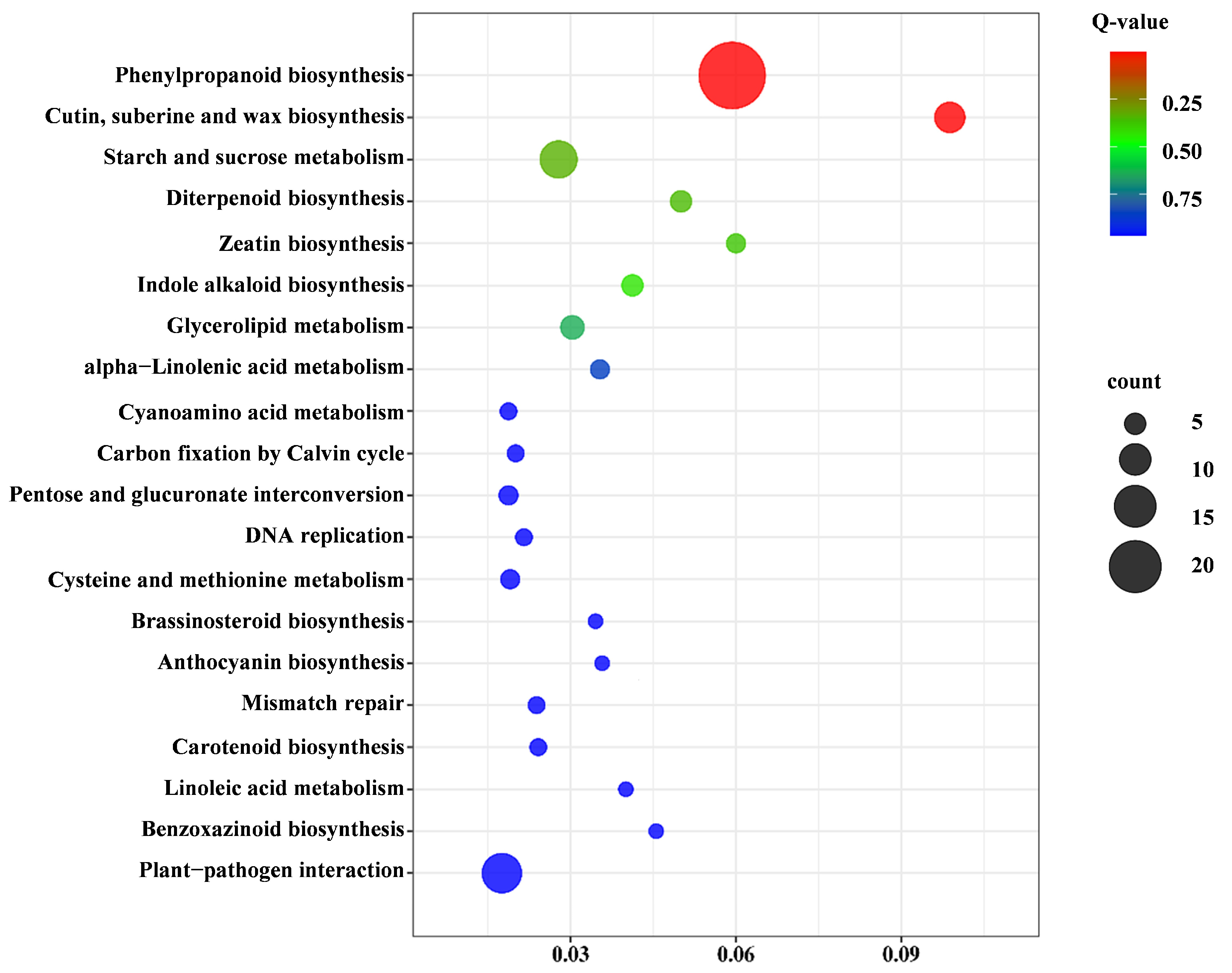
2.3.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of Pep1 Stress Response
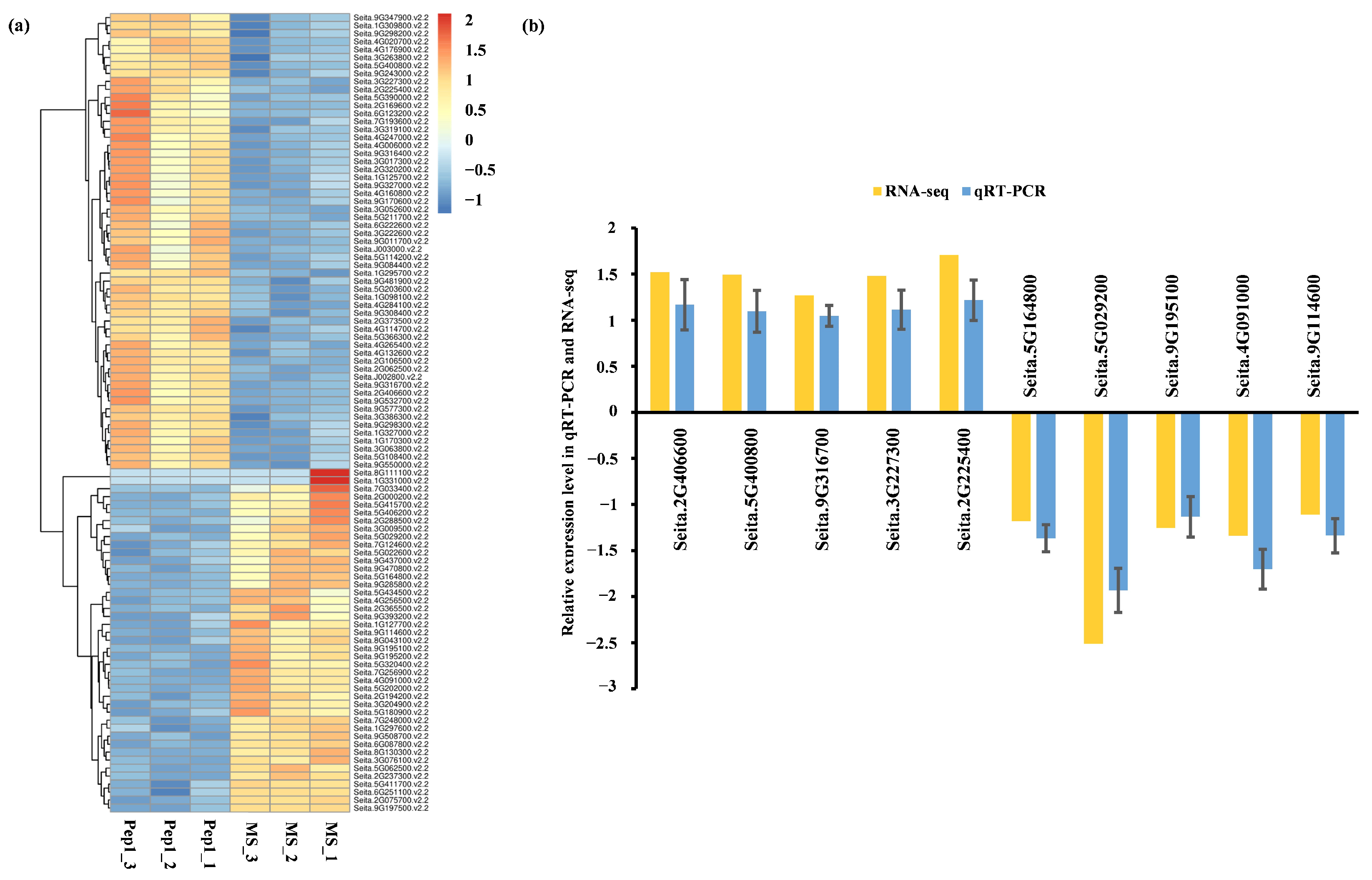
2.4. qRT-PCR Validation and Functional Analysis of Pep1-Responsive DEGs
2.4.1. qRT-PCR Validation
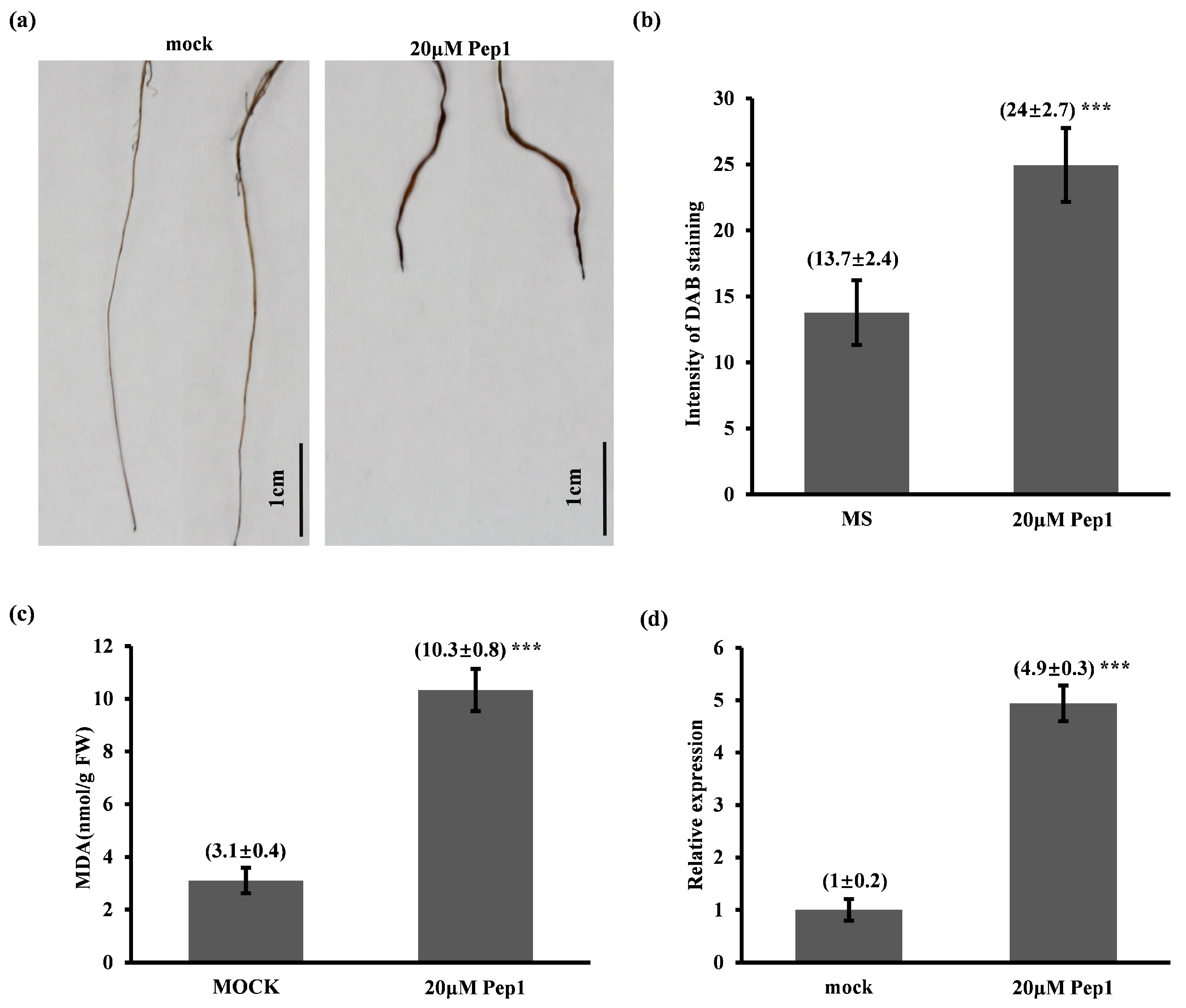
2.4.2. Pep1 Activates ROS-Mediated Immune Response and Pathogenesis-Related Protein 1 (PR1) Expression in Foxtail Millet Seedlings
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA-Seq Analysis
4.3. GO and KEGG Analysis
4.4. Validation of the DEGs by Quantitative Real-Time PCR
4.5. DAB Staining and Malondialdehyde (MDA) Content
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matsubayashi, Y.; Sakagami, Y. Peptide Hormones in Plants. Annu. Rev. Plant Biol. 2006, 57, 649–674. [Google Scholar] [CrossRef] [PubMed]
- Motomitsu, A.; Sawa, S.; Ishida, T. Plant Peptide Hormone Signalling. Essays Biochem. 2015, 58, 115–131. [Google Scholar] [PubMed]
- Matsubayashi, Y. Posttranslationally Modified Small-Peptide Signals in Plants. Annu. Rev. Plant Biol. 2014, 65, 385–413. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of Pattern Recognition Receptor Signalling in Plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Huffaker, A.; Bryan, A.C.; Tax, F.E.; Ryan, C.A. PEPR2 Is a Second Receptor for the Pep1 and Pep2 Peptides and Contributes to Defense Responses in Arabidopsis. Plant Cell 2010, 22, 508–522. [Google Scholar] [CrossRef]
- Huffaker, A.; Dafoe, N.J.; Schmelz, E.A. ZmPep1, an Ortholog of Arabidopsis Elicitor Peptide 1, Regulates Maize Innate Immunity and Enhances Disease Resistance1. Plant Physiol. 2011, 155, 1325–1338. [Google Scholar] [CrossRef]
- Bartels, S.; Boller, T. Quo Vadis, Pep? Plant Elicitor Peptides at the Crossroads of Immunity, Stress, and Development. J. Exp. Bot. 2015, 66, 5183–5193. [Google Scholar] [CrossRef]
- Huffaker, A.; Pearce, G.; Veyrat, N.; Erb, M.; Turlings, T.C.J.; Sartor, R.; Shen, Z.; Briggs, S.P.; Vaughan, M.M.; Alborn, H.T.; et al. Plant Elicitor Peptides Are Conserved Signals Regulating Direct and Indirect Antiherbivore Defense. Proc. Natl. Acad. Sci. USA 2013, 110, 5707–5712. [Google Scholar] [CrossRef]
- Tintor, N.; Ross, A.; Kanehara, K.; Yamada, K.; Fan, L.; Kemmerling, B.; Nürnberger, T.; Tsuda, K.; Saijo, Y. Layered Pattern Receptor Signaling via Ethylene and Endogenous Elicitor Peptides during Arabidopsis Immunity to Bacterial Infection. Proc. Natl. Acad. Sci. USA 2013, 110, 6211–6216. [Google Scholar] [CrossRef]
- Klauser, D.; Desurmont, G.A.; Glauser, G.; Vallat, A.; Flury, P.; Boller, T.; Turlings, T.C.J.; Bartels, S. The Arabidopsis Pep-PEPR System Is Induced by Herbivore Feeding and Contributes to JA-Mediated Plant Defence against Herbivory. J. Exp. Bot. 2015, 66, 5327–5336. [Google Scholar] [CrossRef]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious Roots and Lateral Roots: Similarities and Differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liao, H. Engineering Crop Nutrient Efficiency for Sustainable Agriculture. J. Integr. Plant Biol. 2017, 59, 710–735. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Xiang, D.; Zhu, J.; Li, Y.; Mao, C. Molecular Mechanisms of Root Development in Rice. Rice 2019, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Orman-Ligeza, B.; Parizot, B.; Gantet, P.P.; Beeckman, T.; Bennett, M.J.; Draye, X. Post-Embryonic Root Organogenesis in Cereals: Branching out from Model Plants. Trends Plant Sci. 2013, 18, 459–467. [Google Scholar] [CrossRef]
- de Dorlodot, S.; Forster, B.; Pagès, L.; Price, A.; Tuberosa, R.; Draye, X. Root System Architecture: Opportunities and Constraints for Genetic Improvement of Crops. Trends Plant Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef]
- Jing, Y.; Zheng, X.; Zhang, D.; Shen, N.; Wang, Y.; Yang, L.; Fu, A.; Shi, J.; Zhao, F.; Lan, W.; et al. Danger-Associated Peptides Interact with PIN-Dependent Local Auxin Distribution to Inhibit Root Growth in Arabidopsis. Plant Cell 2019, 31, 1767–1787. [Google Scholar] [CrossRef]
- Xiang, D.; Meng, F.; Wang, A.; Wu, Y.; Wang, Z.; Zheng, S.; Mao, C. Root-Secreted Peptide OsPEP1 Regulates Primary Root Elongation in Rice. Plant J. 2021, 107, 480–492. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, H.; Li, X.; Shen, H.; Gao, J.; Hou, S.; Zhang, B.; Mayes, S.; Bennett, M.; Ma, J.; et al. A Mini Foxtail Millet with an Arabidopsis-like Life Cycle as a C4 Model System. Nat. Plants 2020, 6, 1167–1178. [Google Scholar] [CrossRef]
- Tang, S.; Li, L.; Wang, Y.; Chen, Q.; Zhang, W.; Jia, G.; Zhi, H.; Zhao, B.; Diao, X. Genotype-Specific Physiological and Transcriptomic Responses to Drought Stress in Setaria Italica (an Emerging Model for Panicoideae Grasses). Sci. Rep. 2017, 7, 10009. [Google Scholar] [CrossRef]
- Jia, X.; Gao, H.; Zhang, L.; Tang, W.; Wei, G.; Sun, J.; Xiong, W. Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis. Biomolecules 2024, 14, 958. [Google Scholar] [CrossRef]
- Yu, Y.; Guo, D.-D.; Min, D.-H.; Cao, T.; Ning, L.; Jiang, Q.-Y.; Sun, X.-J.; Zhang, H.; Tang, W.-S.; Gao, S.-Q.; et al. Foxtail Millet MYB-like Transcription Factor SiMYB16 Confers Salt Tolerance in Transgenic Rice by Regulating Phenylpropane Pathway. Plant Physiol. Biochem. 2023, 195, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Khan, N.U.; Dai, S.; Qin, N.; Han, Z.; Guo, B.; Li, J. Transcriptome Analysis and Identification of the Low Potassium Stress-Responsive Gene SiSnRK2.6 in Foxtail Millet (Setaria italica L.). Theor. Appl. Genet. 2024, 137, 22. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Felix, G.; Boller, T. A Single Locus Determines Sensitivity to Bacterial Flagellin in Arabidopsis Thaliana. Plant J. 1999, 18, 277–284. [Google Scholar] [CrossRef]
- Dhar, S.; Kim, H.; Segonzac, C.; Lee, J.-Y. The Danger-Associated Peptide PEP1 Directs Cellular Reprogramming in the Arabidopsis Root Vascular System. Mol. Cells 2021, 44, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, L.; Li, Y.; Hou, J.; Huang, J.; Liang, W. Involvement of ABA- and H2O2-Dependent Cytosolic Glucose-6-Phosphate Dehydrogenase in Maintaining Redox Homeostasis in Soybean Roots under Drought Stress. Plant Physiol. Biochem. 2016, 107, 126–136. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Bauer, Z.; Boller, T. Both the Extracellular Leucine-Rich Repeat Domain and the Kinase Activity of FLS2 Are Required for Flagellin Binding and Signaling in Arabidopsis. Plant Cell 2001, 13, 1155–1164. [Google Scholar] [CrossRef]
- Albert, M.; Jehle, A.K.; Mueller, K.; Eisele, C.; Lipschis, M.; Felix, G. Arabidopsis Thaliana Pattern Recognition Receptors for Bacterial Elongation Factor Tu and Flagellin Can Be Combined to Form Functional Chimeric Receptors. J. Biol. Chem. 2010, 285, 19035–19042. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Morea, F.A.; Savatin, D.V.; Dejonghe, W.; Kumar, R.; Luo, Y.; Adamowski, M.; Van den Begin, J.; Dressano, K.; Pereira de Oliveira, G.; Zhao, X.; et al. Danger-Associated Peptide Signaling in Arabidopsis Requires Clathrin. Proc. Natl. Acad. Sci. USA 2016, 113, 11028–11033. [Google Scholar] [CrossRef]
- Araya, T.; von Wirén, N.; Takahashi, H. CLE Peptide Signaling and Nitrogen Interactions in Plant Root Development. Plant Mol. Biol. 2016, 91, 607–615. [Google Scholar] [CrossRef]
- Shen, N.; Jiang, C.; Jiang, A. Arabidopsis Plasma Membrane H+-ATPase Interacts with Auxin to Regulate Danger-Associated Peptide Pep1-Induced Root Growth Inhibition. Biochem. Biophys. Res. Commun. 2024, 696, 149507. [Google Scholar] [CrossRef]
- Huffaker, A.; Pearce, G.; Ryan, C.A. An Endogenous Peptide Signal in Arabidopsis Activates Components of the Innate Immune Response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.; Paiva, N. Stress-Induced Phenylpropanoid Metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Ranf, S.; Eschen-Lippold, L.; Pecher, P.; Lee, J.; Scheel, D. Interplay between Calcium Signalling and Early Signalling Elements during Defence Responses to Microbe- or Damage-Associated Molecular Patterns. Plant J. 2011, 68, 100–113. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone Crosstalk in Plant Disease and Defense: More Than Just JASMONATE-SALICYLATE Antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.D. Primary Metabolism and Plant Defense—Fuel for the Fire. Mol. Plant-Microbe Interact. J. 2009, 22, 487–497. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP Transcription Factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP Transcription Factor Family—An Update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.F.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.G.; et al. Reactive Oxygen Species Produced by NADPH Oxidase Regulate Plant Cell Growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Tsukagoshi, H. Control of Root Growth and Development by Reactive Oxygen Species. Curr. Opin. Plant Biol. 2016, 29, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N Terminus of Bacterial Elongation Factor Tu Elicits Innate Immunity in Arabidopsis Plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Fernando, R.L.; Nettleton, D.; Southey, B.R.; Dekkers, J.C.M.; Rothschild, M.F.; Soller, M. Controlling the Proportion of False Positives in Multiple Dependent Tests. Genetics 2004, 166, 611–619. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, H.; Xie, X.; Fu, Q.; Zheng, S.; Liu, X.; Zhu, S. Transcriptome Analysis of DAMP-Induced Root Growth Regulation and Defense in Foxtail Millet. Int. J. Mol. Sci. 2025, 26, 5175. https://doi.org/10.3390/ijms26115175
Ye H, Xie X, Fu Q, Zheng S, Liu X, Zhu S. Transcriptome Analysis of DAMP-Induced Root Growth Regulation and Defense in Foxtail Millet. International Journal of Molecular Sciences. 2025; 26(11):5175. https://doi.org/10.3390/ijms26115175
Chicago/Turabian StyleYe, Hao, Xinyu Xie, Qiongfang Fu, Sheng Zheng, Xunyan Liu, and Shan Zhu. 2025. "Transcriptome Analysis of DAMP-Induced Root Growth Regulation and Defense in Foxtail Millet" International Journal of Molecular Sciences 26, no. 11: 5175. https://doi.org/10.3390/ijms26115175
APA StyleYe, H., Xie, X., Fu, Q., Zheng, S., Liu, X., & Zhu, S. (2025). Transcriptome Analysis of DAMP-Induced Root Growth Regulation and Defense in Foxtail Millet. International Journal of Molecular Sciences, 26(11), 5175. https://doi.org/10.3390/ijms26115175




