Validation of Targeted Relationships of Novel circRNA803/lncRNA MSTRG.19726–oar-let-7a–CPEB1 ceRNA Networks, Key to Follicle Development in Single-Litter and Multi-Litter Sheep Based on Whole-Transcriptome Sequencing
Abstract
1. Introduction
2. Results
2.1. Quality Control and Comparative Analysis to Reference Genome
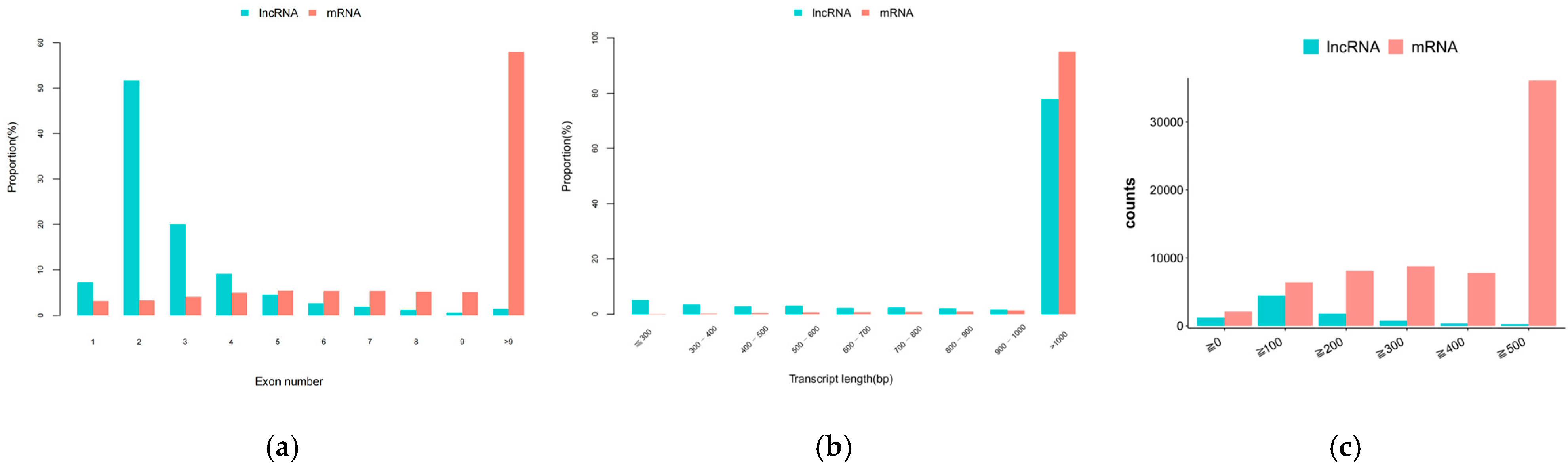
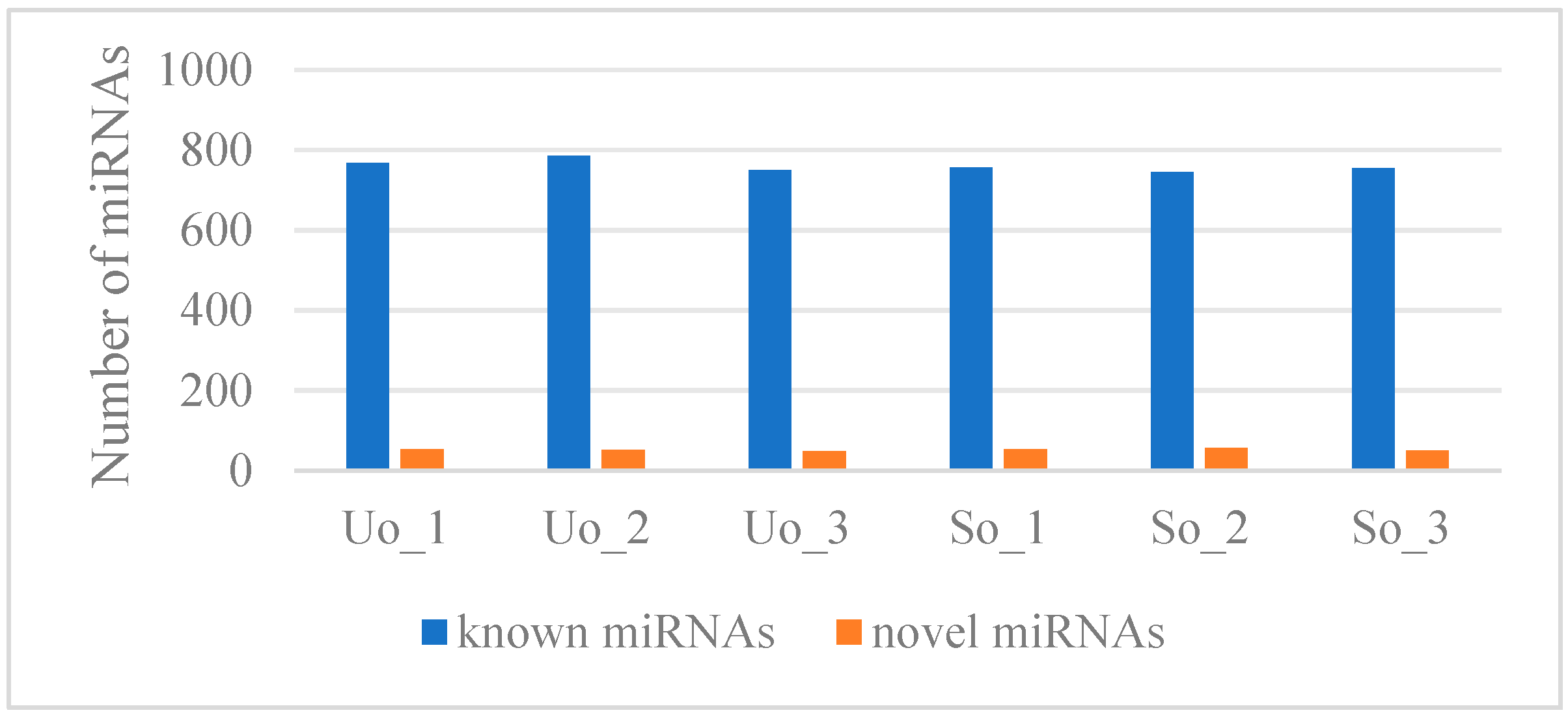
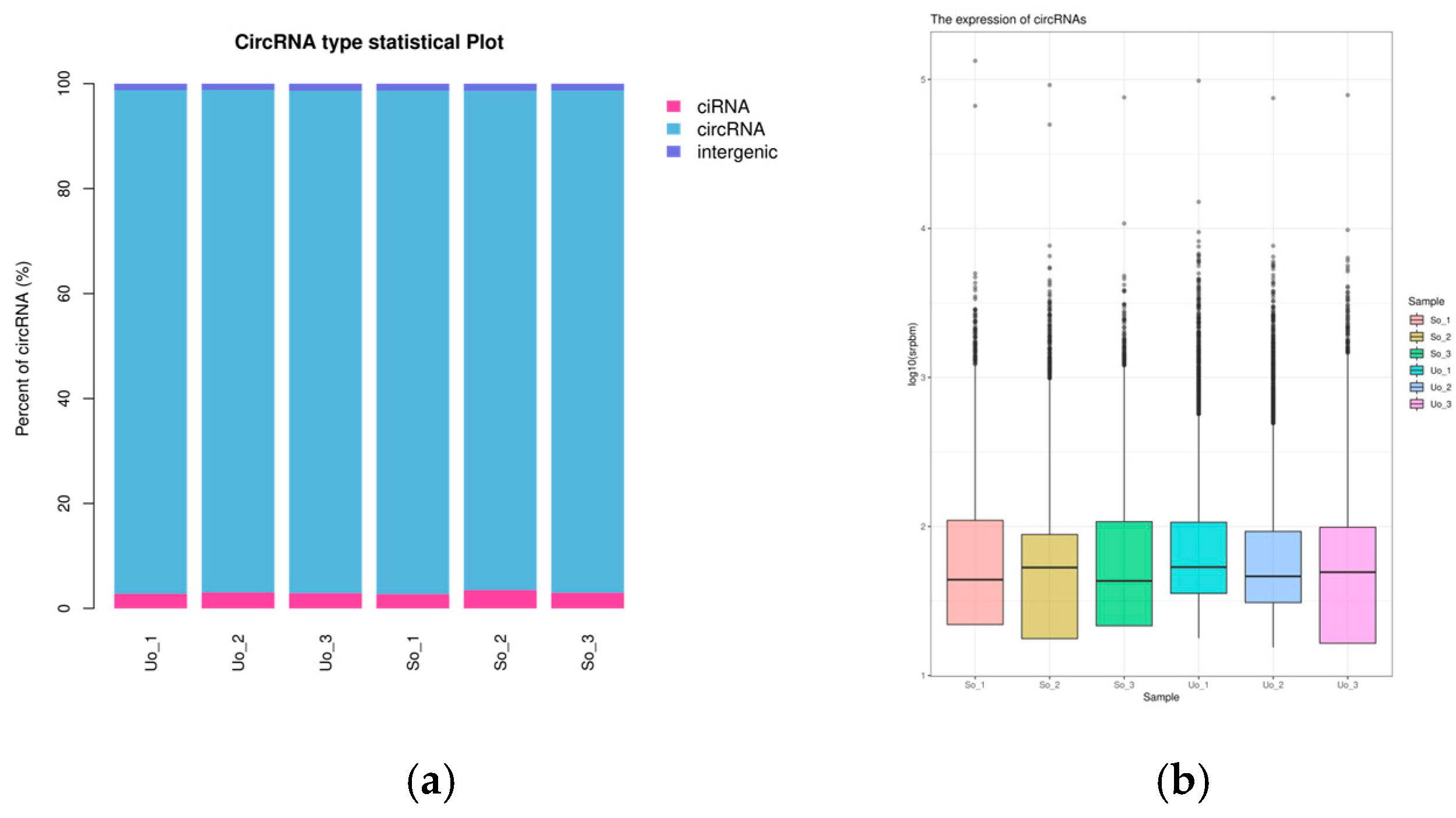
2.2. Screening and Identification of lncRNAs, circRNAs, and miRNAs
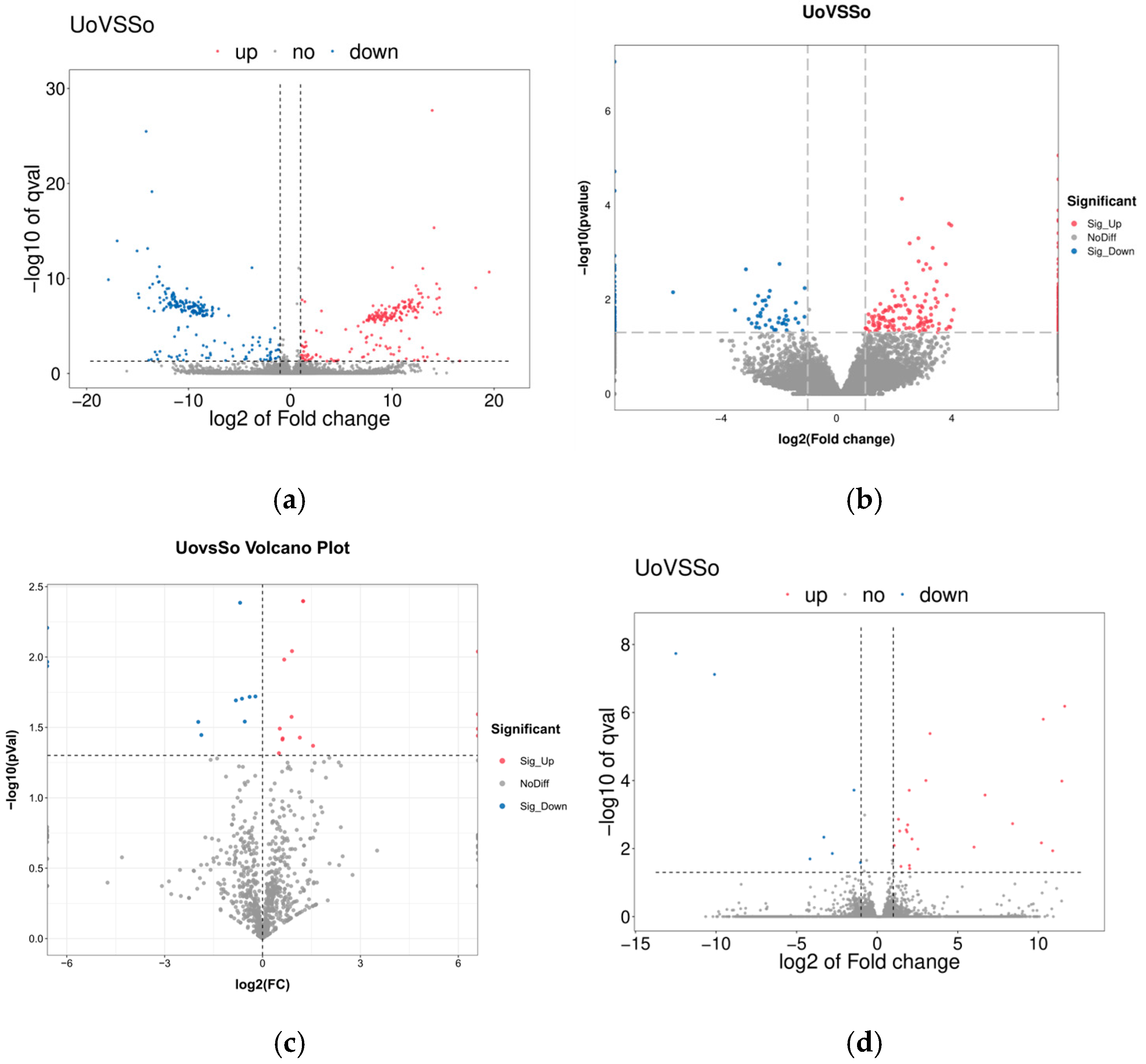
2.3. Differential Expression Analysis and Target Gene Prediction
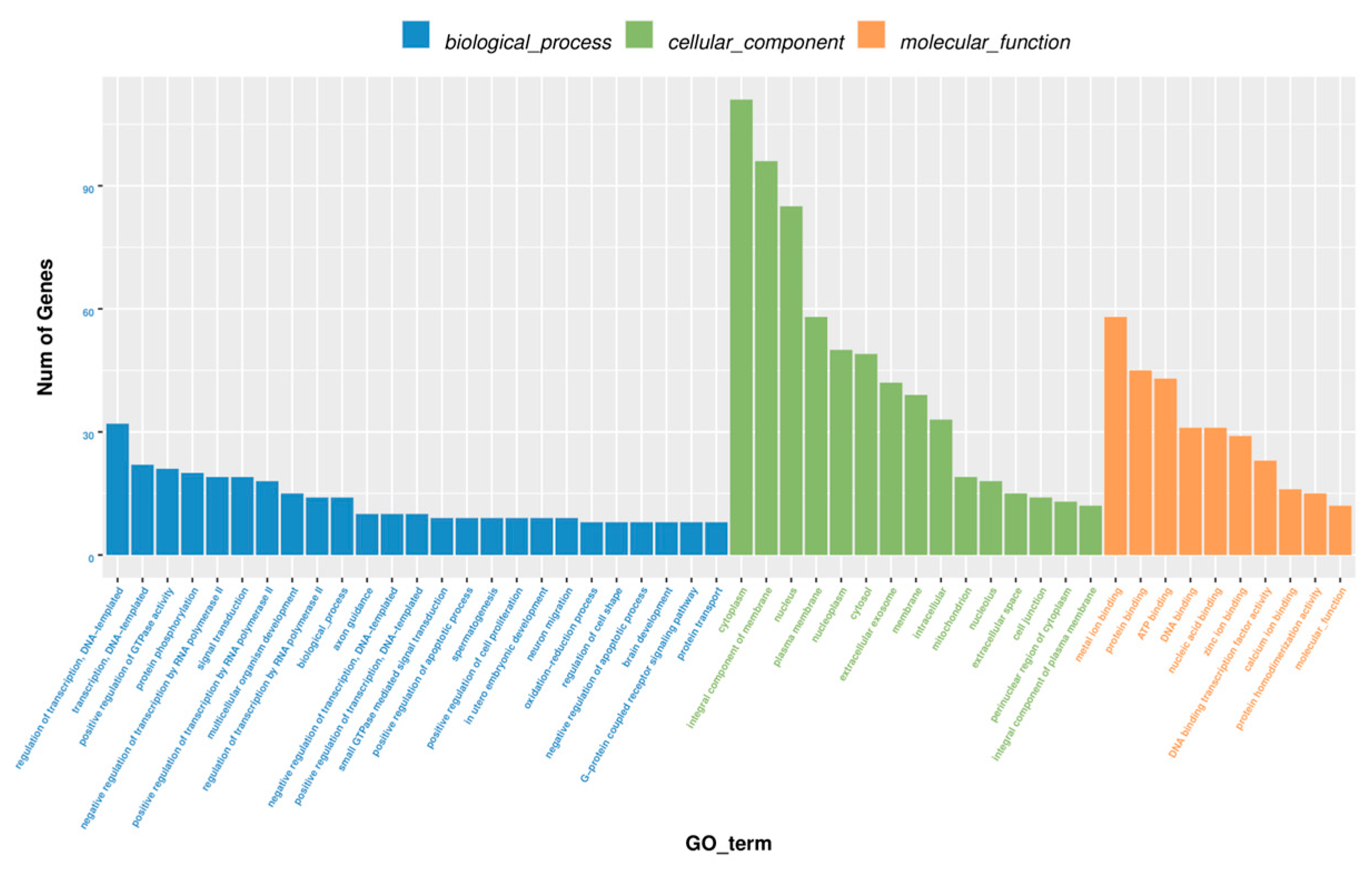
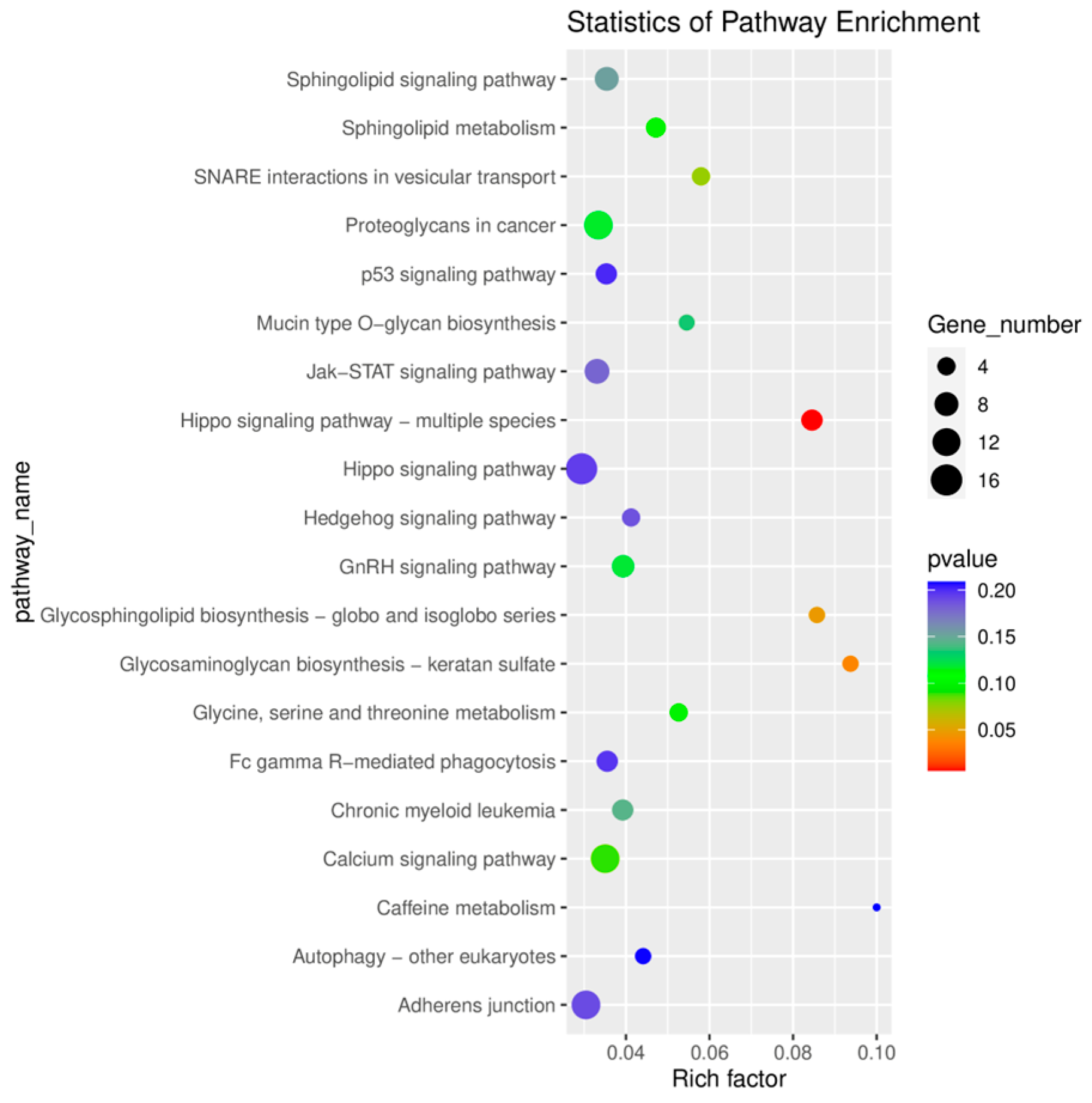
2.4. GO and KEGG Enrichment Analysis
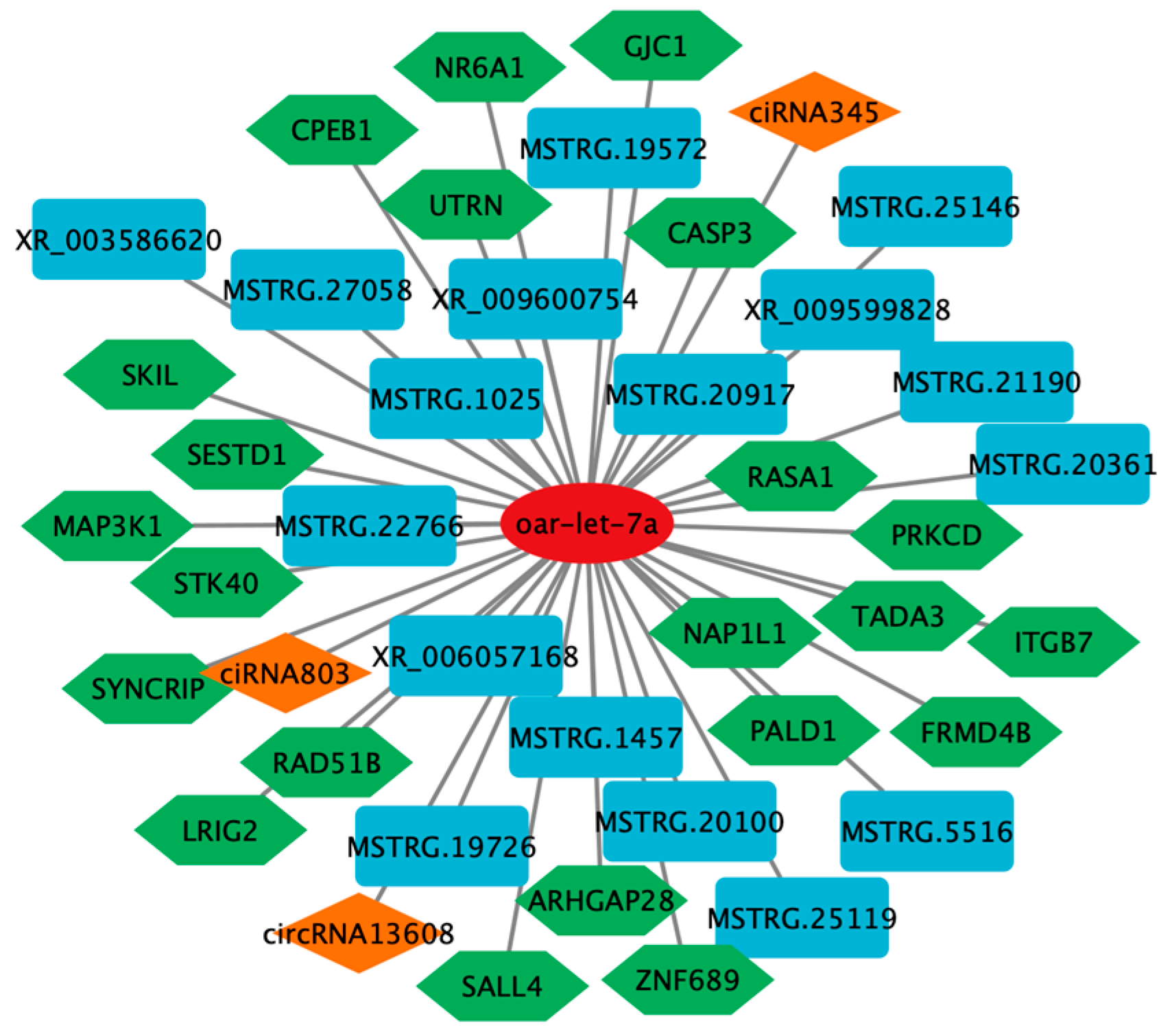
2.5. ceRNA Network Analysis
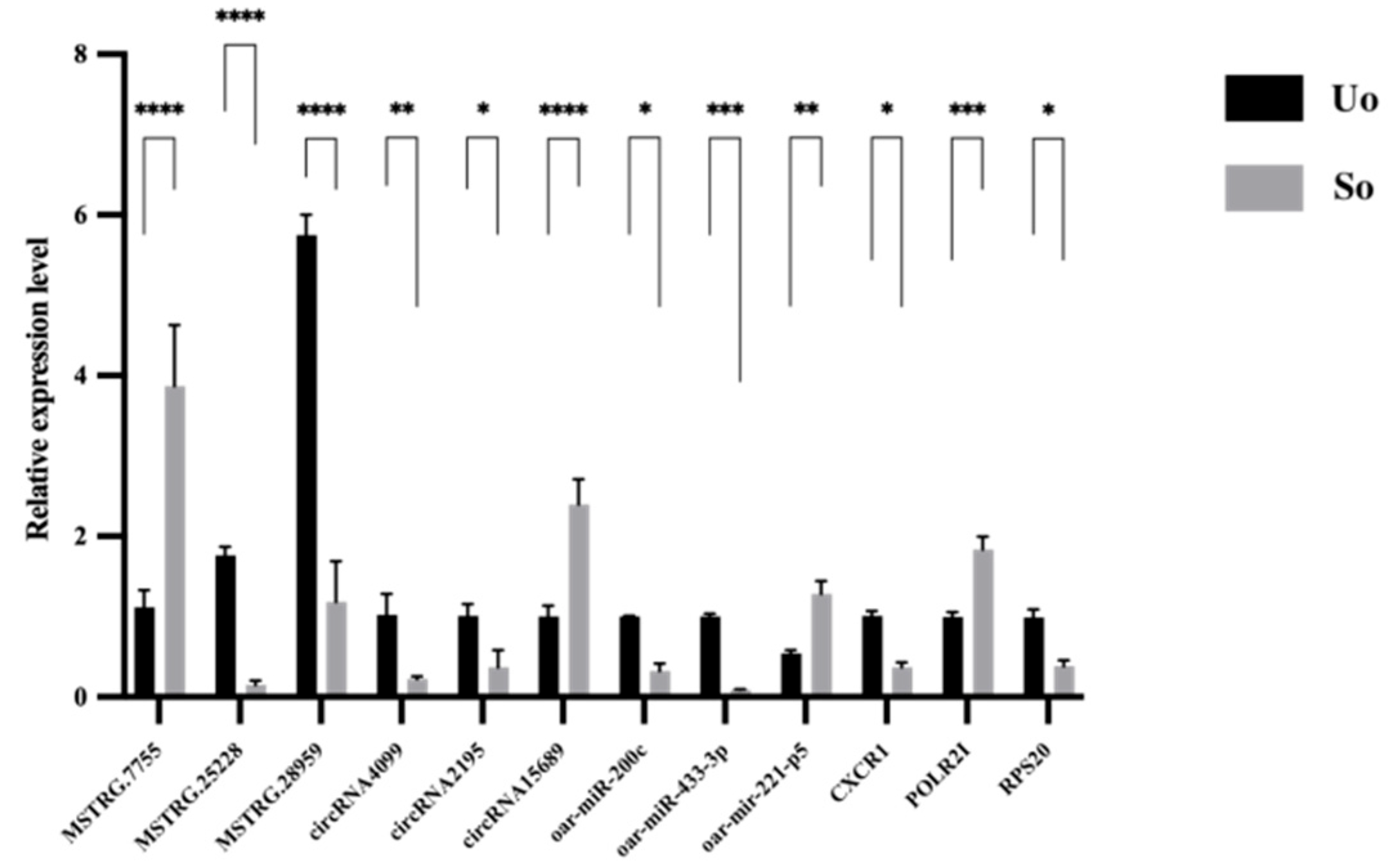
2.6. qRT-PCR Validation of Sequencing Data
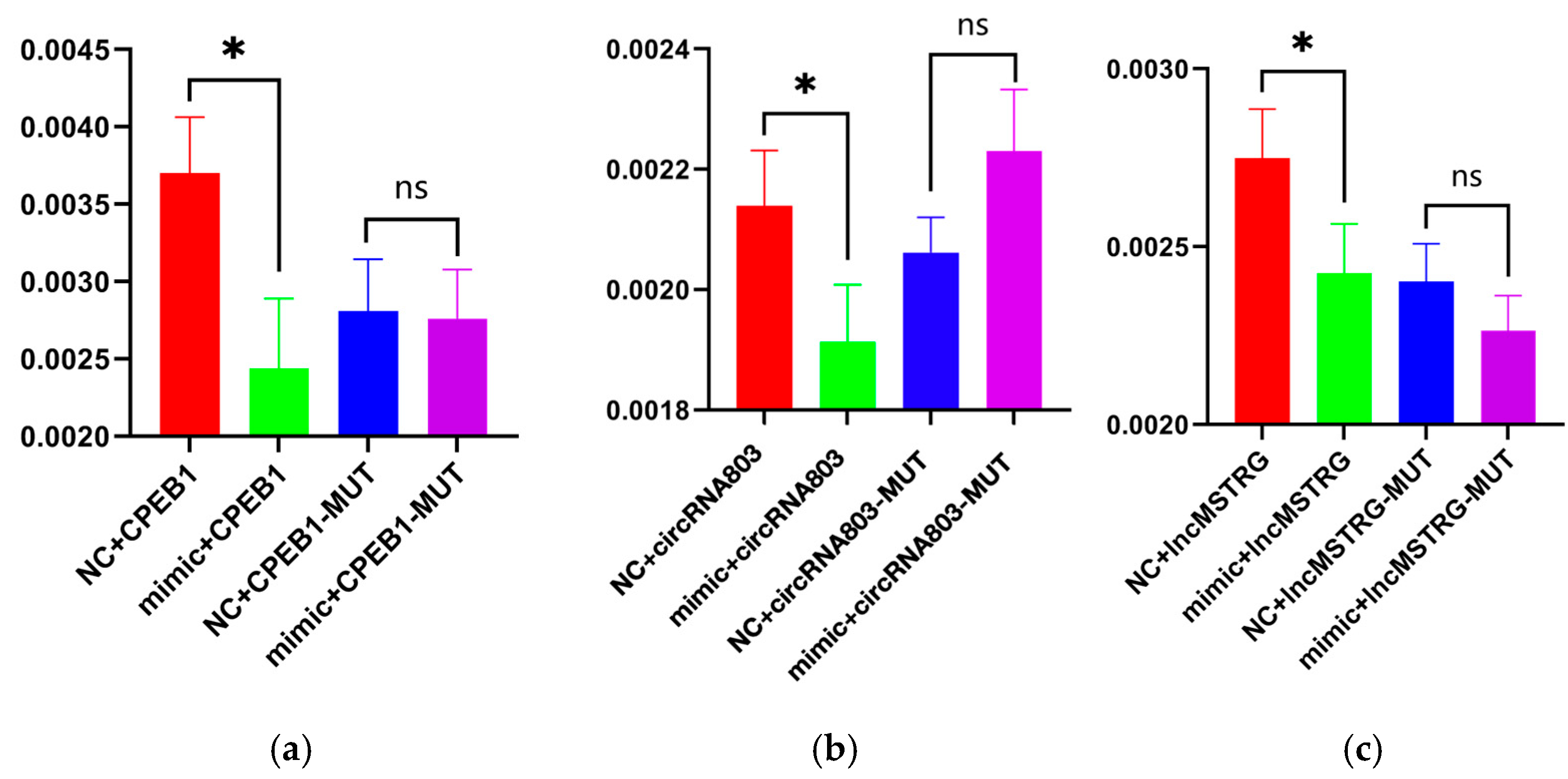
2.7. Double Luciferase Reporter Gene Assay
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Total RNA Extraction and Library Construction
4.3. Quality Control and Reference Genome Alignment
4.4. Differential Expression Analysis and Target Gene Prediction
4.5. ceRNA Network Construction and Bioinformatics Analysis
4.6. RNA-Seq Data Validation Using qRT-PCR
4.7. Cell Culture
4.8. Cell Transfection
- NC: CPEB1 + NC-mimic, circRNA803 + NC-mimic, and lncRNA MSTRG.19726 + NC-mimic;
- Mimic: CPEB1 + mimic, circRNA803 + mimic, and lncRNA MSTRG.19726 + mimic;
- NC + MUT: CPEB1-MUT + NC-mimic, circRNA803-MUT + NC-mimic, and
- lncRNA MSTRG.19726-MUT + NC-mimic;
- Mimic + MUT: CPEB1-MUT + mimic, circRNA803-MUT + mimic, and lncRNA MSTRG.19726-MUT + mimic.
4.9. Double Luciferase Reporter Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DE | differentially expressed |
| ncRNA | non-coding RNA |
| ceRNA | competing endogenous RNA |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Ma, N.; Tie, C.; Yu, B.; Zhang, W.; Wan, J. Identifying lncRNA–miRNA–mRNA networks to investigate Alzheimer’s disease pathogenesis and therapy strategy. Aging 2020, 12, 2897–2920. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Jiang, C.; Zhang, H.; Shi, X.; Ai, X.; Li, R.; Dong, J.; Wang, J.; Zhao, X.; Yu, H. LncRNA-mediated ceRNA networks provide novel potential biomarkers for peanut drought tolerance. Physiol. Plant. 2022, 174, e13610. [Google Scholar] [CrossRef]
- Yang, T.; Qiu, L.; Jiang, Y.; Bai, H.; Bi, Y.; Wang, Z.; Chen, G.; Chang, G. Identification, biogenesis, and function prediction of a novel circRNA_3238 of chicken. Anim. Biotechnol. 2023, 34, 2527–2536. [Google Scholar] [CrossRef] [PubMed]
- Niu, X. Analysis of Ovarian lncRNA Expression and Regulatory Network in Perfumed Pigs During Estrus. Doctoral Thesis, Guizhou University, Guiyang, China, 2023. [Google Scholar]
- Meng, C.; Wang, Y.; Yao, Y.; Xi, G.; Niu, J.; Solangsizhu; Guo, M.; Xu, Y. lncRNA-MSTRG.7889.1 Competitively Binds to bta-miR-146a Targeting Smad4 to Regulate Apoptosis of Yak Granulosa Cells. Sci. Agric. Sin. 2024, 57, 797–809. [Google Scholar]
- Sadeghi, M.; Bahrami, A.; Hasankhani, A.; Kioumarsi, H.; Nouralizadeh, R.; Abdulkareem, S.A.; Ghafouri, F.; Barkema, H.W. lncRNA-miRNA-mRNA ceRNA Network Involved in Sheep Prolificacy: An Integrated Approach. Genes 2022, 13, 1295. [Google Scholar] [CrossRef]
- Zhao, L.; Pan, Y.; Wang, M.; Wang, J.; Wang, Y.; Han, X.; Wang, J.; Zhang, T.; Zhao, T.; He, H.; et al. Integrated analysis of the expression profiles of the lncRNA-miRNA-mRNA ceRNA network in granulosa and cumulus cells from yak ovaries. BMC Genom. 2022, 23, 633. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, Y.; Yuan, Z.; Wu, Y.; Zhao, Z.; Wu, C.; Hou, J.; Zhang, M. Genome-Wide Identification of mRNAs, lncRNAs, and Proteins, and Their Relationship With Sheep Fecundity. Front. Genet. 2021, 12, 750947. [Google Scholar] [CrossRef]
- Yang, F.; Yao, X.; Li, K.; Li, X.; Tang, L.; You, P.; Cai, Q.; Wang, F. Differential expression analysis of lncRNA and mRNA in ovaries of different sheep breeds during estrus. J. Nanjing Agric. Univ. 2023, 46, 1142–1152. [Google Scholar]
- Liu, A.; Chen, X.; Liu, M.; Zhang, L.; Ma, X.; Tian, S. Differential Expression and Functional Analysis of circRNA in the Ovaries of Low and High Fecundity Hanper Sheep. Animals 2021, 11, 1863. [Google Scholar] [CrossRef]
- Ma, S. Association Between Novel Ploymorphisms in LEPR Gene and Litter Size in Mongolia and Ujimoin Sheep Breeds. Master’s Thesis, Inner Mongolia Agricultural University, Huhhot, China, 2022. [Google Scholar]
- Zhu, L.; Li, N.; Sun, L.; Zheng, D.; Shao, G. Non-coding RNAs: The key detectors and regulators in cardiovascular disease. Genomics 2021, 113 Pt 2, 1233–1246. [Google Scholar] [CrossRef]
- Yang, D.; Wang, Y.; Zheng, Y.; Dai, F.; Liu, S.; Yuan, M.; Deng, Z.; Bao, A.; Cheng, Y. Silencing of lncRNA UCA1 inhibited the pathological progression in PCOS mice through the regulation of PI3K/AKT signaling pathway. J. Ovarian Res. 2021, 14, 48. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Yu, H.; Luo, G.; Wang, M.; Zhou, C.; Zhang, L.; Hou, B.; Zhang, C.; Liu, M.; Xu, D. The Interaction of lncRNA XLOC-2222497, AKR1C1, and Progesterone in Porcine Endometrium and Pregnancy. Int. J. Mol. Sci. 2020, 21, 3232. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, Y.; Xu, X.; Xiang, H.; Zhang, Z.; Chen, B.; Hao, Y.; Wei, Z.; Zhou, P.; Chen, D. Increased New lncRNA–mRNA Gene Pair Levels in Human Cumulus Cells Correlate With Oocyte Maturation and Embryo Development. Reprod. Sci. 2015, 22, 1008–1014. [Google Scholar] [CrossRef]
- Yerushalmi, G.M.; Salmon-Divon, M.; Yung, Y.; Maman, E.; Kedem, A.; Ophir, L.; Elemento, O.; Coticchio, G.; Dal Canto, M.; Mignini Renzinu, M.; et al. Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol. Hum. Reprod. 2014, 20, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.L. Construction of the ceRNA Network of Bovine circRNA and the Functional Mechanism of Regulating the Development of Bovine Skeletal Muscle Cells. Ph.D. Thesis, Northwest A&F University, Xianyang, China, 2021. [Google Scholar]
- Liu, M.; Du, S.; Yang, J.; Huang, X.; Guo, Y.; Li, Y.; Guo, K.; Cao, S. Mechanism and Research Progress of lncRNAs Copetitive Endogenous Molecules. China Anim. Husb. Vet. Med. 2021, 48, 3604–3613. [Google Scholar]
- Duan, B.; Xu, X. How to Break a Fever: A Feedback Circuit for Body Temperature Control. Neuron 2019, 103, 179–181. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Chu, M. Research Progress of Non-coding RNA in Animal Reproduction. China Herbiv. Sci. 2023, 43, 39–44. [Google Scholar]
- Plewes, M.R.; Hou, X.; Zhang, P.; Liang, A.; Hua, G.; Wood, J.R.; Cupp, A.S.; Lv, X.; Wang, C.; Davis, J.S. Yes-associated protein 1 is required for proliferation and function of bovine granulosa cells in vitro†. Biol. Reprod 2019, 101, 1001–1017. [Google Scholar] [CrossRef]
- Tay, J.; Richter, D.J. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell 2001, 1, 201–213. [Google Scholar] [CrossRef]
- Racki, W.J.; Richter, J.D. CPEB controls oocyte growth and follicle development in the mouse. Development 2006, 133, 4527–4537. [Google Scholar] [CrossRef]
- Ke, H.; Tang, S.; Guo, T.; Hou, D.; Jiao, X.; Li, S.; Luo, W.; Xu, B.; Zhao, S.; Li, G.; et al. Landscape of pathogenic mutations in premature ovarian insufficiency. Nat. Med. 2023, 29, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Franciosi, F.; Daldello, E.M.; Luong, X.G.; Althoff, P.; Wang, X.; Conti, M. CPEB1-dependent disruption of the mRNA translation program in oocytes during maternal aging. Nat. Commun. 2023, 14, 416. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Tsukahara, T.; Inoue, K. Localization of c-mos mRNA around the animal pole in the zebrafish oocyte with Zor-1/Zorba. Biosci. Trends 2009, 3, 96–104. [Google Scholar] [PubMed]
- Zhang, Y.; Sheets, M.D. Analyses of zebrafish and Xenopus oocyte maturation reveal conserved and diverged features of translational regulation of maternal cyclin B1 mRNA. BMC Dev. Biol. 2009, 9, 7. [Google Scholar] [CrossRef]
- Nishimura, Y.; Kano, K.; Naito, K. Porcine CPEB1 is involved in Cyclin B translation and meiotic resumption in porcine oocytes. Anim Sci J 2010, 81, 444–452. [Google Scholar] [CrossRef]
- Yao, X.; Li, P.; Jiang, X.; Meng, J.; Huang, Y.; Liu, Y.; Chen, J.; Zhao, M.; Lv, L. Research of the Genes Expression Associated with Follicular Development in Bovine Dominant and Subordinate Follicle Granulose Cells. Acta Vet. Zootech. Sinica 2014, 45, 1957–1963. [Google Scholar]
- Zhang, H. Study on the Regulation Proliferation of CPEB1 Gene in Goat Ovarian Granulosa Cells. Master’s Thesis, Yangzhou University, Yangzhou, China, 2022. [Google Scholar]
- Zhang, J.; Wang, X.; Li, W.; He, J.; Bai, H.; Chen, J.; Ren, Q.; Wei, C.; Xing, B. Target Gene Prediction and Bioinformatics Analysis of ssc-let-7a. Acta Ecol. Anim. Domastici 2020, 41, 14–21. [Google Scholar]
- Zhao, Y. Effects of Bovine Follicular Fluid Low-Density Small Extracellular Vesicles-Derived let-7i on Apoptosis of Granulosa Cells. Master’s Thesis, Northwest A&F University, Xianyang, China, 2022. [Google Scholar]
- Liu, A. Differential Expression and Functional Prediction of RNA in Ovaries of Sheep with Low and High Litter Sizes. Ph.D. Thesis, Hebei Agricultural University, Baoding, China, 2021. [Google Scholar]
- Dai, T. Screening of Key miRNAs and Function of Let-7b Granulosa Cells in Kisspeptin-Treated Ovarian Granulosa Cells in Tan Sheep. Master’s Thesis, Ningxia University, Yinchuan, China, 2023. [Google Scholar]
- Marcel, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, R.; Zhang, Y.; Zhang, J.; Luo, Z.; Zhang, J.; Chen, L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [PubMed]
| Sample | Raw Data | Valid Data | Mapped Reads | Q20% | Q30% | GC Content% |
|---|---|---|---|---|---|---|
| Uo_1 | 92,231,148 | 89,013,622 | 83,650,569 (93.98%) | 99.81 | 98.31 | 44.50 |
| Uo_2 | 95,944,996 | 92,841,144 | 86,837,283 (93.53%) | 99.81 | 98.24 | 46.00 |
| Uo_3 | 89,602,454 | 86,517,238 | 81,071,259 (93.71%) | 99.79 | 98.16 | 46.00 |
| So_1 | 74,106,726 | 71,910,234 | 66,210,782 (92.07%) | 99.85 | 98.05 | 45.00 |
| So_2 | 90,402,358 | 87,502,414 | 81,890,808 (93.59%) | 99.82 | 97.99 | 44.50 |
| So_3 | 88,799,682 | 86,496,168 | 81,411,487 (94.12%) | 99.86 | 98.02 | 45.00 |
| Sample | Raw Data | Valid Data | Mapped Reads | Q20% | Q30% | GC Content% |
|---|---|---|---|---|---|---|
| Uo_1 | 10,900,124 | 6,614,309 | 6,564,040 (99.24%) | 99.26 | 97.16 | 51.08 |
| Uo_2 | 11,914,260 | 6,735,685 | 6,679,105 (99.16%) | 99.36 | 97.49 | 51.20 |
| Uo_3 | 11,923,394 | 7,321,518 | 7,265,874 (99.24%) | 99.33 | 97.42 | 51.10 |
| So_1 | 11,979,084 | 7,224,597 | 7,153,073 (99.01%) | 99.26 | 97.11 | 51.19 |
| So_2 | 11,528,577 | 6,679,309 | 6,629,214 (99.25%) | 99.29 | 97.16 | 51.05 |
| So_3 | 12,785,226 | 5,824,807 | 5,763,647 (98.95%) | 99.16 | 97.11 | 51.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, B.; Wang, A.; Liu, H.; Liu, X.; Jiang, H. Validation of Targeted Relationships of Novel circRNA803/lncRNA MSTRG.19726–oar-let-7a–CPEB1 ceRNA Networks, Key to Follicle Development in Single-Litter and Multi-Litter Sheep Based on Whole-Transcriptome Sequencing. Int. J. Mol. Sci. 2025, 26, 5161. https://doi.org/10.3390/ijms26115161
Gu B, Wang A, Liu H, Liu X, Jiang H. Validation of Targeted Relationships of Novel circRNA803/lncRNA MSTRG.19726–oar-let-7a–CPEB1 ceRNA Networks, Key to Follicle Development in Single-Litter and Multi-Litter Sheep Based on Whole-Transcriptome Sequencing. International Journal of Molecular Sciences. 2025; 26(11):5161. https://doi.org/10.3390/ijms26115161
Chicago/Turabian StyleGu, Bo, Anqi Wang, Hang Liu, Xudong Liu, and Huaizhi Jiang. 2025. "Validation of Targeted Relationships of Novel circRNA803/lncRNA MSTRG.19726–oar-let-7a–CPEB1 ceRNA Networks, Key to Follicle Development in Single-Litter and Multi-Litter Sheep Based on Whole-Transcriptome Sequencing" International Journal of Molecular Sciences 26, no. 11: 5161. https://doi.org/10.3390/ijms26115161
APA StyleGu, B., Wang, A., Liu, H., Liu, X., & Jiang, H. (2025). Validation of Targeted Relationships of Novel circRNA803/lncRNA MSTRG.19726–oar-let-7a–CPEB1 ceRNA Networks, Key to Follicle Development in Single-Litter and Multi-Litter Sheep Based on Whole-Transcriptome Sequencing. International Journal of Molecular Sciences, 26(11), 5161. https://doi.org/10.3390/ijms26115161





