hnRNPH1: A Multifaceted Regulator in RNA Processing and Disease Pathogenesis
Abstract
1. Introduction
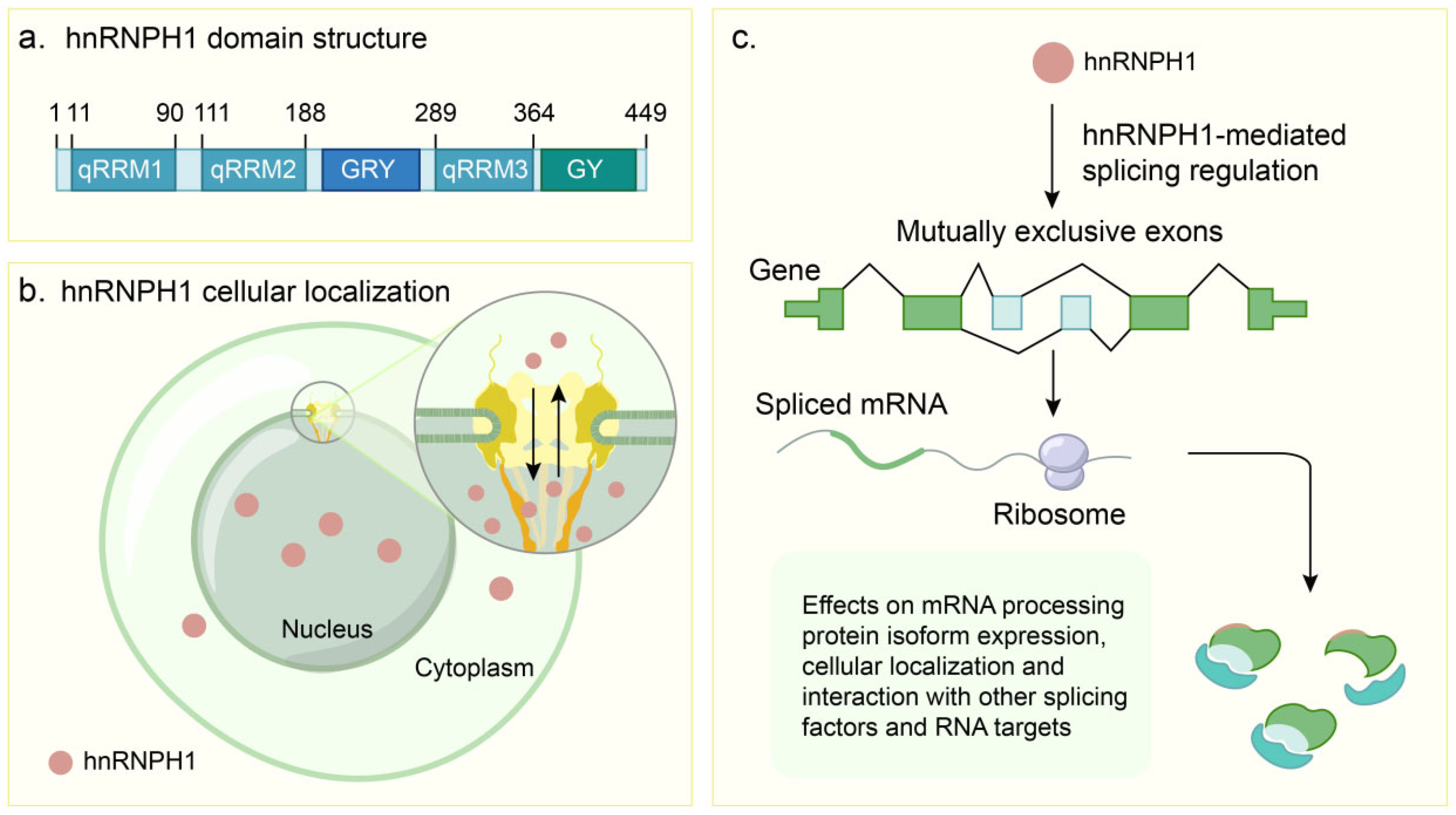
2. Structural Basis and Molecular Functions of hnRNPH1
2.1. Structural Architecture
| Domain | Position | Structural Hallmarks | Binding Specificity | Core Functions |
|---|---|---|---|---|
| qRRM1 | N-terminus (Approx. aa 11–90) | Classical RRM fold (β/α, β/β, α/β). Proline linker affects dynamics. | G-rich sequences, G-tracts, potential G4 structures. | RNA binding. Regulation of RNA processing, alternative splicing [28,31]. |
| qRRM2 | After qRRM1 (Approx. aa 111–188) | Classical RRM fold (β/α, β/β, α/β). Proline linker affects dynamics. Characteristic RGLP motif. | G-rich sequences, G-tracts, potential G4 structures. | RNA binding. Regulation of RNA processing, alternative splicing [27,38]. |
| GYR | Between qRRM2 and qRRM3 (Approx. aa 190–260 region). Overlapping NLS region. | Low-complexity domain (LCD). Gly/Tyr/Arg-rich. Forms hydrogel in vitro. Phase separation driver. | Interacts with other LCD proteins. | Drives phase separation. Protein–protein interactions. Essential for physiological splicing function [14,34]. |
| qRRM3 | After qRRM2 (Approx. aa 289–364) | Classical RRM fold (β/α, β/β, α/β). | G-rich sequences, G-tracts, potential G4 structures. | RNA binding. Regulation of RNA processing (splicing, 3′ end). Involved in cancer fusion proteins [14,27]. |
| NLS | Close to/overlapping GYR (Approx. aa 205–213) | Putative nuclear localization signal. Includes YDPP motif. | Recognized by karyopherin receptor complex (transportin). | Mediates nuclear import/shuttling. Mutations affect localization [35,36]. |
| GY | C-terminus (Approx. aa 340–449) | Low-complexity domain (LCD). Glycine-rich. Does not form hydrogel in vitro. | Undefined RNA binding. | Can activate transcription in assays. Component of cancer fusion proteins [14,34]. |
2.2. Core Molecular Mechanisms of hnRNPH1
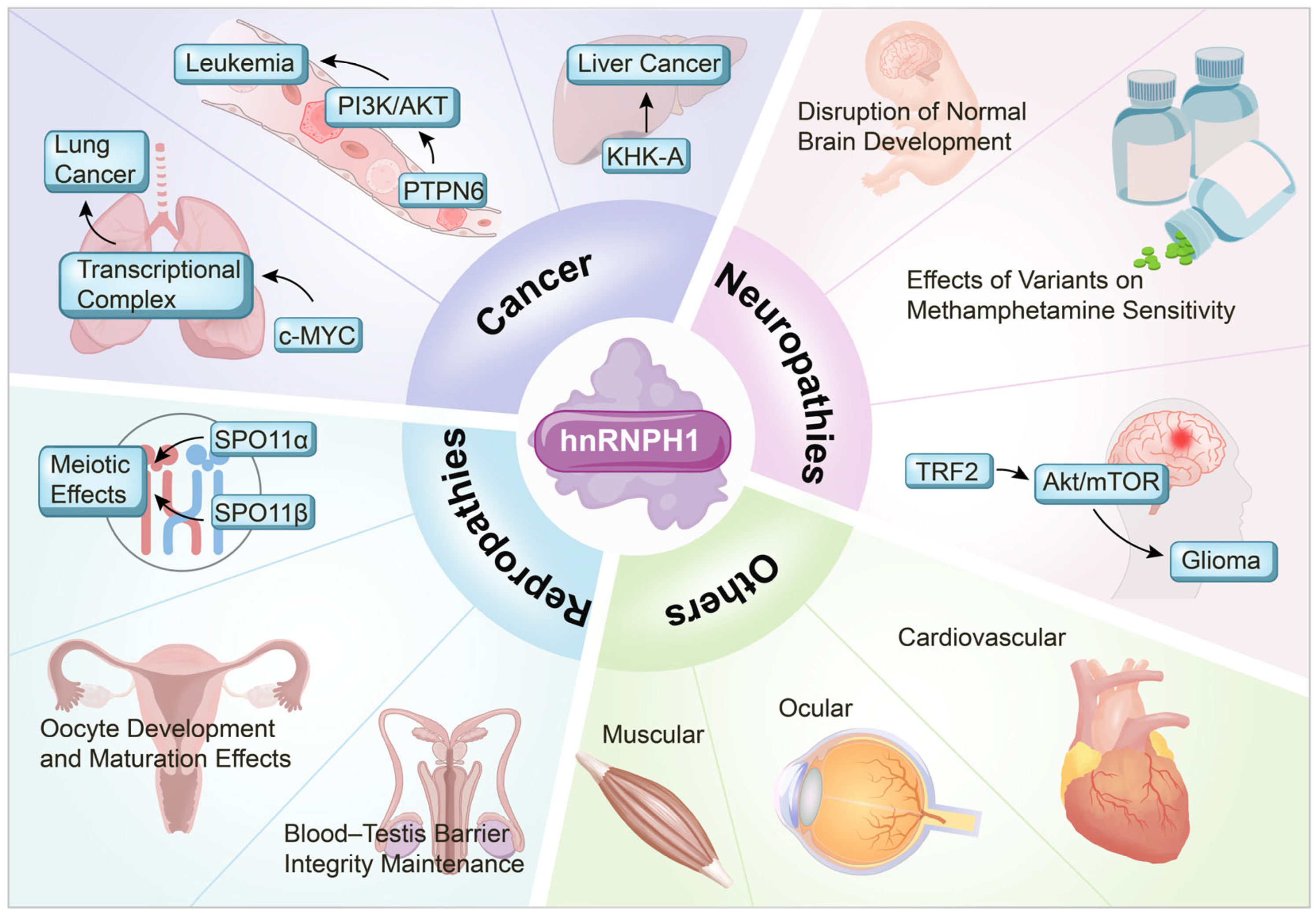
3. Role of hnRNPH1 in Disease Pathogenesis
3.1. Cancer
3.2. Neurological Disorders
3.3. Reproductive System Disorders
3.4. hnRNPH1 in Non-Cancer Pathologies
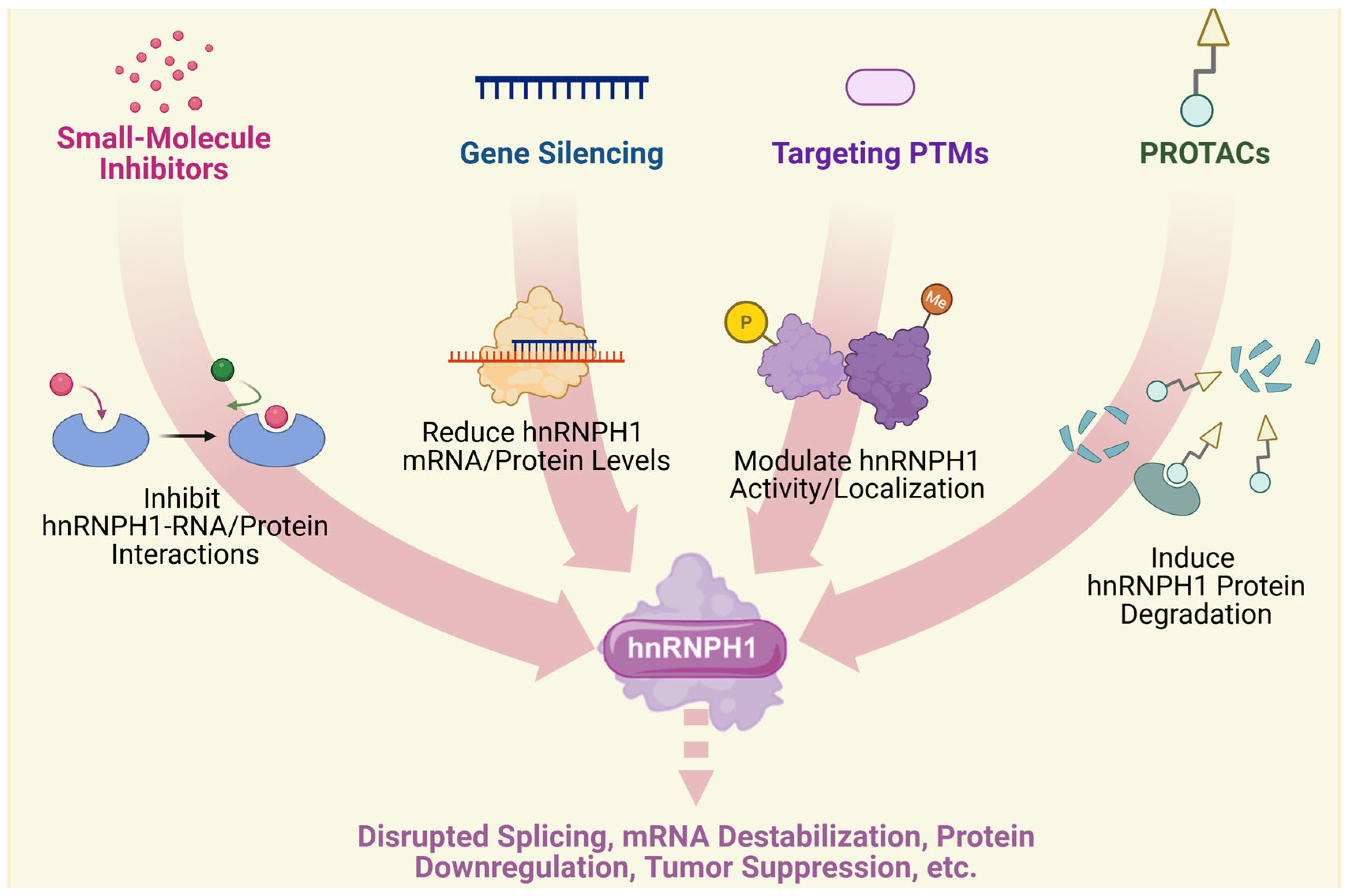
4. Therapeutic Implications
4.1. Current Therapeutic Strategies
4.2. Challenges and Optimizing Strategies
5. Prospects
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Full Name |
| AI | Artificial Intelligence |
| ALS/FTLD | Amyotrophic Lateral Sclerosis/Frontotemporal Lobar Degeneration |
| AR | Androgen Receptor |
| AS | Alternative Splicing |
| ASD | Autism Spectrum Disorder |
| ASO | Antisense Oligonucleotide |
| AUC | Area Under the Curve |
| BBB | Blood–Brain Barrier |
| BCP-ALL | B-Cell Precursor Acute Lymphoblastic Leukemia |
| BTB | Blood–Testis Barrier |
| CML | Chronic Myeloid Leukemia |
| CPA | Conditioned Place Aversion |
| CPP | Conditioned Place Preference |
| CRC | Colorectal Cancer |
| DM1 | Myotonic Dystrophy Type 1 |
| EAE | Experimental Autoimmune Encephalomyelitis |
| eoHM | Early-Onset High Myopia |
| ER | Endoplasmic Reticulum |
| ES | Ewing Sarcoma |
| G4 | G-quadruplex |
| G4PA | G-quadruplex Proximity Aptamer |
| G-rich | Guanine-rich |
| GY | Glycine-rich region |
| GYR | Glycine-Tyrosine-Arginine-rich region |
| HAR | High-Altitude Retinopathy |
| HCC | Hepatocellular Carcinoma |
| HCV | Hepatitis C Virus |
| hnRNP | Heterogeneous Nuclear Ribonucleoprotein |
| hnRNPH1 | Heterogeneous Nuclear Ribonucleoprotein H1 |
| HTS | High-Throughput Screening |
| iPSC | Induced Pluripotent Stem Cell |
| JAK | Janus Kinase |
| KAT | Lysine Acetyltransferase |
| LCD | Low-Complexity Domain |
| LLPS | Liquid–Liquid Phase Separation |
| lncRNA | Long Non-coding RNA |
| LoF | Loss-of-Function |
| MCL | Mantle Cell Lymphoma |
| mESC | Mouse Embryonic Stem Cell |
| ML | Machine Learning |
| MS | Multiple Sclerosis |
| NLS | Nuclear Localization Signal |
| NMD | Nonsense-Mediated Decay |
| NPC | Nasopharyngeal Carcinoma |
| NSCLC | Non-Small Cell Lung Cancer |
| NTD | N-Terminal Domain |
| OTSCC | Oral Tongue Squamous Cell Carcinoma |
| PAH | Pulmonary Arterial Hypertension |
| PASMC | Pulmonary Artery Smooth Muscle Cell |
| PCOS | Polycystic Ovary Syndrome |
| PNA | Peptide Nucleic Acid |
| POF | Premature Ovarian Failure |
| POI | Primary Ovarian Insufficiency |
| PPI | Protein–Protein Interaction |
| pre-mRNA | Precursor Messenger RNA |
| PRI | Protein-RNA Interaction |
| PROTAC | Proteolysis-Targeting Chimera |
| PS | Phosphorothioate |
| PTM | Post-Translational Modification |
| qRRM | Quasi-RNA Recognition Motif |
| RA | Rheumatoid Arthritis |
| RBP | RNA-Binding Protein |
| RBD | RNA Recognition Domain |
| rG4 | RNA G-quadruplex |
| RNAi | RNA Interference |
| RNP | Ribonucleoprotein |
| ROC | Receiver Operating Characteristic |
| RRM | RNA Recognition Motif |
| RSTS | Rubinstein–Taybi Syndrome |
| SBDD | Structure-Based Drug Design |
| sgRNA | Single Guide RNA |
| shRNA | Short Hairpin RNA |
| siRNA | Small Interfering RNA |
| SLE | Systemic Lupus Erythematosus |
| TDS | Testicular Dysgenesis Syndrome |
| TfR | Transferrin Receptor |
| UTR | Untranslated Region |
| WGCNA | Weighted Gene Co-expression Network Analysis |
| WT | Wild-Type |
| 2′-OMe | 2′-O-methyl |
References
- Gao, C.; Wang, Y. mRNA Metabolism in Cardiac Development and Disease: Life After Transcription. Physiol. Rev. 2020, 100, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Furlan, M.; de Pretis, S.; Pelizzola, M. Dynamics of transcriptional and post-transcriptional regulation. Brief. Bioinform. 2021, 22, bbaa389. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.; Ho, U.Y.; James, A.; De Souza, P.; Roberts, T.L. RNA metabolism and links to inflammatory regulation and disease. Cell. Mol. Life Sci. CMLS 2022, 79, 21. [Google Scholar] [CrossRef]
- Kumar, S.; Mohapatra, T. Deciphering Epitranscriptome: Modification of mRNA Bases Provides a New Perspective for Post-transcriptional Regulation of Gene Expression. Front. Cell Dev. Biol. 2021, 9, 628415. [Google Scholar] [CrossRef]
- Liu, J.; Cao, X. RBP-RNA interactions in the control of autoimmunity and autoinflammation. Cell Res. 2023, 33, 97–115. [Google Scholar] [CrossRef]
- Goswami, B.; Nag, S.; Ray, P.S. Fates and functions of RNA-binding proteins under stress. Wiley Interdiscip. Rev. RNA 2024, 15, e1825. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Ma, C.; Du, C.; Huang, Y.; Xu, H.; Li, C.; Cheng, X.; Hao, R.; Xu, Y. RNA-Binding Proteins in the Regulation of Adipogenesis and Adipose Function. Cells 2022, 11, 2357. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Q.; Wang, H.; Yang, X.; Mu, H. Alternative splicing and related RNA binding proteins in human health and disease. Signal Transduct. Target. Ther. 2024, 9, 26. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Z.; Jiang, P.; Kong, D.; Yu, Z.; Shi, D.; Han, Y.; Chen, E.; Zheng, W.; Sun, J.; et al. Phase separation-competent FBL promotes early pre-rRNA processing and translation in acute myeloid leukaemia. Nat. Cell Biol. 2024, 26, 946–961. [Google Scholar] [CrossRef]
- Yang, X.; Stentenbach, M.; A Hughes, L.; Siira, S.J.; Lau, K.; Hothorn, M.; Martinou, J.-C.; Rackham, O.; Filipovska, A. The Vsr-like protein FASTKD4 regulates the stability and polyadenylation of the MT-ND3 mRNA. Nucleic Acids Res. 2024, 53, gkae1261. [Google Scholar] [CrossRef]
- Wang, J.; Sun, D.; Wang, M.; Cheng, A.; Zhu, Y.; Mao, S.; Ou, X.; Zhao, X.; Huang, J.; Gao, Q.; et al. Multiple functions of heterogeneous nuclear ribonucleoproteins in the positive single-stranded RNA virus life cycle. Front. Immunol. 2022, 13, 989298. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.; Holcik, M. Diverse roles of heterogeneous nuclear ribonucleoproteins in viral life cycle. Front. Virol. 2022, 2, 1044652. [Google Scholar] [CrossRef]
- Chen, K.; Luo, M.; Lv, Y.; Luo, Z.; Yang, H. Undervalued and novel roles of heterogeneous nuclear ribonucleoproteins in autoimmune diseases: Resurgence as potential biomarkers and targets. Wiley Interdiscip. Rev. RNA 2023, 14, e1806. [Google Scholar] [CrossRef]
- Kim, G.H.; Kwon, I. Distinct roles of hnRNPH1 low-complexity domains in splicing and transcription. Proc. Natl. Acad. Sci. USA 2021, 118, e2109668118. [Google Scholar] [CrossRef]
- Neckles, C.; Boer, R.E.; Aboreden, N.; Cross, A.M.; Walker, R.L.; Kim, B.-H.; Kim, S.; Schneekloth, J.S.; Caplen, N.J. HNRNPH1-dependent splicing of a fusion oncogene reveals a targetable RNA G-quadruplex interaction. RNA 2019, 25, 1731–1750. [Google Scholar] [CrossRef]
- Liu, M.; Yang, L.; Liu, X.; Nie, Z.; Zhang, X.; Lu, Y.; Pan, Y.; Wang, X.; Luo, J. HNRNPH1 Is a Novel Regulator of Cellular Proliferation and Disease Progression in Chronic Myeloid Leukemia. Front. Oncol. 2021, 11, 682859. [Google Scholar] [CrossRef]
- Jiang, F.; Lang, X.; Chen, N.; Jin, L.; Liu, L.; Wei, X.; Pan, J.; Yu, F.; Blake, A.; Xiao, S. A novel HNRNPH1::ERG rearrangement in aggressive acute myeloid leukemia. Genes Chromosomes Cancer 2022, 61, 503–508. [Google Scholar] [CrossRef]
- Ruan, Q.T.; Yazdani, N.; Blum, B.C.; Beierle, J.A.; Lin, W.; Coelho, M.A.; Fultz, E.K.; Healy, A.F.; Shahin, J.R.; Kandola, A.K.; et al. A Mutation in Hnrnph1 That Decreases Methamphetamine-Induced Reinforcement, Reward, and Dopamine Release and Increases Synaptosomal hnRNP H and Mitochondrial Proteins. J. Neurosci. 2020, 40, 107–130. [Google Scholar] [CrossRef]
- Feng, S.; Li, J.; Wen, H.; Liu, K.; Gui, Y.; Wen, Y.; Wang, X.; Yuan, S. hnRNPH1 recruits PTBP2 and SRSF3 to modulate alternative splicing in germ cells. Nat. Commun. 2022, 13, 3588. [Google Scholar] [CrossRef]
- Gillentine, M.A.; Wang, T.; Hoekzema, K.; Rosenfeld, J.; Liu, P.; Guo, H.; Kim, C.N.; De Vries, B.B.A.; Vissers, L.E.L.M.; Nordenskjold, M.; et al. Rare deleterious mutations of HNRNP genes result in shared neurodevelopmental disorders. Genome Med. 2021, 13, 63. [Google Scholar] [CrossRef]
- Nahalkova, J. On the interface of aging, cancer, and neurodegeneration with SIRT6 and L1 retrotransposon protein interaction network. Ageing Res. Rev. 2024, 101, 102496. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Xia, W.; Wang, S.; Dai, G.; Jiao, W.; Guo, N.; Li, H.; Zhang, G. Etodolac improves collagen induced rheumatoid arthritis in rats by inhibiting synovial inflammation, fibrosis and hyperplasia. Mol. Biomed. 2021, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Penumutchu, S.R.; Chiu, L.-Y.; Meagher, J.L.; Hansen, A.L.; Stuckey, J.A.; Tolbert, B.S. Differential Conformational Dynamics Encoded by the Linker between Quasi RNA Recognition Motifs of Heterogeneous Nuclear Ribonucleoprotein H. J. Am. Chem. Soc. 2018, 140, 11661–11673. [Google Scholar] [CrossRef]
- Korff, A.; Yang, X.; O’donovan, K.; Gonzalez, A.; Teubner, B.J.; Nakamura, H.; Messing, J.; Yang, F.; Carisey, A.F.; Wang, Y.-D.; et al. A murine model of hnRNPH2-related neurodevelopmental disorder reveals a mechanism for genetic compensation by Hnrnph1. J. Clin. Investig. 2024, 134, e160309. [Google Scholar] [CrossRef]
- Larizza, L.; Calzari, L.; Alari, V.; Russo, S. Genes for RNA-binding proteins involved in neural-specific functions and diseases are downregulated in Rubinstein-Taybi iNeurons. Neural Regen. Res. 2022, 17, 5–14. [Google Scholar] [CrossRef]
- GeneCards. HHNRNPH1 Gene—Heterogeneous Nuclear Ribonucleoprotein H1. HNRH1 Protein. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=HNRNPH1 (accessed on 23 March 2025).
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Vo, T.; Brownmiller, T.; Hall, K.; Jones, T.L.; Choudhari, S.; Grammatikakis, I.; Ludwig, K.R.; Caplen, N.J. HNRNPH1 destabilizes the G-quadruplex structures formed by G-rich RNA sequences that regulate the alternative splicing of an oncogenic fusion transcript. Nucleic Acids Res. 2022, 50, 6474–6496. [Google Scholar] [CrossRef]
- Sun, J.; Bie, X.M.; Wang, N.; Zhang, X.S.; Gao, X.-Q. Genome-wide identification and expression analysis of YTH domain-containing RNA-binding protein family in common wheat. BMC Plant Biol. 2020, 20, 351. [Google Scholar] [CrossRef]
- Dominguez, C.; Fisette, J.-F.; Chabot, B.; Allain, F.H.-T. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat. Struct. Mol. Biol. 2010, 17, 853–861. [Google Scholar] [CrossRef]
- Gautrey, H.; Jackson, C.; Dittrich, A.-L.; Browell, D.; Lennard, T.; Tyson-Capper, A. SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol. 2015, 12, 1139–1151. [Google Scholar] [CrossRef]
- von Appen, A.; LaJoie, D.; Johnson, I.E.; Trnka, M.J.; Pick, S.M.; Burlingame, A.L.; Ullman, K.S.; Frost, A. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature 2020, 582, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Grese, Z.R.; Bastos, A.C.; Mamede, L.D.; French, R.L.; Miller, T.M.; Ayala, Y.M. Specific RNA interactions promote TDP-43 multivalent phase separation and maintain liquid properties. EMBO Rep. 2021, 22, e53632. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zou, X.; Deng, J.; Sun, B.; Li, Y.; Zhao, H.; Zhang, X.; Yuan, X.; Zhao, X.; Zou, F. hnRNPH1 maintains mitochondrial homeostasis by establishing NRF1/DRP1 retrograde signaling under mitochondrial stress. Cell Death Differ. 2025, 32, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Van Dusen, C.M.; Yee, L.; McNally, L.M.; McNally, M.T. A glycine-rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Mol. Cell Biol. 2010, 30, 2552–2562. [Google Scholar] [CrossRef]
- Lee, B.J.; Cansizoglu, A.E.; Süel, K.E.; Louis, T.H.; Zhang, Z.; Chook, Y.M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006, 126, 543–558. [Google Scholar] [CrossRef]
- Kreienkamp, H.; Wagner, M.; Weigand, H.; McConkie-Rossell, A.; McDonald, M.; Keren, B.; Mignot, C.; Gauthier, J.; Soucy, J.-F.; Michaud, J.L.; et al. Variant-specific effects define the phenotypic spectrum of HNRNPH2-associated neurodevelopmental disorders in males. Hum. Genet. 2022, 141, 257–272. [Google Scholar] [CrossRef]
- Brownmiller, T.; Caplen, N.J. The HNRNPF/H RNA binding proteins and disease. Wiley Interdiscip. Rev. RNA 2023, 14, e1788. [Google Scholar] [CrossRef]
- Lin, J.Q.; Khuperkar, D.; Pavlou, S.; Makarchuk, S.; Patikas, N.; Lee, F.C.; Zbiegly, J.M.; Kang, J.; Field, S.F.; Bailey, D.M.; et al. HNRNPH1 regulates the neuroprotective cold-shock protein RBM3 expression through poison exon exclusion. EMBO J. 2023, 42, e113168. [Google Scholar] [CrossRef]
- Chen, X.; Yang, H.T.; Zhang, B.; Phillips, J.W.; Cheng, D.; Rigo, F.; Witte, O.N.; Xing, Y.; Black, D.L. The RNA-binding proteins hnRNP H and F regulate splicing of a MYC-dependent HRAS exon in prostate cancer cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2220190120. [Google Scholar] [CrossRef]
- Wang, M.; Ma, T.; Wang, H.; Liu, J.; Chen, Y.; Shim, W.B.; Ma, Z. The RNA binding protein FgRbp1 regulates specific pre-mRNA splicing via interacting with U2AF23 in Fusarium. Nat. Commun. 2021, 12, 2661. [Google Scholar] [CrossRef]
- Pritts, J.D.; Oluyadi, A.A.; Huang, W.; Shimberg, G.D.; Kane, M.A.; Wilks, A.; Michel, S.L.J. Understanding RNA Binding by the Nonclassical Zinc Finger Protein CPSF30, a Key Factor in Polyadenylation during Pre-mRNA Processing. Biochemistry 2021, 60, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ye, R.; Cao, C.; Lv, Z.; Wang, C.; Xie, X.; Chen, X.; Yao, X.; Tian, S.; Yan, L.; et al. BCAS2 and hnRNPH1 orchestrate alternative splicing for DNA double-strand break repair and synapsis in meiotic prophase I. Cell. Mol. Life Sci. CMLS 2024, 81, 449. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Z.; Chen, C.; Guo, T.; Zhao, S.; Zhao, J.; Zhang, W.; Qi, Y.; Zhang, J.; Wang, Y.; et al. MACC1 enhances an oncogenic RNA splicing of IRAK1 through interacting with HNRNPH1 in lung adenocarcinoma. J. Cell. Physiol. 2024, 239, e31426. [Google Scholar] [CrossRef]
- Cesari, E.; Loiarro, M.; Naro, C.; Pieraccioli, M.; Farini, D.; Pellegrini, L.; Pagliarini, V.; Bielli, P.; Sette, C. Combinatorial control of Spo11 alternative splicing by modulation of RNA polymerase II dynamics and splicing factor recruitment during meiosis. Cell Death Dis. 2020, 11, 240. [Google Scholar] [CrossRef]
- Yamazaki, T.; Liu, L.; Conlon, E.G.; Manley, J.L. Burkitt lymphoma-related TCF3 mutations alter TCF3 alternative splicing by disrupting hnRNPH1 binding. RNA Biol. 2020, 17, 1383–1390. [Google Scholar] [CrossRef]
- Xi, S.; Cai, H.; Lu, J.; Zhang, Y.; Yu, Y.; Chen, F.; Huang, Q.; Wang, F.; Chen, Z. The pseudogene PRELID1P6 promotes glioma progression via the hnHNPH1-Akt/mTOR axis. Oncogene 2021, 40, 4453–4467. [Google Scholar] [CrossRef]
- Deng, Q.; Wu, M.; Deng, J. USP36 promotes tumor growth of non-small cell lung cancer via increasing KHK-A expression by regulating c-MYC–hnRNPH1/H2 axis. Hum. Cell Off. J. Hum. Cell Res. Soc. 2022, 35, 694–704. [Google Scholar] [CrossRef]
- Pararajalingam, P.; Coyle, K.M.; Arthur, S.E.; Thomas, N.; Alcaide, M.; Meissner, B.; Boyle, M.; Qureshi, Q.; Grande, B.M.; Rushton, C.; et al. Coding and noncoding drivers of mantle cell lymphoma identified through exome and genome sequencing. Blood 2020, 136, 572–584. [Google Scholar] [CrossRef]
- Li, Y.; Bakke, J.; Finkelstein, D.; Zeng, H.; Wu, J.; Chen, T. HNRNPH1 is required for rhabdomyosarcoma cell growth and survival. Oncogenesis 2018, 7, 9. [Google Scholar] [CrossRef]
- Liang, L.; Li, Y.; Ying, B.; Huang, X.; Liao, S.; Yang, J.; Liao, G. Mutation-associated transcripts reconstruct the prognostic features of oral tongue squamous cell carcinoma. Int. J. Oral Sci. 2023, 15, 1. [Google Scholar] [CrossRef]
- Nasrullah, U.; Stanke, K.; Recknagel, V.; Bozkurt, S.; Wurzel, P.; Gauer, S.; Imre, G.; Münch, C.; Pfeilschifter, J.; Eberhardt, W. The E3 Ligase TRIM25 Impairs Apoptotic Cell Death in Colon Carcinoma Cells via Destabilization of Caspase-7 mRNA: A Possible Role of hnRNPH1. Cells 2023, 12, 201. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Fujiya, M.; Konishi, H.; Murakami, Y.; Iwama, T.; Sasaki, T.; Kunogi, T.; Sakatani, A.; Ando, K.; Ueno, N.; et al. Heterogenous Nuclear Ribonucleoprotein H1 Promotes Colorectal Cancer Progression through the Stabilization of mRNA of Sphingosine-1-Phosphate Lyase 1. Int. J. Mol. Sci. 2020, 21, 4514. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, J.; Zeng, C.; Qin, D.; Zhang, Z.; Lv, Q.; Li, J.; Huang, W. HNRNPH1 stabilizes FLOT2 mRNA in a non-canonical m6A-dependent manner to promote malignant progression in nasopharyngeal carcinoma. Cell. Oncol. 2024, 47, 2279–2295. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, Q.; Ju, X.; Hu, Z.; Xia, L.; Deng, Y.; Zhao, P.; Zhang, M.; Shao, Y.; Huang, S.; et al. HNRNPH1-stabilized LINC00662 promotes ovarian cancer progression by activating the GRP78/p38 pathway. Oncogene 2021, 40, 4770–4782. [Google Scholar] [CrossRef]
- Parisi, S.; Castaldo, D.; Piscitelli, S.; D’ambrosio, C.; Divisato, G.; Passaro, F.; Avolio, R.; Castellucci, A.; Gianfico, P.; Masullo, M.; et al. Identification of RNA-binding proteins that partner with Lin28a to regulate Dnmt3a expression. Sci. Rep. 2021, 11, 2345. [Google Scholar] [CrossRef]
- Gonzalez, A.; Kim, H.J.; Freibaum, B.D.; Fung, H.Y.J.; Brautigam, C.A.; Taylor, J.P.; Chook, Y.M. A new Karyopherin-beta2 binding PY-NLS epitope of HNRNPH2 linked to neurodevelopmental disorders. Structure 2023, 31, 924–934.e4. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Sun, L.; Wang, Z.; Hao, C.; Wang, W. Eukaryotic RNA binding protein hnRNPH1 suppresses influenza A virus replication through interaction with virus NS1 protein. Emerg. Microbes Infect. 2025, 14, 2477645. [Google Scholar] [CrossRef]
- Luo, E.; Nathanson, J.L.; Tan, F.E.; Schwartz, J.L.; Schmok, J.C.; Shankar, A.; Markmiller, S.; Yee, B.A.; Sathe, S.; Pratt, G.A.; et al. Large-scale tethered function assays identify factors that regulate mRNA stability and translation. Nat. Struct. Mol. Biol. 2020, 27, 989–1000. [Google Scholar] [CrossRef]
- Larizza, L.; Alari, V.; Calzari, L.; Russo, S. Interconnected Gene Networks Underpin the Clinical Overlap of HNRNPH1-Related and Rubinstein-Taybi Intellectual Disability Syndromes. Front. Neurosci. 2021, 15, 745684. [Google Scholar] [CrossRef]
- Ruan, Q.T.; Lynch, W.B.; Cole, R.H.; Rieger, M.A.; Beierle, J.A.; Yao, E.J.; Cox, J.W.; Kandola, A.; Richardson, K.T.; Chen, M.M.; et al. Cacna2d2 is an hnRNP H target of the striatal mRNA targetome and regulates methamphetamine behavior. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bryant, C.D.; Healy, A.F.; Ruan, Q.T.; Coehlo, M.A.; Lustig, E.; Yazdani, N.; Luttik, K.P.; Tran, T.; Swancy, I.; Brewin, L.W.; et al. Sex-dependent effects of an Hnrnph1 mutation on fentanyl addiction-relevant behaviors but not antinociception in mice. Genes Brain Behav. 2021, 20, e12711. [Google Scholar] [CrossRef] [PubMed]
- Fultz, E.K.; Coelho, M.A.; Lieberman, D.; Jimenez-Chavez, C.L.; Bryant, C.D.; Szumlinski, K.K. Hnrnph1 is a novel regulator of alcohol reward. Drug Alcohol. Depend. 2021, 220, 108518. [Google Scholar] [CrossRef] [PubMed]
- Valori, C.F.; Neumann, M. Contribution of RNA/DNA Binding Protein Dysfunction in Oligodendrocytes in the Pathogenesis of the Amyotrophic Lateral Sclerosis/Frontotemporal Lobar Degeneration Spectrum Diseases. Front. Neurosci. 2021, 15, 724891. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; James, D.; Huang, J.; Lee, M. The Emerging Role of the RNA-Binding Protein SFPQ in Neuronal Function and Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 7151. [Google Scholar] [CrossRef]
- Feng, S.; Wen, H.; Liu, K.; Xiong, M.; Li, J.; Gui, Y.; Lv, C.; Zhang, J.; Ma, X.; Wang, X.; et al. hnRNPH1 establishes Sertoli-germ cell crosstalk through cooperation with PTBP1 and AR, and is essential for male fertility in mice. Development 2023, 150, dev201040. [Google Scholar] [CrossRef]
- Arandel, L.; Matloka, M.; Klein, A.F.; Rau, F.; Sureau, A.; Ney, M.; Cordier, A.; Kondili, M.; Polay-Espinoza, M.; Naouar, N.; et al. Reversal of RNA toxicity in myotonic dystrophy via a decoy RNA-binding protein with high affinity for expanded CUG repeats. Nat. Biomed. Eng. 2022, 6, 207–220. [Google Scholar] [CrossRef]
- Sznajder, L.J.; Khan, M.; Ciesiołka, A.; Tadross, M.; Nutter, C.A.; Taylor, K.; Pearson, C.E.; Lewis, M.H.; Hines, R.M.; Swanson, M.S.; et al. Autism-related traits in myotonic dystrophy type 1 model mice are due to MBNL sequestration and RNA mis-splicing of autism-risk genes. Nat. Neurosci. 2025. [Google Scholar] [CrossRef]
- Paul, S.; Dansithong, W.; Kim, D.; Rossi, J.; Webster, N.J.G.; Comai, L.; Reddy, S. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. EMBO J. 2006, 25, 4271–4283. [Google Scholar] [CrossRef]
- Ruffenach, G.; Medzikovic, L.; Sun, W.; Hong, J.; Eghbali, M. Functions of RNA-Binding Proteins in Cardiovascular Disease. Cells 2023, 12, 2794. [Google Scholar] [CrossRef]
- Wu, W.; Chen, A.; Lin, S.; Wang, Q.; Lian, G.; Luo, L.; Xie, L. The identification and verification of hub genes associated with pulmonary arterial hypertension using weighted gene co-expression network analysis. BMC Pulm. Med. 2022, 22, 474. [Google Scholar] [CrossRef]
- Ouyang, J.; Li, S.; Sun, W.; Xiao, X.; Wang, Y.; Jiang, Y.; Zhang, Q. Variants in HNRNPH1 are associated with high myopia in humans and ocular coloboma in zebrafish. Clin. Genet. 2022, 102, 424–433. [Google Scholar] [CrossRef]
- Su, T.; Gu, C.; Draga, D.; Zhou, C.; Lhamo, T.; Zheng, Z.; Qiu, Q. Integrative analysis of miRNA-mRNA network in high altitude retinopathy by bioinformatics analysis. Biosci. Rep. 2021, 41, BSR20200776. [Google Scholar] [CrossRef]
- Marklein, B.; Jenning, M.; Konthur, Z.; Häupl, T.; Welzel, F.; Nonhoff, U.; Krobitsch, S.; Mulder, D.M.; Koenders, M.I.; Joshua, V.; et al. The citrullinated/native index of autoantibodies against hnRNP-DL predicts an individual “window of treatment success” in RA patients. Arthritis Res. Ther. 2021, 23, 239. [Google Scholar] [CrossRef]
- Cardamone, G.; Paraboschi, E.M.; Soldà, G.; Cantoni, C.; Supino, D.; Piccio, L.; Duga, S.; Asselta, R. Not only cancer: The long non-coding RNA MALAT1 affects the repertoire of alternatively spliced transcripts and circular RNAs in multiple sclerosis. Hum. Mol. Genet. 2019, 28, 1414–1428. [Google Scholar] [CrossRef]
- Lee, J.; Liao, P.-C.; Young, K.-C.; Chang, C.L.; Chen, S.S.L.; Chang, T.-T.; Lai, M.-D.; Wang, S.-W. Identification of hnRNPH1, NF45, and C14orf166 as novel host interacting partners of the mature hepatitis C virus core protein. J. Proteome Res. 2011, 10, 4522–4534. [Google Scholar] [CrossRef]
- Velayutham, S.; Seal, T.; Danthurthy, S.; Zaias, J.; Smalley, K.S.M.; Minond, D. In Vivo Acute Toxicity Studies of Novel Anti-Melanoma Compounds Downregulators of hnRNPH1/H2. Biomolecules 2023, 13, 349. [Google Scholar] [CrossRef]
- Micaelli, M.; Vedove, A.D.; Cerofolini, L.; Vigna, J.; Sighel, D.; Zaccara, S.; Bonomo, I.; Poulentzas, G.; Rosatti, E.F.; Cazzanelli, G.; et al. Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N6-Methyladenosine-Modified RNAs. ACS Pharmacol. Transl. Sci. 2022, 5, 872–891. [Google Scholar] [CrossRef]
- Borgelt, L.; Li, F.; Hommen, P.; Lampe, P.; Hwang, J.; Goebel, G.L.; Sievers, S.; Wu, P. Trisubstituted Pyrrolinones as Small-Molecule Inhibitors Disrupting the Protein–RNA Interaction of LIN28 and Let-7. ACS Med. Chem. Lett. 2021, 12, 893–898. [Google Scholar] [CrossRef]
- Vaidya, A.; Moore, S.; Chatterjee, S.; Guerrero, E.; Kim, M.; Farbiak, L.; Dilliard, S.A.; Siegwart, D.J. Expanding RNAi to Kidneys, Lungs, and Spleen via Selective ORgan Targeting (SORT) siRNA Lipid Nanoparticles. Adv. Mater. 2024, 36, e2313791. [Google Scholar] [CrossRef]
- Ayyar, V.S.; Song, D.; Zheng, S.; Carpenter, T.; Heald, D.L. Minimal Physiologically Based Pharmacokinetic-Pharmacodynamic (mPBPK-PD) Model of N-Acetylgalactosamine–Conjugated Small Interfering RNA Disposition and Gene Silencing in Preclinical Species and Humans. J. Pharmacol. Exp. Ther. 2021, 379, 134–146. [Google Scholar] [CrossRef]
- Lenard, A.J.; Hutten, S.; Zhou, Q.; Usluer, S.; Zhang, F.; Bourgeois, B.M.R.; Dormann, D.; Madl, T. Phosphorylation Regulates CIRBP Arginine Methylation, Transportin-1 Binding and Liquid-Liquid Phase Separation. Front. Mol. Biosci. 2021, 8, 689687. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, H.; Zhao, X.; Luo, Q.; Wang, Q.; Tan, K.; Wang, Z.; Jiang, J.; Cui, J.; Du, E.; et al. Arginine methylation of METTL14 promotes RNA N6-methyladenosine modification and endoderm differentiation of mouse embryonic stem cells. Nat. Commun. 2021, 12, 3780. [Google Scholar] [CrossRef]
- Chandra, A.; Ananda, H.; Singh, N.; Qamar, I. Identification of a novel and potent small molecule inhibitor of SRPK1: Mechanism of dual inhibition of SRPK1 for the inhibition of cancer progression. Aging 2021, 13, 163–180. [Google Scholar] [CrossRef]
- Zhao, H.; Lau, H.L.; Zhang, K.; Kwok, C.K. Selective recognition of RNA G-quadruplex in vitro and in cells by L-aptamer-D-oligonucleotide conjugate. Nucleic Acids Res. 2024, 52, 13544–13560. [Google Scholar] [CrossRef]
- Park, S.; Kim, S.H.; Dezhbord, M.; Lee, E.-H.; Jeon, Y.; Jung, D.; Gu, S.H.; Yu, C.; Lee, S.H.; Kim, S.C.; et al. Cell-permeable peptide nucleic acid antisense oligonucleotide platform targeting human betacoronaviruses. Front. Microbiol. 2023, 14, 1258091. [Google Scholar] [CrossRef]
- Wu, P. Inhibition of RNA-binding proteins with small molecules. Nature reviews. Chemistry 2020, 4, 441–458. [Google Scholar]
- Singh, V.; Singh, A.; Liu, A.J.; Fuchs, S.Y.; Sharma, A.K.; Spiegelman, V.S. RNA Binding Proteins as Potential Therapeutic Targets in Colorectal Cancer. Cancers 2024, 16, 3502. [Google Scholar] [CrossRef]
- Sarma, P.; Shekhar, N.; Prajapat, M.; Avti, P.; Kaur, H.; Kumar, S.; Singh, S.; Kumar, H.; Prakash, A.; Dhibar, D.P.; et al. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). J. Biomol. Struct. Dyn. 2021, 39, 2724–2732. [Google Scholar] [CrossRef]
- Wu, X.; Ramesh, R.; Wang, J.; Zheng, Y.; Armaly, A.M.; Wei, L.; Xing, M.; Roy, S.; Lan, L.; Gao, F.P.; et al. Small Molecules Targeting the RNA-Binding Protein HuR Inhibit Tumor Growth in Xenografts. J. Med. Chem. 2023, 66, 2032–2053. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Song, J.; Cheng, X.; Zhou, C.; Huang, S.; Zhao, W.; Zong, Z.; Yang, L. Targeting the “tumor microenvironment”: RNA-binding proteins in the spotlight in colorectal cancer therapy. Int. Immunopharmacol. 2024, 131, 111876. [Google Scholar] [CrossRef]
- Fan, Q.; Yang, Z.; Li, Y.; Cheng, Y.; Li, Y. Polycatechol Mediated Small Interfering RNA Delivery for the Treatment of Ulcerative Colitis. Adv. Funct. Mater. 2021, 31, 2101646. [Google Scholar] [CrossRef]
- Supe, S.; Upadhya, A.; Tripathi, S.; Dighe, V.; Singh, K. Liposome-polyethylenimine complexes for the effective delivery of HuR siRNA in the treatment of diabetic retinopathy. Drug Deliv. Transl. Res. 2023, 13, 1675–1698. [Google Scholar] [CrossRef]
- Du, Y.; Wang, Q.; Shi, L.; Li, T. G-Quadruplex-Proximized Aptamers (G4PA) Efficiently Targeting Cell-Surface Transferrin Receptors for Targeted Cargo Delivery. Nano Lett. 2022, 22, 6328–6333. [Google Scholar] [CrossRef]
- Geczy, R.; Thommandru, B.; Swaminathan, M.; Zhao, R.; Tan, K.; Low, K.; Balgi, A.; Park, S.; Watt, E.; Thada, V.; et al. Lipid Nanoparticle-Mediated Gene Editing of Human Primary T Cells and Off-Target Analysis of the CRISPR-Cas9 Indels. Blood 2023, 142 (Suppl. S1), 6833. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, J.K.W.; Zhang, Q.; Hughes, N.W.; Xia, Q.; Winslow, M.M.; Cong, L. Microbial single-strand annealing proteins enable CRISPR gene-editing tools with improved knock-in efficiencies and reduced off-target effects. Nucleic Acids Res. 2021, 49, e36. [Google Scholar] [CrossRef]
- Bhat, A.A.; Nisar, S.; Mukherjee, S.; Saha, N.; Yarravarapu, N.; Lone, S.N.; Masoodi, T.; Chauhan, R.; Maacha, S.; Bagga, P.; et al. Integration of CRISPR/Cas9 with artificial intelligence for improved cancer therapeutics. J. Transl. Med. 2022, 20, 534. [Google Scholar] [CrossRef]
- Chang, Y.; Kang, Z.; Bei, J.; Chou, S.-J.; Lu, M.-Y.J.; Su, Y.-L.; Lin, S.-W.; Wang, H.-H.; Lin, S.; Chang, C.-J. Generation of TRIM28 Knockout K562 Cells by CRISPR/Cas9 Genome Editing and Characterization of TRIM28-Regulated Gene Expression in Cell Proliferation and Hemoglobin Beta Subunits. Int. J. Mol. Sci. 2022, 23, 6839. [Google Scholar] [CrossRef]
- Zavileyskiy, L.G.; Pervouchine, D.D. Post-transcriptional Regulation of Gene Expression via Unproductive Splicing. Acta Nat. 2024, 16, 4–13. [Google Scholar] [CrossRef]
- Noblejas-Lopez, M.D.M.; Tébar-García, D.; López-Rosa, R.; Alcaraz-Sanabria, A.; Cristóbal-Cueto, P.; Pinedo-Serrano, A.; Rivas-García, L.; Galán-Moya, E.M. TACkling Cancer by Targeting Selective Protein Degradation. Pharmaceutics 2023, 15, 2442. [Google Scholar] [CrossRef]
- Li, R.; Liu, M.; Yang, Z.; Li, J.; Gao, Y.; Tan, R. Proteolysis-Targeting Chimeras (PROTACs) in Cancer Therapy: Present and Future. Molecules 2022, 27, 8828. [Google Scholar] [CrossRef]
- Kargbo, R.B. PROTAC Compounds Targeting Androgen Receptor for Cancer Therapeutics: Prostate Cancer and Kennedy’s Disease. ACS Med. Chem. Lett. 2020, 11, 1092–1093. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Elhassan, R.M.; Hou, X.; Fang, H. Recent Advances in Small Molecule PROTACs for the Treatment of Cancer. Curr. Med. Chem. 2021, 28, 4893–4909. [Google Scholar] [CrossRef] [PubMed]
- Kargbo, R.B. Degradation of Janus Kinase for Potential Application in Immune Response Therapeutics. ACS Med. Chem. Lett. 2021, 12, 316–317. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Yan, C.; Gao, H.; Liu, T. Small-molecule PROTACs: Novel agents for cancer therapy. Future Med. Chem. 2020, 12, 915–938. [Google Scholar] [CrossRef]
- Nathani, A.; Aare, M.; Sun, L.; Li, Y.; Singh, M. Abstract 7578: BRD4 protein degradation for p53 mutant diffuse intrinsic pontine glioma (DIPG) via various delivery approaches for ARV-825 by using camel milk exosomes. Cancer Res. 2024, 84 (Suppl. S6), 7578. [Google Scholar] [CrossRef]
- Li, Z.; Lim, S.L.; Tao, Y.; Li, X.; Xie, Y.; Yang, C.; Zhang, Z.; Jiang, Y.; Zhang, X.; Cao, X.; et al. PROTAC Bromodomain Inhibitor ARV-825 Displays Anti-Tumor Activity in Neuroblastoma by Repressing Expression of MYCN or c-Myc. Front. Oncol. 2020, 10, 574525. [Google Scholar] [CrossRef]
- Yang, L.; Jing, Y.; Xia, X.; Yin, X. ARV-825 Showed Antitumor Activity against BRD4-NUT Fusion Protein by Targeting the BRD4. J. Oncol. 2023, 2023, 9904143. [Google Scholar] [CrossRef]
- Qin, A.; Jin, H.; Song, Y.; Gao, Y.; Chen, Y.-F.; Zhou, L.-N.; Wang, S.-S.; Lu, X.-S. The therapeutic effect of the BRD4-degrading PROTAC A1874 in human colon cancer cells. Cell Death Dis. 2020, 11, 805. [Google Scholar] [CrossRef]
- Qian, H.; Zhu, M.; Tan, X.; Zhang, Y.; Liu, X.; Yang, L. Super-enhancers and the super-enhancer reader BRD4: Tumorigenic factors and therapeutic targets. Cell Death Discov. 2023, 9, 470. [Google Scholar] [CrossRef]
- Virgilio, A.; Benigno, D.; Aliberti, C.; Vellecco, V.; Bucci, M.; Esposito, V.; Galeone, A. Improving the Biological Properties of Thrombin-Binding Aptamer by Incorporation of 8-Bromo-2′-Deoxyguanosine and 2′-Substituted RNA Analogues. Int. J. Mol. Sci. 2023, 24, 15529. [Google Scholar] [CrossRef]
- Anderson, B.A.; Freestone, G.C.; Low, A.; De-Hoyos, C.L.; Drury, W.J., III; Østergaard, M.E.; Migawa, M.T.; Fazio, M.; Wan, W.B.; Berdeja, A.; et al. Towards next generation antisense oligonucleotides: Mesylphosphoramidate modification improves therapeutic index and duration of effect of gapmer antisense oligonucleotides. Nucleic Acids Res. 2021, 49, 9026–9041. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, Y.; He, A.; Guan, B.; He, S.; Zhang, C.; Kang, Z.; Gong, Y.; Li, X.; Zhou, L. Identification of the Six-RNA-Binding Protein Signature for Prognosis Prediction in Bladder Cancer. Front. Genet. 2020, 11, 992. [Google Scholar] [CrossRef] [PubMed]
- Shiina, T.; Shimizu, Y. Temperature-Dependent Alternative Splicing of Precursor mRNAs and Its Biological Significance: A Review Focused on Post-Transcriptional Regulation of a Cold Shock Protein Gene in Hibernating Mammals. Int. J. Mol. Sci. 2020, 21, 7599. [Google Scholar] [CrossRef]
- Schieweck, R.; Ninkovic, J.; Kiebler, M.A. RNA-binding proteins balance brain function in health and disease. Physiol. Rev. 2021, 101, 1309–1370. [Google Scholar] [CrossRef]
- Zhao, Y.; Mir, C.; Garcia-Mayea, Y.; Paciucci, R.; Kondoh, H.; Lleonart, M. RNA-binding proteins: Underestimated contributors in tumorigenesis. Semin. Cancer Biol. 2022, 86 Pt 3, 431–444. [Google Scholar] [CrossRef]
- Kosti, A.; De Araujo, P.R.; Li, W.-Q.; Guardia, G.D.A.; Chiou, J.; Yi, C.; Ray, D.; Meliso, F.; Li, Y.M.; Delambre, T.; et al. The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation. Genome Biol. 2020, 21, 195. [Google Scholar] [CrossRef]
- Dong, Y.; Xiao, Y.; Shi, Q.; Jiang, C. Dysregulated lncRNA-miRNA-mRNA Network Reveals Patient Survival-Associated Modules and RNA Binding Proteins in Invasive Breast Carcinoma. Front. Genet. 2020, 10, 1284. [Google Scholar] [CrossRef]
- Qiu, Y.; Ching, W.; Zou, Q. Prediction of RNA-binding protein and alternative splicing event associations during epithelial-mesenchymal transition based on inductive matrix completion. Brief. Bioinform. 2021, 22, bbaa440. [Google Scholar] [CrossRef]
- Schisa, J.A.; Elaswad, M.T. An Emerging Role for Post-translational Modifications in Regulating RNP Condensates in the Germ Line. Front. Mol. Biosci. 2021, 8, 658020. [Google Scholar] [CrossRef]
- Hamey, J.J.; Nguyen, A.; Wilkins, M.R. Discovery of Arginine Methylation, Phosphorylation, and Their Co-occurrence in Condensate-Associated Proteins in Saccharomyces cerevisiae. J. Proteome Res. 2021, 20, 2420–2434. [Google Scholar] [CrossRef]
- Kang, D.; Lee, Y.; Lee, J. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers 2020, 12, 2699. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yan, K.; Zhang, J.; Liu, B. iDRNA-ITF: Identifying DNA- and RNA-binding residues in proteins based on induction and transfer framework. Brief. Bioinform. 2022, 23, bbac236. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Huang, C.; Wang, X.; Tong, D. The RNA-binding protein GRSF1 promotes hepatocarcinogenesis via competitively binding to YY1 mRNA with miR-30e-5p. J. Exp. Clin. Cancer Res. 2022, 41, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Chen, K.; Chen, Y.; Liu, J.; Zhang, X.; Zhou, Y.; Liu, Q.; Wang, B.; Chen, T.; Cao, X. RNA-binding protein ZCCHC4 promotes human cancer chemoresistance by disrupting DNA-damage-induced apoptosis. Signal Transduct. Target. Ther. 2022, 7, 240. [Google Scholar] [CrossRef]
- Kim, S.; Ju, J.-S.; Kang, M.-H.; Won, J.E.; Kim, Y.H.; Raninga, P.V.; Khanna, K.K.; Győrffy, B.; Pack, C.-G.; Han, H.-D.; et al. RNA-binding protein NONO contributes to cancer cell growth and confers drug resistance as a theranostic target in TNBC. Theranostics 2020, 10, 7974–7992. [Google Scholar] [CrossRef]
- Okholm, T.L.H.; Sathe, S.; Park, S.S.; Kamstrup, A.B.; Rasmussen, A.M.; Shankar, A.; Chua, Z.M.; Fristrup, N.; Nielsen, M.M.; Vang, S.; et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 2020, 12, 112. [Google Scholar] [CrossRef]
- Chen, P.; Xu, J.; Cui, Z.; Wu, S.; Xie, T.; Zhang, X. Multi-omics analysis of N6-methyladenosine reader IGF2BP3 as a promising biomarker in pan-cancer. Front. Immunol. 2023, 14, 1071675. [Google Scholar] [CrossRef]
- Zhou, W.; Jie, Q.; Pan, T.; Shi, J.; Jiang, T.; Zhang, Y.; Ding, N.; Xu, J.; Ma, Y.; Li, Y. Single-cell RNA binding protein regulatory network analyses reveal oncogenic HNRNPK-MYC signalling pathway in cancer. Commun. Biol. 2023, 6, 82. [Google Scholar] [CrossRef]
- Li, H.; Meng, X.; You, X.; Zhou, W.; Ouyang, W.; Pu, X.; Zhao, R.; Tang, H. Increased expression of the RNA-binding protein Musashi-2 is associated with immune infiltration and predicts better outcomes in ccRCC patients. Front. Oncol. 2022, 12, 949705. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Li, J.; Zhao, B.; Huang, G.; Li, X.; Xie, Z.; Zhou, Z. The Emerging Role of m6A Modification in Regulating the Immune System and Autoimmune Diseases. Front. Cell Dev. Biol. 2021, 9, 755691. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, Y.; Zhang, J.; Zheng, L. Biogenesis and functions of circular RNAs and their role in diseases of the female reproductive system. Reprod. Biol. Endocrinol. 2020, 18, 104. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Li, H.; Liu, Y.; Wang, X.; Mei, Q.; Xiang, W. N6-Methyladenosine Modifications in the Female Reproductive System: Roles in Gonad Development and Diseases. Int. J. Biol. Sci. 2022, 18, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Glass, Z.; Chen, J.; Haas, M.; Jin, X.; Zhao, X.; Rui, X.; Ye, Z.; Li, Y.; Zhang, F.; et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. USA 2021, 118, e2020401118. [Google Scholar] [CrossRef]
- Dubey, A.K.; Gupta, V.K.; Kujawska, M.; Orive, G.; Kim, N.-Y.; Li, C.-Z.; Mishra, Y.K.; Kaushik, A. Exploring nano-enabled CRISPR-Cas-powered strategies for efficient diagnostics and treatment of infectious diseases. J. Nanostructure Chem. 2022, 12, 833–864. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, Q.; Zhao, H.; Xie, R.; He, X.; Gu, H. Unraveling the Role of RNA-Binding Proteins, with a Focus on RPS5, in the Malignant Progression of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2024, 25, 773. [Google Scholar] [CrossRef]
- Alemi, F.; Poornajaf, Y.; Hosseini, F.; Vahedian, V.; Gharekhani, M.; Shoorei, H.; Taheri, M. Interaction between lncRNAs and RNA-binding proteins (RBPs) influences DNA damage response in cancer chemoresistance. Mol. Biol. Rep. 2024, 51, 308. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Lu, Q.; Tian, L.; Yao, S.; Lai, F.; Liang, Y.; Liu, C.; Lu, Y.; Tian, S.; et al. Nanocarrier-mediated siRNA delivery: A new approach for the treatment of traumatic brain injury-related Alzheimer’s disease. Neural Regen Res. 2025, 20, 2538–2555. [Google Scholar] [CrossRef]
- Song, N.; Tao, R.; Li, H.; Zhang, R.; Huang, Y.; Zhang, L.; Liu, Y.; Yang, D.; Yao, C. Spatially Controlled Co-Delivery of Diagnostic and Therapeutic Agents Using DNA Nanoframeworks for Pancreatic Cancer Precision Therapy. Angew. Chem. Int. Ed. Engl. 2025, 64, e202500566. [Google Scholar] [CrossRef]
- Mallick, R.; Basak, S.; Chowdhury, P.; Bhowmik, P.; Das, R.K.; Banerjee, A.; Paul, S.; Pathak, S.; Duttaroy, A.K. Targeting Cytokine-Mediated Inflammation in Brain Disorders: Developing New Treatment Strategies. Pharmaceuticals 2025, 18, 104. [Google Scholar] [CrossRef]
- Yin, Q.; Zheng, M.; Luo, Q.; Jiang, D.; Zhang, H.; Chen, C. YB-1 as an Oncoprotein: Functions, Regulation, Post-Translational Modifications, and Targeted Therapy. Cells 2022, 11, 1217. [Google Scholar] [CrossRef]
- Guillemin, A.; Kumar, A.; Wencker, M.; Ricci, E.P. Shaping the Innate Immune Response Through Post-Transcriptional Regulation of Gene Expression Mediated by RNA-Binding Proteins. Front. Immunol. 2022, 12, 796012. [Google Scholar] [CrossRef]
- Kelm, J.M.; Pandey, D.S.; Malin, E.; Kansou, H.; Arora, S.; Kumar, R.; Gavande, N.S. PROTAC’ing oncoproteins: Targeted protein degradation for cancer therapy. Mol. Cancer 2023, 22, 62. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Yi, W.; Zhang, L.; Qiu, C.; Sun, N.; He, J.; Feng, P.; Wu, Q.; Wang, G.; Wu, G. hnRNPH1: A Multifaceted Regulator in RNA Processing and Disease Pathogenesis. Int. J. Mol. Sci. 2025, 26, 5159. https://doi.org/10.3390/ijms26115159
Zhu L, Yi W, Zhang L, Qiu C, Sun N, He J, Feng P, Wu Q, Wang G, Wu G. hnRNPH1: A Multifaceted Regulator in RNA Processing and Disease Pathogenesis. International Journal of Molecular Sciences. 2025; 26(11):5159. https://doi.org/10.3390/ijms26115159
Chicago/Turabian StyleZhu, Lijing, Wei Yi, Like Zhang, Chenyue Qiu, Ning Sun, Jingwen He, Ping Feng, Qiong Wu, Guangyi Wang, and Guosheng Wu. 2025. "hnRNPH1: A Multifaceted Regulator in RNA Processing and Disease Pathogenesis" International Journal of Molecular Sciences 26, no. 11: 5159. https://doi.org/10.3390/ijms26115159
APA StyleZhu, L., Yi, W., Zhang, L., Qiu, C., Sun, N., He, J., Feng, P., Wu, Q., Wang, G., & Wu, G. (2025). hnRNPH1: A Multifaceted Regulator in RNA Processing and Disease Pathogenesis. International Journal of Molecular Sciences, 26(11), 5159. https://doi.org/10.3390/ijms26115159





