Ion Channel–Extracellular Matrix Interplay in Colorectal Cancer: A Network-Based Approach to Tumor Microenvironment Remodeling
Abstract
1. Introduction
2. Results
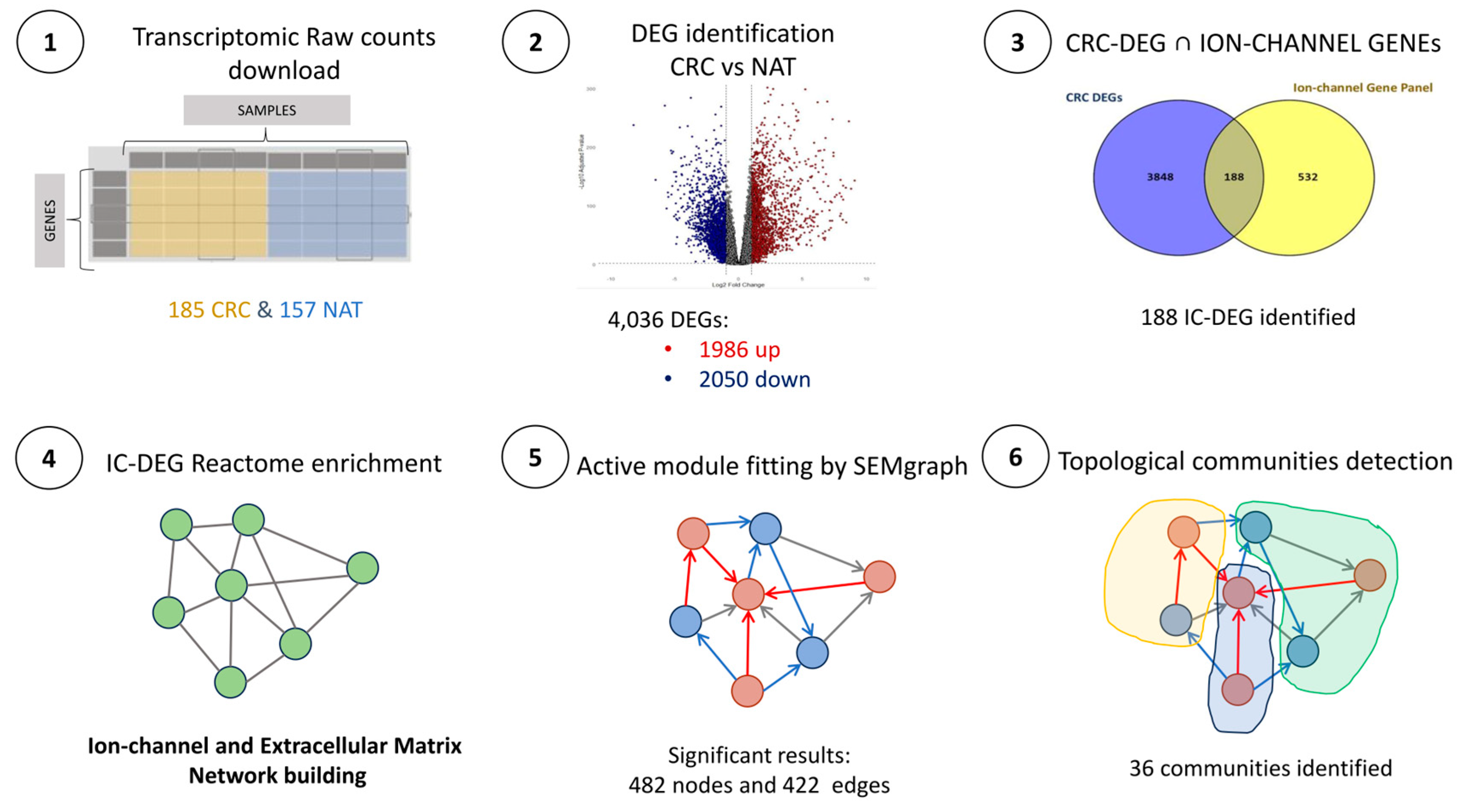
2.1. Dataset Description and Quality Control (QC)
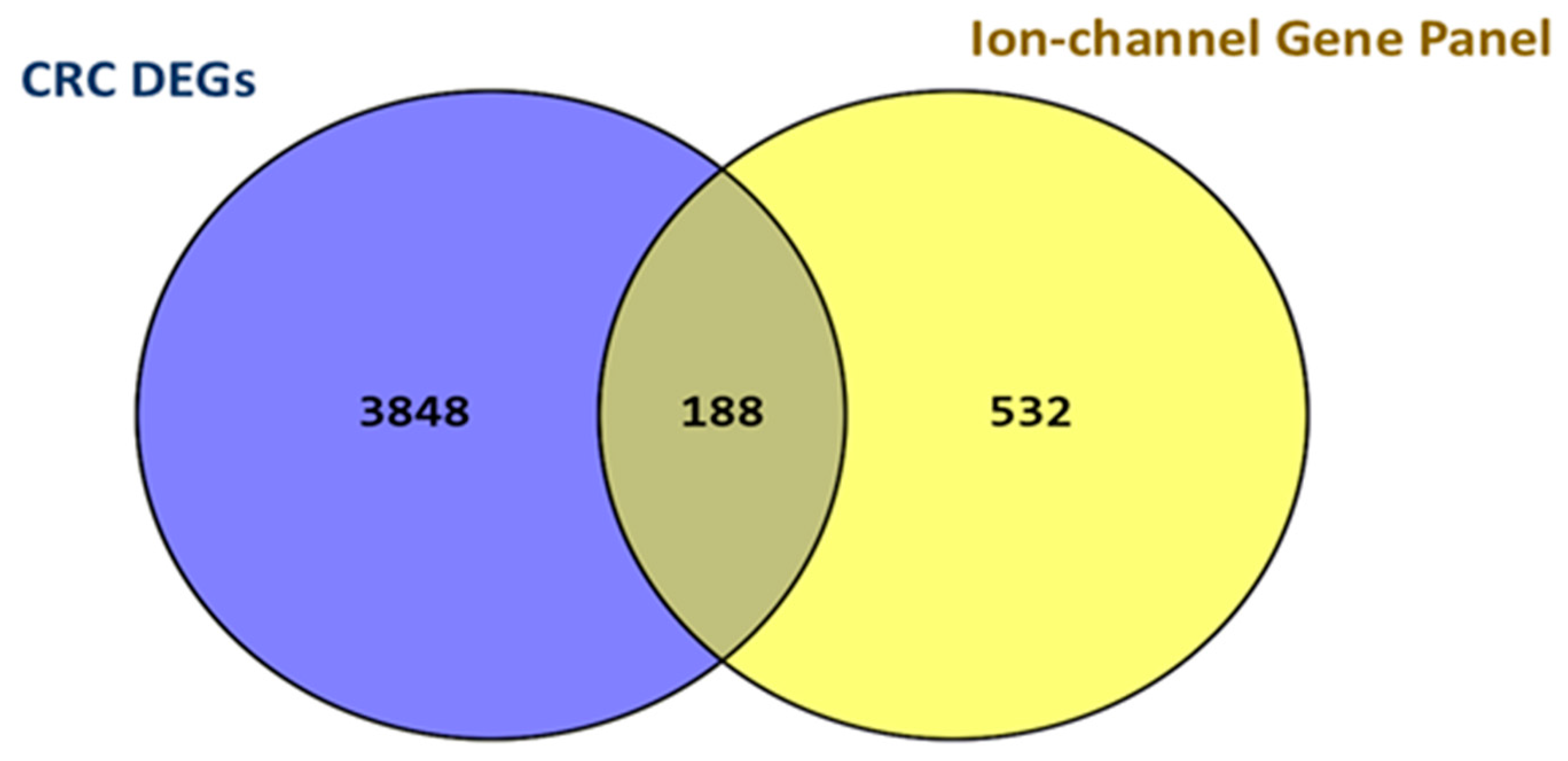
2.2. CRC DEGs Determination and Ion Channel Subset
2.3. Reactome Enrichment Analysis Results
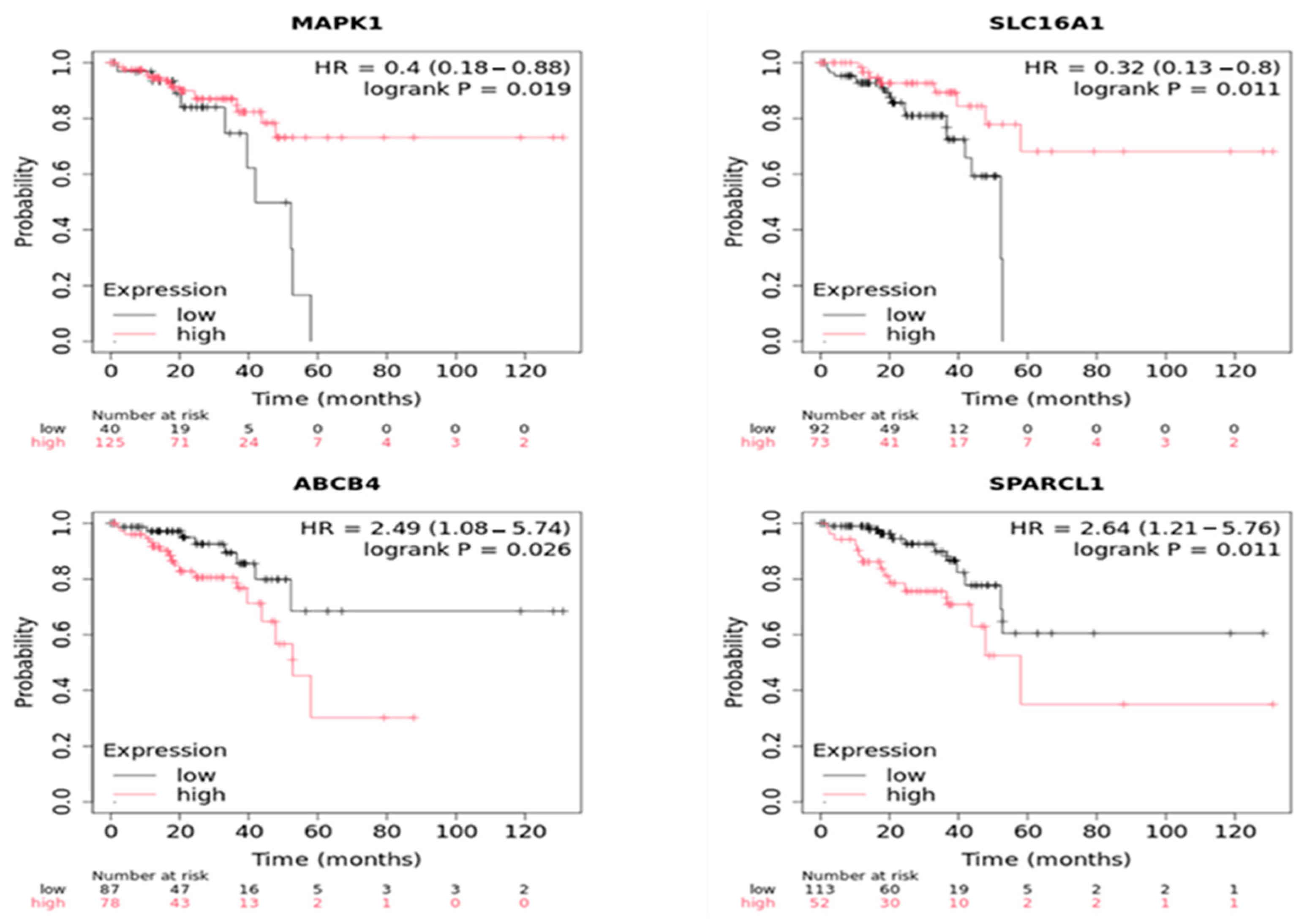
2.4. IC-DEGs Validation in “The Cancer Genome Atlas” Database
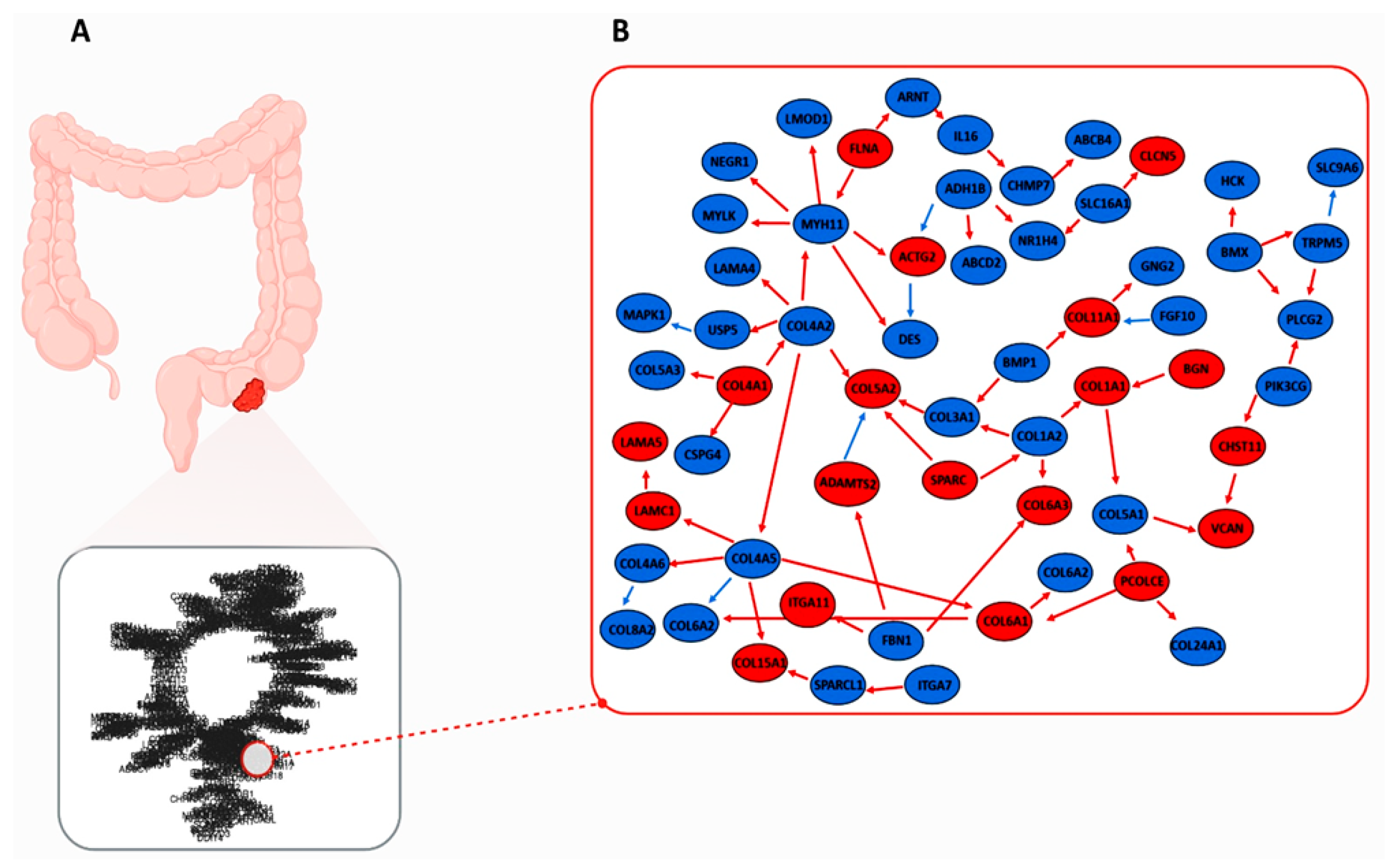
2.5. CRC-IC Module and Statistical Analysis
2.5.1. CRC-IC Module Analysis
2.5.2. Gene Communities Detection Results
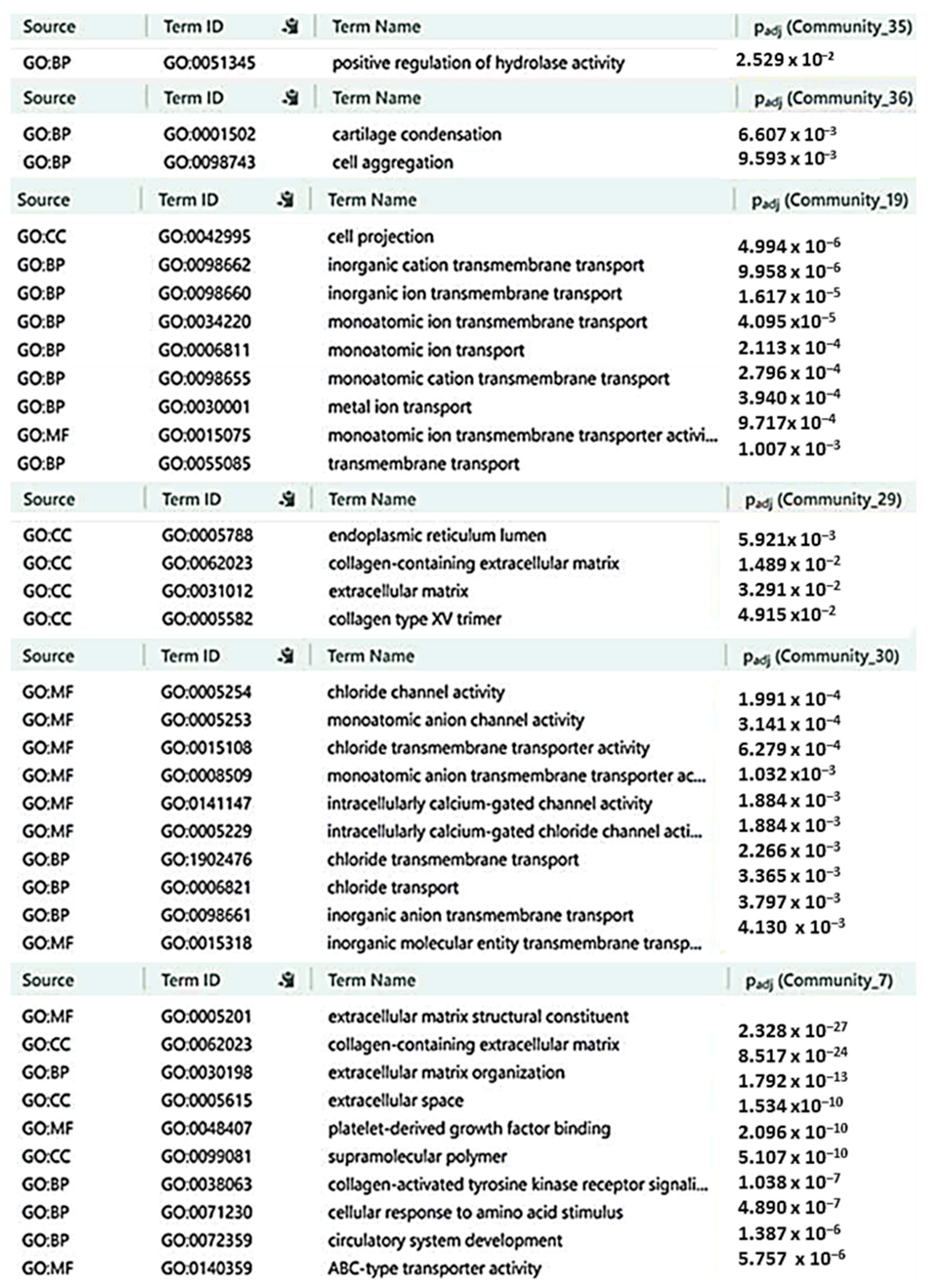
2.6. Characterization of CRC-IC Module Communities
2.7. Statistical External Validation of the Active CRC-IC Module Results
3. Discussion
4. Materials and Methods
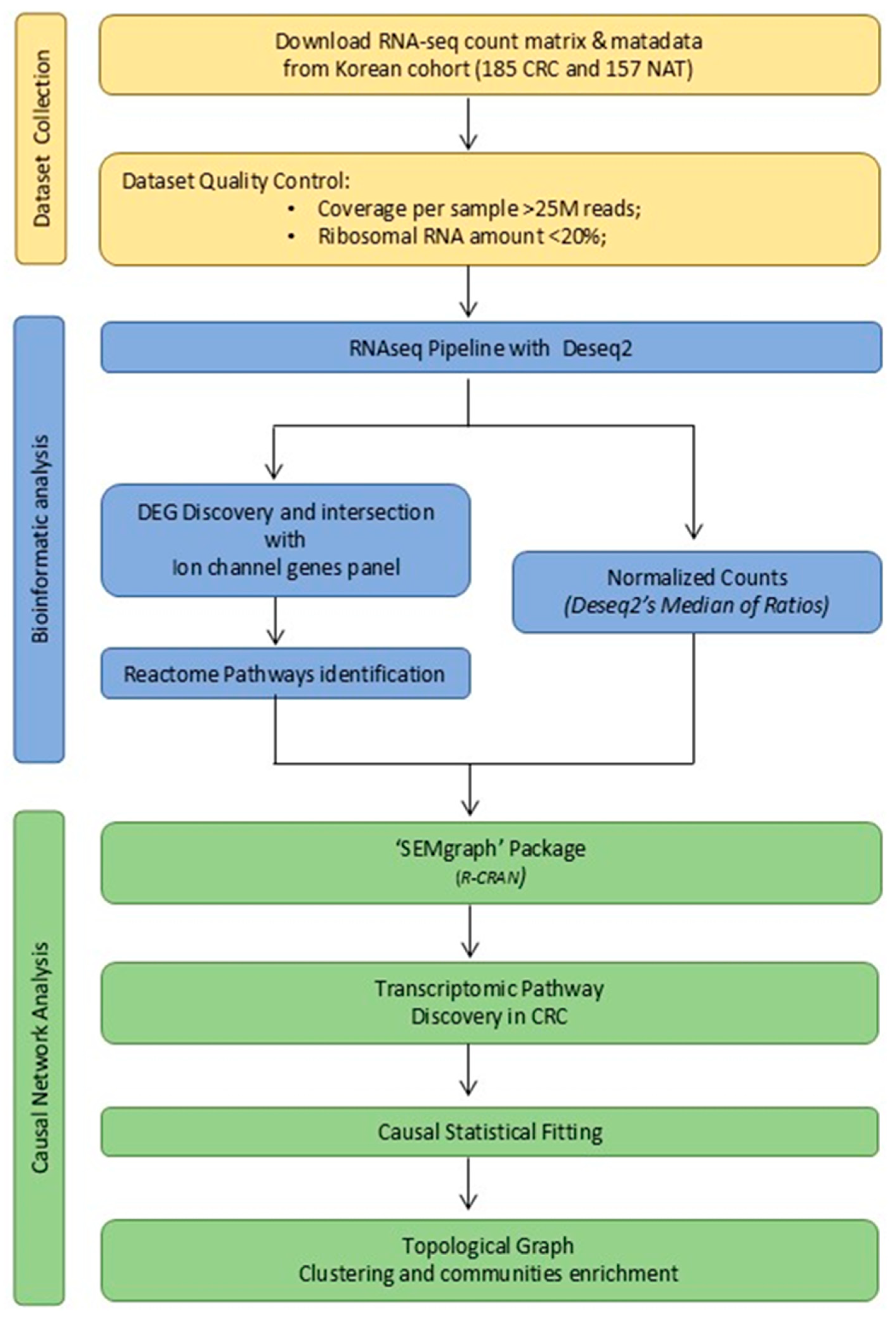
4.1. Data Collection and Study Design
4.2. Dataset Quality Control and Ion-Channel Gene Panel Selection
4.3. IC-DEGs Identification and Normalized Counts Production
4.4. IC-DEGs External Validation
4.5. Reactome Enrichment Analysis
4.6. Statistical Analysis
4.6.1. Analysis of the Active CRC-IC Module
4.6.2. Gene Communities Detection
4.7. Communities’ Enrichment Analysis
4.8. Statistical External Validation of the Active CRC-IC Module
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, L.; Xiao, H.; Jiang, J.; Li, B.; Liu, W.; Huang, W. The KMeansGraphMIL Model. Am. J. Pathol. 2025, 195, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Geng, S.; Luo, H.; Wang, W.; Mo, Y.-Q.; Luo, Q.; Wang, L.; Song, G.-B.; Sheng, J.-P.; Xu, B. Signaling Pathways Involved in Colorectal Cancer: Pathogenesis and Targeted Therapy. Signal Transduct. Target. Ther. 2024, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, X.; Chakravarti, D.; Shalapour, S.; DePinho, R.A. Genetic and Biological Hallmarks of Colorectal Cancer. Genes. Dev. 2021, 35, 787–820. [Google Scholar] [CrossRef]
- Gentile, R.; Feudi, D.; Sallicandro, L.; Biagini, A. Can the Tumor Microenvironment Alter Ion Channels? Unraveling Their Role in Cancer. Cancers 2025, 17, 1244. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, L.; Wang, M.; Zhou, S.; Lu, Y.; Cui, H.; Racanelli, A.C.; Zhang, L.; Ye, T.; Ding, B.; et al. Targeting Fibrosis: Mechanisms and Clinical Trials. Signal Transduct. Target. Ther. 2022, 7, 206. [Google Scholar] [CrossRef]
- Hu, M.; Feng, X.; Liu, Q.; Liu, S.; Huang, F.; Xu, H. The Ion Channels of Endomembranes. Physiol. Rev. 2024, 104, 1335–1385. [Google Scholar] [CrossRef]
- Karska, J.; Kowalski, S.; Saczko, J.; Moisescu, M.G.; Kulbacka, J. Mechanosensitive Ion Channels and Their Role in Cancer Cells. Membranes 2023, 13, 167. [Google Scholar] [CrossRef]
- Rao, V.; Perez-Neut, M.; Kaja, S.; Gentile, S. Voltage-Gated Ion Channels in Cancer Cell Proliferation. Cancers 2015, 7, 849–875. [Google Scholar] [CrossRef]
- Zhu, T.; Zhao, J.; Liu, J.; Tian, S.; Li, S.; Yuan, H. Advances in the Role of Ion Channels in Leukemia. Heliyon 2024, 10, e33452. [Google Scholar] [CrossRef]
- Mierke, C.T. Extracellular Matrix Cues Regulate Mechanosensing and Mechanotransduction of Cancer Cells. Cells 2024, 13, 96. [Google Scholar] [CrossRef]
- Anderson, K.J.; Cormier, R.T.; Scott, P.M. Role of Ion Channels in Gastrointestinal Cancer. World J. Gastroenterol. 2019, 25, 5732–5772. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Deng, Y.; Ye, J.; Luo, Y.; Weng, J.; He, Q.; Liu, F.; Li, M.; Liang, R.; Lin, Y.; et al. Store-Operated Ca2+ Entry as a Key Oncogenic Ca2+ Signaling Driving Tumor Invasion-Metastasis Cascade and Its Translational Potential. Cancer Lett. 2021, 516, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the Extracellular Matrix in Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Zhang, X.; Al-Danakh, A.; Zhu, X.; Feng, D.; Yang, L.; Wu, H.; Li, Y.; Wang, S.; Chen, Q.; Yang, D. Insights into the Mechanisms, Regulation, and Therapeutic Implications of Extracellular Matrix Stiffness in Cancer. Bioeng. Transl. Med. 2025, 10, e10698. [Google Scholar] [CrossRef]
- Lee, S.-A.; Cho, G.-J.; Kim, D.; Kim, D.-H. Biophysical Interplay between Extracellular Matrix Remodeling and Hypoxia Signaling in Regulating Cancer Metastasis. Front. Cell Dev. Biol. 2024, 12, 1335636. [Google Scholar] [CrossRef]
- Ji, C.; McCulloch, C.A. TRPV4 Integrates Matrix Mechanosensing with Ca 2+ Signaling to Regulate Extracellular Matrix Remodeling. FEBS J. 2021, 288, 5867–5887. [Google Scholar] [CrossRef]
- Dzobo, K.; Dandara, C. The Extracellular Matrix: Its Composition, Function, Remodeling, and Role in Tumorigenesis. Biomimetics 2023, 8, 146. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of Extracellular Matrix Remodelling in Tumour Progression and Metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Toss, M.S.; Miligy, I.M.; Gorringe, K.L.; AlKawaz, A.; Mittal, K.; Aneja, R.; Ellis, I.O.; Green, A.R.; Roxanis, I.; Rakha, E.A. Geometric Characteristics of Collagen Have Independent Prognostic Significance in Breast Ductal Carcinoma in Situ: An Image Analysis Study. Mod. Pathol. 2019, 32, 1473–1485. [Google Scholar] [CrossRef]
- Maqoud, F.; Zizzo, N.; Attimonelli, M.; Tinelli, A.; Passantino, G.; Antonacci, M.; Ranieri, G.; Tricarico, D. Immunohistochemical, Pharmacovigilance, and Omics Analyses Reveal the Involvement of ATP-Sensitive K+ Channel Subunits in Cancers: Role in Drug–Disease Interactions. Front. Pharmacol. 2023, 14, 1115543. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Farina, L.; Paci, P. Network-Based Approaches to Explore Complex Biological Systems towards Network Medicine. Genes 2018, 9, 437. [Google Scholar] [CrossRef] [PubMed]
- Beran, T.N.; Violato, C. Structural Equation Modeling in Medical Research: A Primer. BMC Res. Notes 2010, 3, 267. [Google Scholar] [CrossRef] [PubMed]
- Guido, D.; Maqoud, F.; Aloisio, M.; Mallardi, D.; Ura, B.; Gualandi, N.; Cocca, M.; Russo, F. Transcriptomic Module Discovery of Diarrhea-Predominant Irritable Bowel Syndrome: A Causal Network Inference Approach. Int. J. Mol. Sci. 2024, 25, 9322. [Google Scholar] [CrossRef]
- Kohestani, H.; Giuliani, A. Organization Principles of Biological Networks: An Explorative Study. Biosystems 2016, 141, 31–39. [Google Scholar] [CrossRef]
- Chartrand, G.; Zhang, P. Chromatic Graph Theory; Chapman and Hall/CRC: New York, NY, USA, 2019; ISBN 9780429438868. [Google Scholar]
- van Kesteren, E.-J. Structural Equations with Latent Variables: Computational Solutions for Modern Data Problems. Ph.D. Thesis, Utrecht University, Utrecht, The Netherlands, 2021. [Google Scholar]
- Lee, J.; Kim, J.-H.; Chu, H.B.K.; Oh, S.-T.; Kang, S.-B.; Lee, S.; Kim, D.-W.; Oh, H.-K.; Park, J.-H.; Kim, J.; et al. Comprehensive RNA-Sequencing Analysis of Colorectal Cancer in a Korean Cohort. Mol. Cells 2024, 47, 100033. [Google Scholar] [CrossRef]
- Grassi, M.; Palluzzi, F.; Tarantino, B. SEMgraph: An R Package for Causal Network Inference of High-Throughput Data with Structural Equation Models. Bioinformatics 2022, 38, 4829–4830. [Google Scholar] [CrossRef]
- Felmlee, M.A.; Jones, R.S.; Rodriguez-Cruz, V.; Follman, K.E.; Morris, M.E. Monocarboxylate Transporters (SLC16): Function, Regulation, and Role in Health and Disease. Pharmacol. Rev. 2020, 72, 466–485. [Google Scholar] [CrossRef]
- Sticova, E.; Jirsa, M. ABCB4 Disease: Many Faces of One Gene Deficiency. Ann. Hepatol. 2020, 19, 126–133. [Google Scholar] [CrossRef]
- Hu, H.; Wang, M.; Guan, X.; Yuan, Z.; Liu, Z.; Zou, C.; Wang, G.; Gao, X.; Wang, X. Loss of ABCB4 Attenuates the Caspase-Dependent Apoptosis Regulating Resistance to 5-Fu in Colorectal Cancer. Biosci. Rep. 2018, 38, BSR20171428. [Google Scholar] [CrossRef]
- Franchi, M.; Piperigkou, Z.; Mastronikolis, N.S.; Karamanos, N. Extracellular Matrix Biomechanical Roles and Adaptation in Health and Disease. FEBS J. 2024, 291, 430–440. [Google Scholar] [CrossRef]
- Haworth, A.S.; Brackenbury, W.J. Emerging Roles for Multifunctional Ion Channel Auxiliary Subunits in Cancer. Cell Calcium 2019, 80, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.J.; Huang, X. Ion Channels in Cancer: Orchestrators of Electrical Signaling and Cellular Crosstalk. Rev. Physiol. Biochem. Pharmacol. 2022, 183, 103–133. [Google Scholar] [PubMed]
- Altamura, C.; Gavazzo, P.; Pusch, M.; Desaphy, J.-F. Ion Channel Involvement in Tumor Drug Resistance. J. Pers. Med. 2022, 12, 210. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, X.; Zhao, W.; Xu, J.; Zhang, H.; Huang, T.; Wu, C.; Yang, J.; Tang, C.; Ye, Q.; et al. A Comprehensive Analysis of TRP-Related Gene Signature, and Immune Infiltration in Patients with Colorectal Cancer. Discov. Oncol. 2024, 15, 357. [Google Scholar] [CrossRef]
- Li, X.; Wu, D.; Li, Q.; Gu, J.; Gao, W.; Zhu, X.; Yin, W.; Zhu, R.; Zhu, L.; Jiao, N. Host-Microbiota Interactions Contributing to the Heterogeneous Tumor Microenvironment in Colorectal Cancer. Physiol. Genom. 2024, 56, 221–234. [Google Scholar] [CrossRef]
- Potier-Cartereau, M.; Raoul, W.; Weber, G.; Mahéo, K.; Rapetti-Mauss, R.; Gueguinou, M.; Buscaglia, P.; Goupille, C.; Le Goux, N.; Abdoul-Azize, S.; et al. Potassium and Calcium Channel Complexes as Novel Targets for Cancer Research. Rev. Physiol. Biochem. Pharmacol. 2022, 183, 157–176. [Google Scholar]
- Maeda, T.; Suzuki, A.; Koga, K.; Miyamoto, C.; Maehata, Y.; Ozawa, S.; Hata, R.-I.; Nagashima, Y.; Nabeshima, K.; Miyazaki, K.; et al. TRPM5 Mediates Acidic Extracellular PH Signaling and TRPM5 Inhibition Reduces Spontaneous Metastasis in Mouse B16-BL6 Melanoma Cells. Oncotarget 2017, 8, 78312–78326. [Google Scholar] [CrossRef]
- Li, L.; Kanemitsu, K.; Ohnishi, K.; Yamada, R.; Yano, H.; Fujiwara, Y.; Miyamoto, Y.; Mikami, Y.; Hibi, T.; Baba, H.; et al. CXCL10 Expression in Human Colorectal Cancer Tissue and Its Correlation With Serum Levels of CXCL10. Cancer Genom.-Proteom. 2024, 21, 54–64. [Google Scholar] [CrossRef]
- Waldner, M.J.; Foersch, S.; Neurath, M.F. Interleukin-6—A Key Regulator of Colorectal Cancer Development. Int. J. Biol. Sci. 2012, 8, 1248–1253. [Google Scholar] [CrossRef]
- Atreya, R.; Mudter, J.; Finotto, S.; Müllberg, J.; Jostock, T.; Wirtz, S.; Schütz, M.; Bartsch, B.; Holtmann, M.; Becker, C.; et al. Blockade of Interleukin 6 Trans Signaling Suppresses T-Cell Resistance against Apoptosis in Chronic Intestinal Inflammation: Evidence in Crohn Disease and Experimental Colitis In Vivo. Nat. Med. 2000, 6, 583–588. [Google Scholar] [CrossRef]
- Ji, Y.; Wang, Y.; Zou, J.; Liu, G.; Xia, M.; Ren, J.; Wang, D. Methyltransferase DNMT3B Promotes Colorectal Cancer Cell Proliferation by Inhibiting PLCG2. Acta Biochim. Biophys. Sin. 2024, 56, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Chen, Y.; Zhang, L.; Guo, X.; Wei, X.; Shao, Y.; Wang, D.; Wu, B. Effect of CHST11, a Novel Biomarker, on the Biological Functionalities of Clear Cell Renal Cell Carcinoma. Sci. Rep. 2024, 14, 7704. [Google Scholar] [CrossRef] [PubMed]
- Hope, C.; Emmerich, P.B.; Papadas, A.; Pagenkopf, A.; Matkowskyj, K.A.; Van De Hey, D.R.; Payne, S.N.; Clipson, L.; Callander, N.S.; Hematti, P.; et al. Versican-Derived Matrikines Regulate Batf3–Dendritic Cell Differentiation and Promote T Cell Infiltration in Colorectal Cancer. J. Immunol. 2017, 199, 1933–1941. [Google Scholar] [CrossRef] [PubMed]
- Papadas, A.; Asimakopoulos, F. Versican in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1272, 55–72. [Google Scholar]
- Wang, J.; Jiang, Y.-H.; Yang, P.-Y.; Liu, F. Increased Collagen Type V A2 (COL5A2) in Colorectal Cancer Is Associated with Poor Prognosis and Tumor Progression. Onco. Targets Ther. 2021, 14, 2991–3002. [Google Scholar] [CrossRef]
- Albrethsen, J.; Knol, J.C.; Piersma, S.R.; Pham, T.V.; de Wit, M.; Mongera, S.; Carvalho, B.; Verheul, H.M.W.; Fijneman, R.J.A.; Meijer, G.A.; et al. Subnuclear Proteomics in Colorectal Cancer. Mol. Cell. Proteom. 2010, 9, 988–1005. [Google Scholar] [CrossRef]
- Chuang, J.; Wang, C.; Guo, Y.; Valenzuela, V.; Wu, J.; Fakih, M. MAP2K1 Mutations in Advanced Colorectal Cancer Predict Poor Response to Anti-EGFR Therapy and to Vertical Targeting of MAPK Pathway. Clin. Color. Cancer 2021, 20, 72–78. [Google Scholar] [CrossRef]
- Mauri, G.; Patelli, G.; Gori, V.; Lauricella, C.; Mussolin, B.; Amatu, A.; Bencardino, K.; Tosi, F.; Bonazzina, E.; Bonoldi, E.; et al. Corrigendum: Case Report: MAP2K1 K57N Mutation Is Associated with Primary Resistance to Anti-EGFR Monoclonal Antibodies in Metastatic Colorectal Cancer. Front. Oncol. 2023, 13, 1147497. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Oliveros, J.C. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 5 March 2020).
- Edgar, R. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.gtexportal.org/home (accessed on 6 March 2025).
- Available online: https://www.cancer.gov/ccg/research/genome-sequencing/tcga/using-tcga-data/citing (accessed on 6 March 2025).
- Available online: https://www.cancer.gov/ccg/research/genome-sequencing/target/using-target-data/citing (accessed on 6 March 2025).
- Győrffy, B. Integrated Analysis of Public Datasets for the Discovery and Validation of Survival-Associated Genes in Solid Tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef] [PubMed]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- Zhao, T.; Liu, H.; Roeder, K.; Lafferty, J.; Wasserman, L. The Huge Package for High-Dimensional Undirected Graph Estimation in R. J. Mach. Learn. Res. 2012, 13, 1059–1062. [Google Scholar]
- Fisher, R.A. Frequency Distribution of the Values of the Correlation Coefficient in Samples from an Indefinitely Large Population. Biometrika 1915, 10, 507. [Google Scholar] [CrossRef]
- Kou, L.; Markowsky, G.; Berman, L. A Fast Algorithm for Steiner Trees. Acta Inform. 1981, 15, 141–145. [Google Scholar] [CrossRef]
- Pepe, D.; Grassi, M. Investigating Perturbed Pathway Modules from Gene Expression Data via Structural Equation Models. BMC Bioinform. 2014, 15, 132. [Google Scholar] [CrossRef]
- Jankova, J.; van de Geer, S. Confidence Intervals for High-Dimensional Inverse Covariance Estimation. Electron. J. Statist. 2015, 9, 1205–1229. [Google Scholar] [CrossRef]
- Hu, L.; Bentler, P.M. Cutoff Criteria for Fit Indexes in Covariance Structure Analysis: Conventional Criteria versus New Alternatives. Struct. Equ. Model. 1999, 6, 1–55. [Google Scholar] [CrossRef]
- Pons, P.; Latapy, M. Computing Communities in Large Networks Using Random Walks; Springer: Berlin/Heidelberg, Germany, 2005; pp. 284–293. [Google Scholar]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. G:Profiler—Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]


| Category | Value |
|---|---|
| CRC Sample Number | 185 |
| NAT Sample Number | 157 |
| Age (mean ± SD) | 62.3 ± 10.5 years |
| Sex | Male: 112 (60.5%), Female: 73 (39.5%) |
| Disease Grade | Grade I: 22 (11.9%), Grade II: 58 (31.4%), Grade III: 75 (40.5%), Grade IV: 30 (16.2%) |
| Community_ID | Community Gene Number | Intersection with Reduced Graph | Intersection Percentage |
|---|---|---|---|
| 35 | 5 | 5 | 100 |
| 36 | 5 | 4 | 80 |
| 19 | 13 | 8 | 62 |
| 29 | 5 | 3 | 60 |
| 30 | 6 | 3 | 50 |
| 7 | 66 | 32 | 48 |
| 20 | 10 | 2 | 20 |
| 4 | 48 | 1 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terzi, A.; Maqoud, F.; Guido, D.; Mallardi, D.; Aloisio, M.; Ura, B.; Gualandi, N.; Russo, F.; Giannelli, G. Ion Channel–Extracellular Matrix Interplay in Colorectal Cancer: A Network-Based Approach to Tumor Microenvironment Remodeling. Int. J. Mol. Sci. 2025, 26, 5147. https://doi.org/10.3390/ijms26115147
Terzi A, Maqoud F, Guido D, Mallardi D, Aloisio M, Ura B, Gualandi N, Russo F, Giannelli G. Ion Channel–Extracellular Matrix Interplay in Colorectal Cancer: A Network-Based Approach to Tumor Microenvironment Remodeling. International Journal of Molecular Sciences. 2025; 26(11):5147. https://doi.org/10.3390/ijms26115147
Chicago/Turabian StyleTerzi, Alberta, Fatima Maqoud, Davide Guido, Domenica Mallardi, Michelangelo Aloisio, Blendi Ura, Nicolò Gualandi, Francesco Russo, and Gianluigi Giannelli. 2025. "Ion Channel–Extracellular Matrix Interplay in Colorectal Cancer: A Network-Based Approach to Tumor Microenvironment Remodeling" International Journal of Molecular Sciences 26, no. 11: 5147. https://doi.org/10.3390/ijms26115147
APA StyleTerzi, A., Maqoud, F., Guido, D., Mallardi, D., Aloisio, M., Ura, B., Gualandi, N., Russo, F., & Giannelli, G. (2025). Ion Channel–Extracellular Matrix Interplay in Colorectal Cancer: A Network-Based Approach to Tumor Microenvironment Remodeling. International Journal of Molecular Sciences, 26(11), 5147. https://doi.org/10.3390/ijms26115147









