Low-Dose Salinomycin Alters Mitochondrial Function and Reprograms Global Metabolism in Burkitt Lymphoma
Abstract
1. Introduction
2. Results
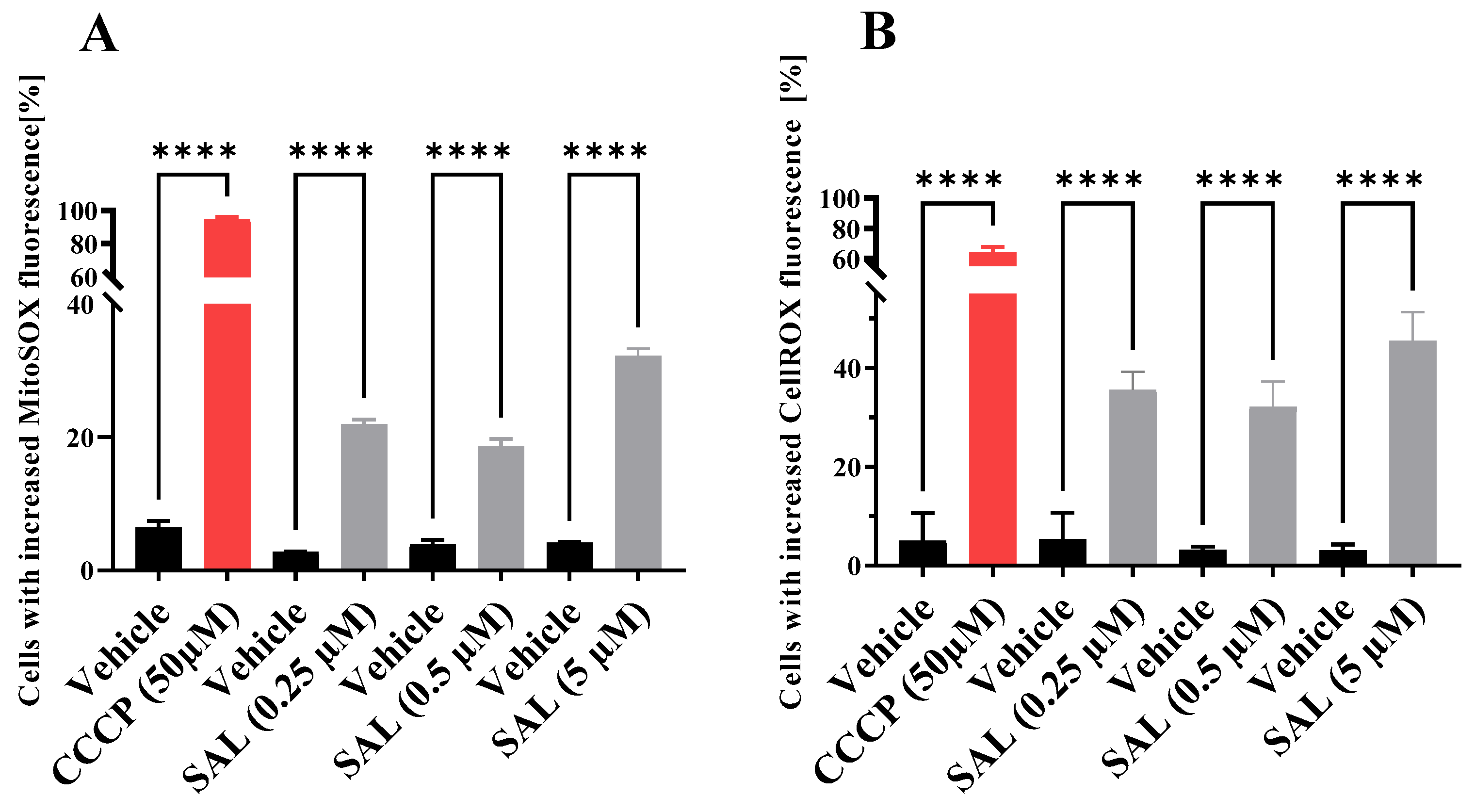
2.1. SAL Causes Mitochondrial Dysfunction and Induces Oxidative Stress in Burkitt Lymphoma Cells
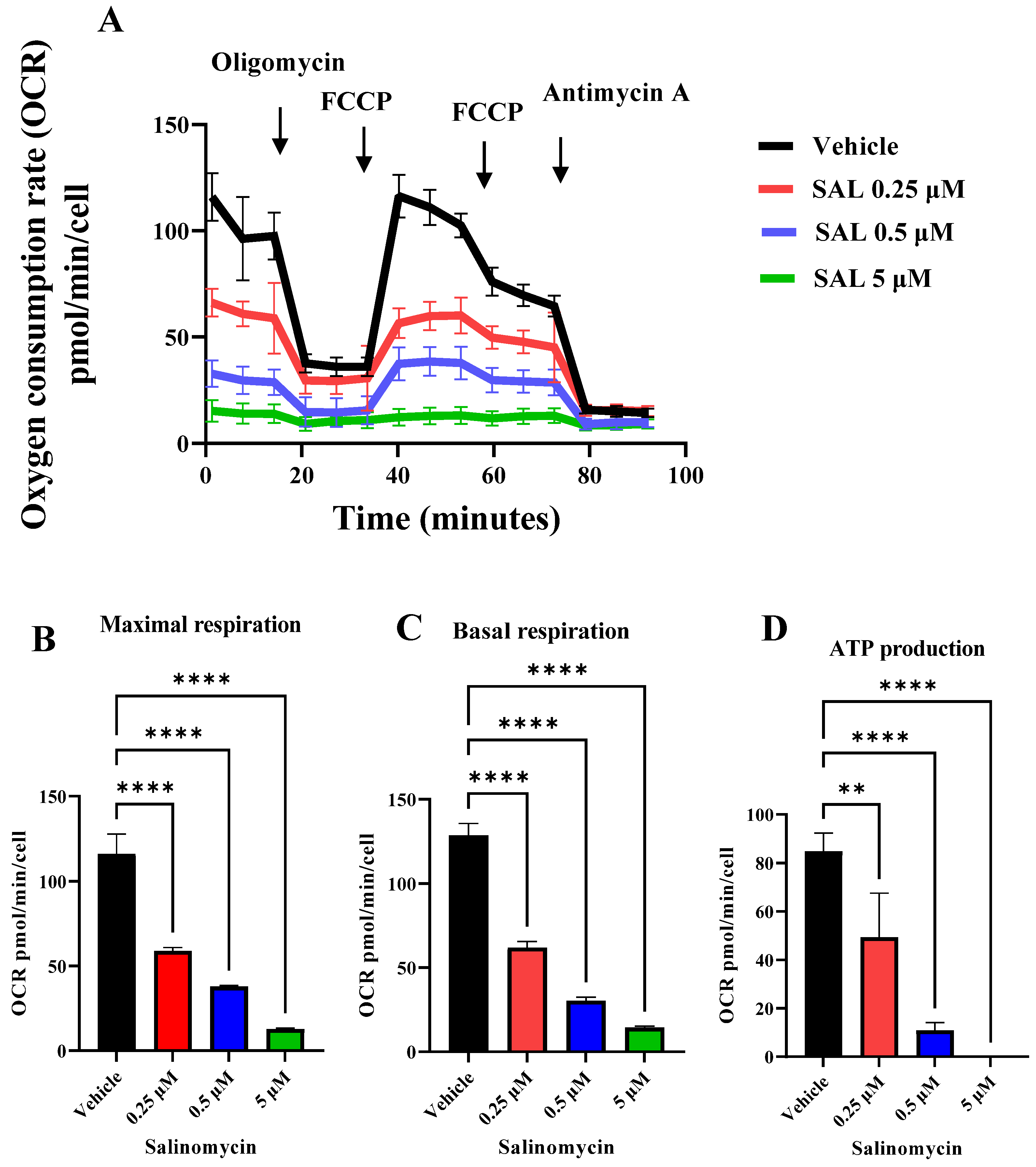
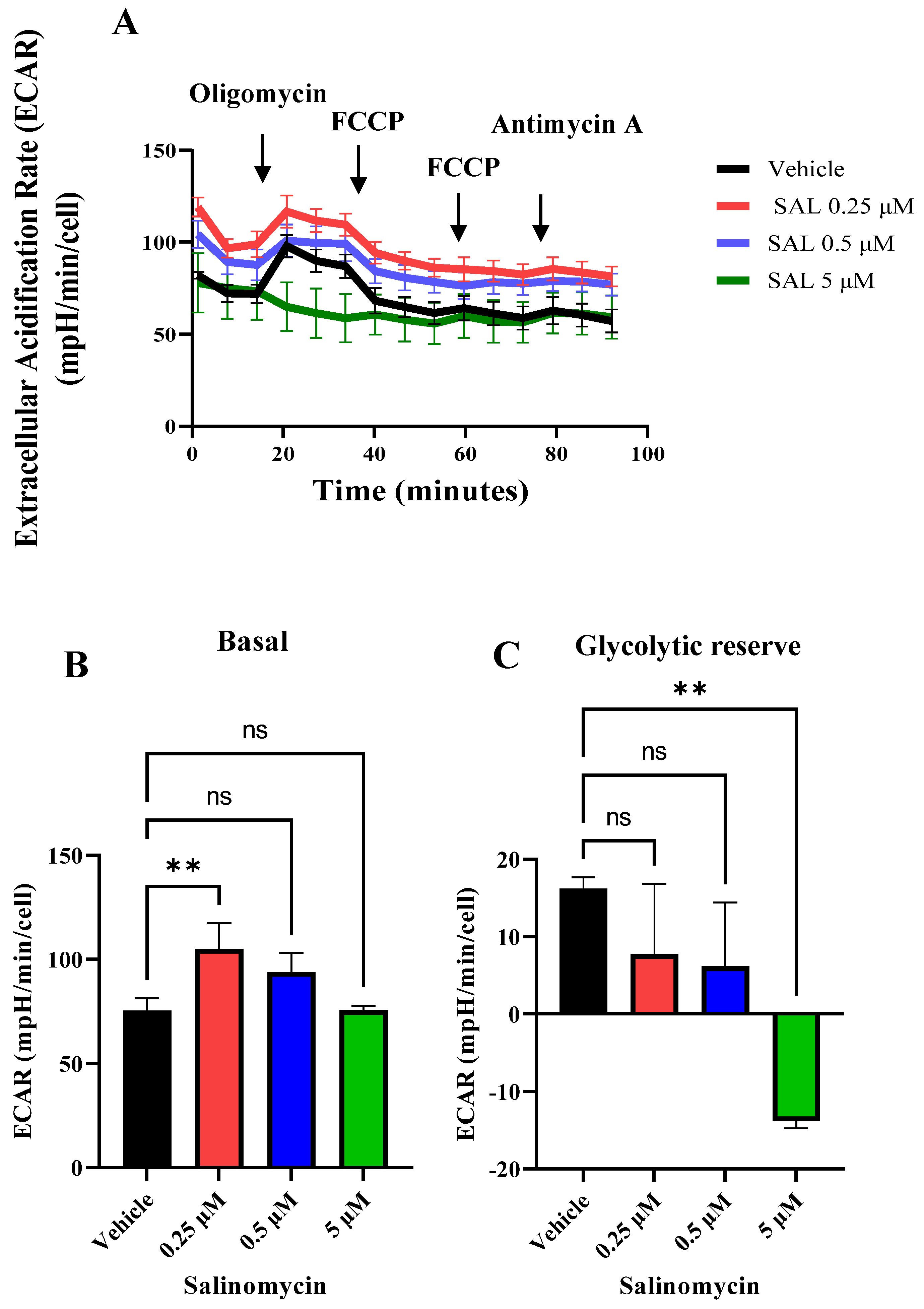
2.2. SAL Suppresses Mitochondrial Respiration and Glycolytic Activity
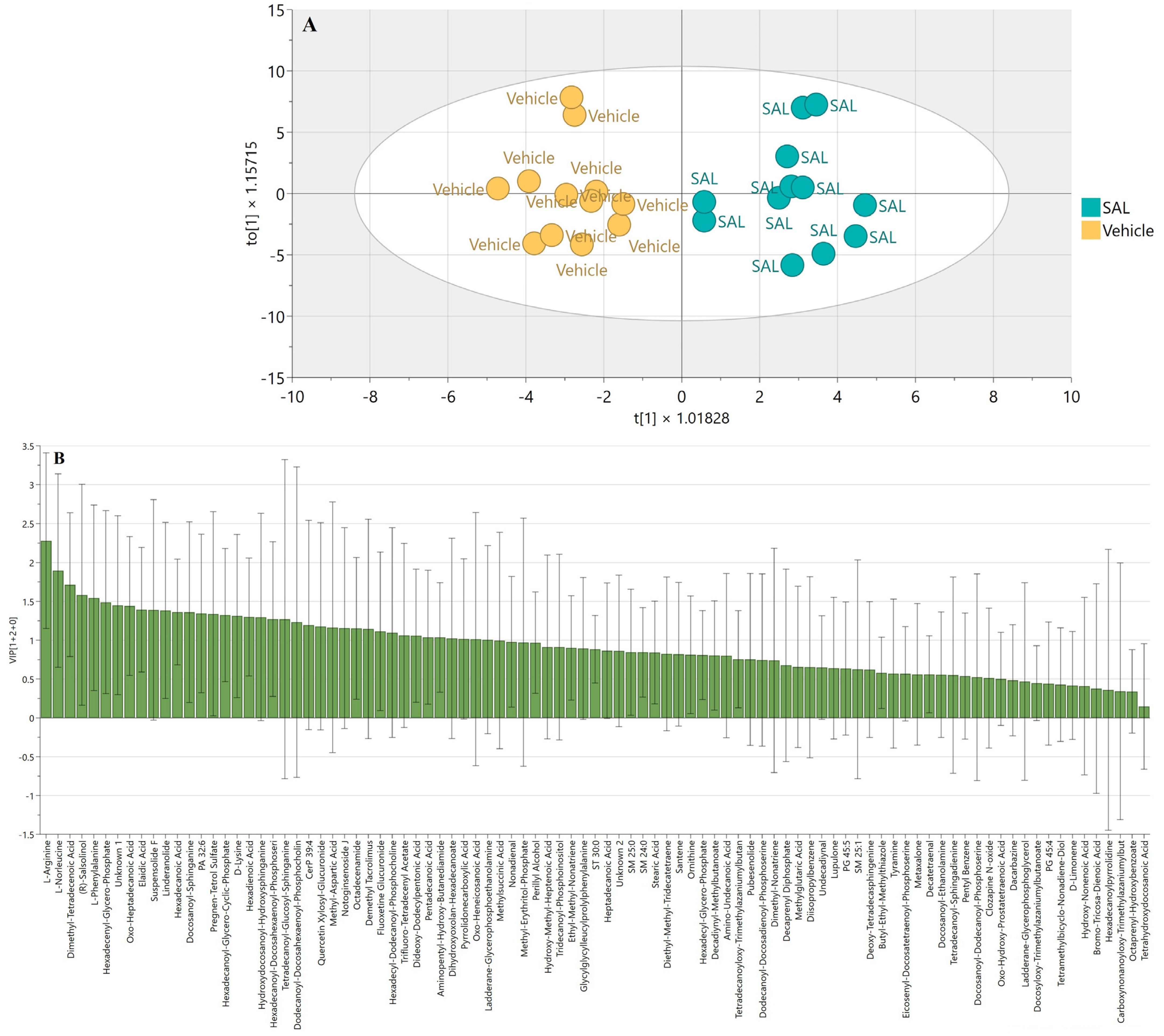
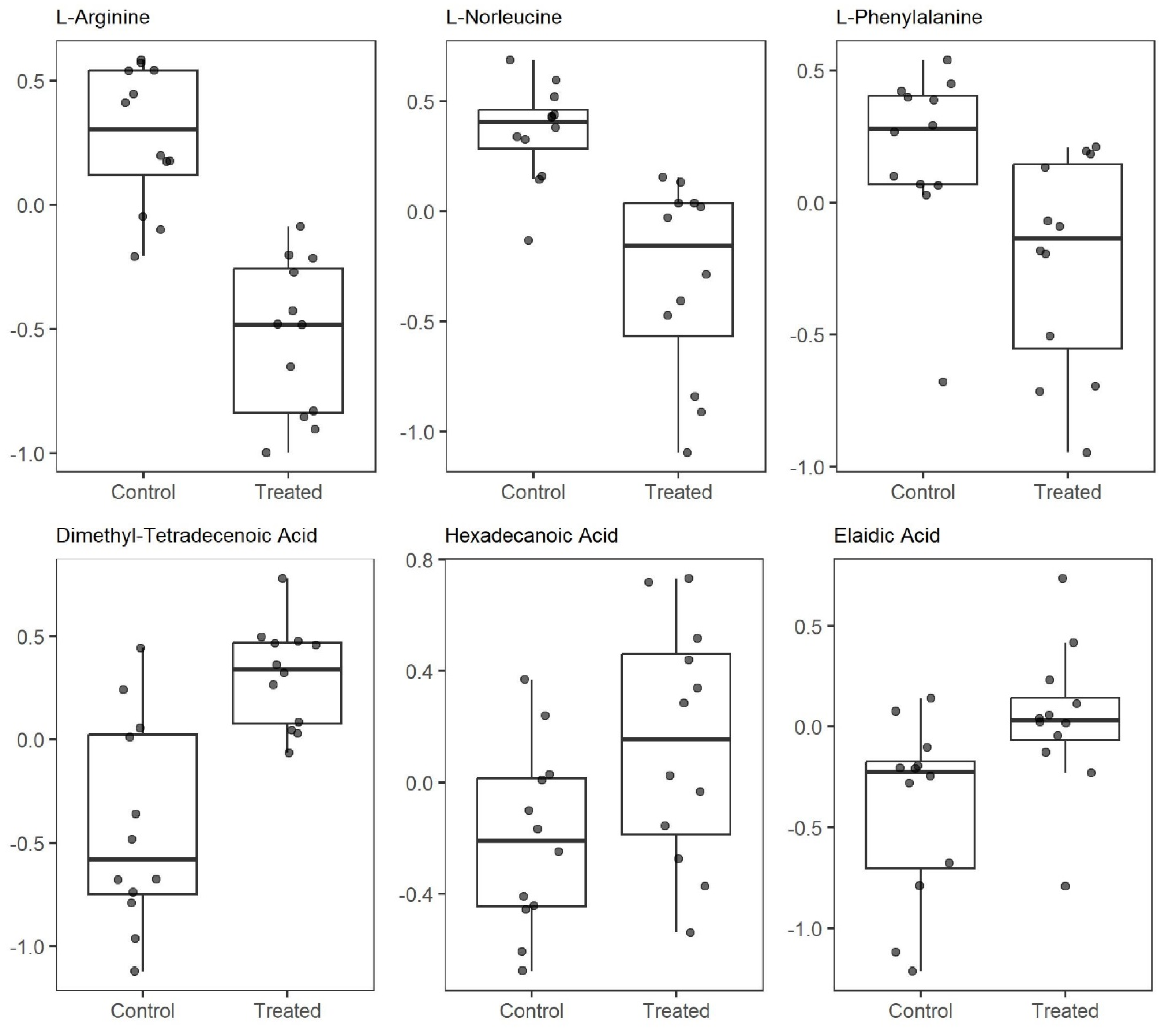
2.3. SAL Reprograms Burkitt Lymphoma Metabolism
3. Discussion
4. Materials and Methods
4.1. Cell Line and Chemicals
4.2. Mitochondrial Membrane Potential (MMP) Assessment
4.3. Assessment of Mitochondrial Superoxide and Global Oxidative Stress
4.4. Measurement of Cellular Bioenergetics Using Seahorse XFe96 Extracellular Flux Analyzer
4.5. Metabolomics Sample Preparation
4.5.1. Sample Overview
4.5.2. Data Treatment
- Missing Value Imputation
- 2.
- Data Transformation
- 3.
- Data Centering
- 4.
- Data Normalization
4.6. Statistical Analysis
4.6.1. Descriptive Statistical Analysis
- Distribution characteristics: kurtosis and skewness—evaluated deviations from normality;
- Gaussian function fit R2—assessed the goodness-of-fit to a normal distribution;
- One-sample Kolmogorov–Smirnov test—statistically tested normality assumptions.
4.6.2. Univariate Statistical Analysis
- One-sample test Kolmogorov–Smirnov—evaluated distribution normality;
- Student’s t-test—applied to normally distributed data;
- Mann–Whitney U test—used for non-normally distributed data or in the presence of outliers;
- Multiple comparison correction: Benjamini–Hochberg procedure—controlled the false discovery rate (FDR).
4.6.3. Multivariate Statistical Analysis
- Unsupervised Analysis:
- Principal Component Analysis (PCA): provided an initial unsupervised overview of data structure and helped identify outliers.
- 2.
- Supervised Modeling:
- Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA): used to enhance group separation and identify discriminant metabolites;
- Model optimization: performed through iterative component selection;
- Validation: conducted using leave-one-out cross-validation (LOOCV);
- Significance testing: assessed via cross-validated ANOVA (CV-ANOVA), with a p < 0.05 considered significant.
- 3.
- Model Quality Assessment:
- Permutation testing: evaluated potential overfitting.
- 4.
- Calculated model parameters:
- R2X/R2Y: goodness of fit of the model;
- Q2: goodness of prediction of the model.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASS1 | Arginosuccinate synthase 1 |
| CCCP | Carbonyl cyanide 3-chlorophenylhydrazone |
| CSC | Cancer stem cell |
| CV-ANOVA | Cross-validated ANOVA |
| DLBCL | Diffuse large B-cell lymphoma |
| DR5 | Death receptor-5 |
| ECAR | Extracellular acidification rate |
| ETC | Electron transport chain |
| FADD | Fas-associated protein with death domain |
| FCCP | Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone |
| LOOCV | Leave-one-out cross-validation |
| MMP | Mitochondrial membrane potential |
| MSEA | Metabolite set enrichment analysis |
| mtROS | Mitochondrial reactive oxygen species |
| OCR | Oxygen consumption rates |
| OPLS-DA | Orthogonal Partial Least Squares Discriminant Analysis |
| OXPHOS | Oxidative phosphorylation |
| PCA | Principal Component Analysis |
| SAL | Salinomycin |
| SLC | Solute carrier |
| TCA | Tricarboxylic acid |
| TMRM | Tetramethylrhodamine methyl ester |
| VIP | Variable Importance in Projection |
References
- Dutton, C.J.; Banks, B.J.; Cooper, C.B. Polyether ionophores. Nat. Prod. Rep. 1995, 12, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Shibuya, M.; Sugawara, H.; Kawaguchi, O.; Hirsoe, C. Salinomycin, a new polyether antibiotic. J. Antibiot. 1974, 27, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Ekinci, I.B.; Chlodowska, A.; Olejnik, M. Ionophore Toxicity in Animals: A Review of Clinical and Molecular Aspects. Int. J. Mol. Sci. 2023, 24, 1696. [Google Scholar] [CrossRef]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; He, L.; Dai, W.Q.; Xu, Y.P.; Wu, D.; Lin, C.L.; Wu, S.M.; Cheng, P.; Zhang, Y.; Shen, M.; et al. Salinomycin inhibits proliferation and induces apoptosis of human hepatocellular carcinoma cells in vitro and in vivo. PLoS ONE 2012, 7, e50638. [Google Scholar] [CrossRef]
- Lu, D.; Choi, M.Y.; Yu, J.; Castro, J.E.; Kipps, T.J.; Carson, D.A. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc. Natl. Acad. Sci. USA 2011, 108, 13253–13257. [Google Scholar] [CrossRef]
- Jangamreddy, J.R.; Ghavami, S.; Grabarek, J.; Kratz, G.; Wiechec, E.; Fredriksson, B.A.; Rao Pariti, R.K.; Cieslar-Pobuda, A.; Panigrahi, S.; Los, M.J. Salinomycin induces activation of autophagy, mitophagy and affects mitochondrial polarity: Differences between primary and cancer cells. Biochim. Biophys. Acta 2013, 1833, 2057–2069. [Google Scholar] [CrossRef]
- Parajuli, B.; Lee, H.G.; Kwon, S.H.; Cha, S.D.; Shin, S.J.; Lee, G.H.; Bae, I.; Cho, C.H. Salinomycin inhibits Akt/NF-kappaB and induces apoptosis in cisplatin resistant ovarian cancer cells. Cancer Epidemiol. 2013, 37, 512–517. [Google Scholar] [CrossRef]
- Kim, W.K.; Kim, J.H.; Yoon, K.; Kim, S.; Ro, J.; Kang, H.S.; Yoon, S. Salinomycin, a p-glycoprotein inhibitor, sensitizes radiation-treated cancer cells by increasing DNA damage and inducing G2 arrest. Investig. New Drugs 2012, 30, 1311–1318. [Google Scholar] [CrossRef]
- Torun, A.; Zdanowicz, A.; Miazek-Zapala, N.; Zapala, P.; Pradhan, B.; Jedrzejczyk, M.; Ciechanowicz, A.; Pilch, Z.; Skorzynski, M.; Slabicki, M.; et al. Potassium/sodium cation carriers robustly up-regulate CD20 antigen by targeting MYC, and synergize with anti-CD20 immunotherapies to eliminate malignant B cells. Haematologica 2024. [Google Scholar] [CrossRef]
- Casan, J.M.L.; Wong, J.; Northcott, M.J.; Opat, S. Anti-CD20 monoclonal antibodies: Reviewing a revolution. Hum. Vaccines Immunother. 2018, 14, 2820–2841. [Google Scholar] [CrossRef] [PubMed]
- Tomita, A. Genetic and Epigenetic Modulation of CD20 Expression in B-Cell Malignancies: Molecular Mechanisms and Significance to Rituximab Resistance. J. Clin. Exp. Hematop. 2016, 56, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Story, P.; Doube, A. A case of human poisoning by salinomycin, an agricultural antibiotic. N. Z. Med. J. 2004, 117, U799. [Google Scholar]
- Naujokat, C.; Steinhart, R. Salinomycin as a drug for targeting human cancer stem cells. J. Biomed. Biotechnol. 2012, 2012, 950658. [Google Scholar] [CrossRef]
- Boehmerle, W.; Muenzfeld, H.; Springer, A.; Huehnchen, P.; Endres, M. Specific targeting of neurotoxic side effects and pharmacological profile of the novel cancer stem cell drug salinomycin in mice. J. Mol. Med. 2014, 92, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Soni, V.; Nagar, A.; Bardiya, R.; Mara, J.; Von Suskil, L.; Rose, S.; Sonawane, C. A Concise Review of Prodigious Salinomycin and Its Derivatives Effective in Treatment of Breast Cancer: (2012–2022). Int. J. Transl. Med. 2023, 3, 217–245. [Google Scholar] [CrossRef]
- Antoszczak, M.; Huczynski, A. Salinomycin and its derivatives—A new class of multiple-targeted “magic bullets”. Eur. J. Med. Chem. 2019, 176, 208–227. [Google Scholar] [CrossRef]
- Versini, A.; Colombeau, L.; Hienzsch, A.; Gaillet, C.; Retailleau, P.; Debieu, S.; Muller, S.; Caneque, T.; Rodriguez, R. Salinomycin Derivatives Kill Breast Cancer Stem Cells by Lysosomal Iron Targeting. Chemistry 2020, 26, 7416–7424. [Google Scholar] [CrossRef]
- Antoszczak, M. A comprehensive review of salinomycin derivatives as potent anticancer and anti-CSCs agents. Eur. J. Med. Chem. 2019, 166, 48–64. [Google Scholar] [CrossRef]
- Boehmerle, W.; Endres, M. Salinomycin induces calpain and cytochrome c-mediated neuronal cell death. Cell Death Dis. 2011, 2, e168. [Google Scholar] [CrossRef]
- Rutkowski, J.; Brzezinski, B. Structures and properties of naturally occurring polyether antibiotics. BioMed Res. Int. 2013, 2013, 162513. [Google Scholar] [CrossRef]
- Versini, A.; Saier, L.; Sindikubwabo, F.; Muller, S.; Caneque, T.; Rodriguez, R. Chemical biology of salinomycin. Tetrahedron 2018, 74, 5585–5614. [Google Scholar] [CrossRef]
- Mai, T.T.; Hamai, A.; Hienzsch, A.; Caneque, T.; Muller, S.; Wicinski, J.; Cabaud, O.; Leroy, C.; David, A.; Acevedo, V.; et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 2017, 9, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Cosialls, E.; Pacreau, E.; Duruel, C.; Ceccacci, S.; Elhage, R.; Desterke, C.; Roger, K.; Guerrera, C.; Ducloux, R.; Souquere, S.; et al. mTOR inhibition suppresses salinomycin-induced ferroptosis in breast cancer stem cells by ironing out mitochondrial dysfunctions. Cell Death Dis. 2023, 14, 744. [Google Scholar] [CrossRef]
- Rokitskaya, T.I.; Firsov, A.M.; Khailova, L.S.; Kotova, E.A.; Antonenko, Y.N. Selectivity of cation transport across lipid membranes by the antibiotic salinomycin. Biochim. Biophys. Acta Biomembr. 2023, 1865, 184182. [Google Scholar] [CrossRef]
- Verdoodt, B.; Vogt, M.; Schmitz, I.; Liffers, S.T.; Tannapfel, A.; Mirmohammadsadegh, A. Salinomycin induces autophagy in colon and breast cancer cells with concomitant generation of reactive oxygen species. PLoS ONE 2012, 7, e44132. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Yu, S.N.; Lee, S.Y.; Chun, S.S.; Choi, Y.L.; Park, Y.M.; Song, C.S.; Chatterjee, B.; Ahn, S.C. Salinomycin-induced apoptosis of human prostate cancer cells due to accumulated reactive oxygen species and mitochondrial membrane depolarization. Biochem. Biophys. Res. Commun. 2011, 413, 80–86. [Google Scholar] [CrossRef]
- Zhu, L.Q.; Zhen, Y.F.; Zhang, Y.; Guo, Z.X.; Dai, J.; Wang, X.D. Salinomycin activates AMP-activated protein kinase-dependent autophagy in cultured osteoblastoma cells: A negative regulator against cell apoptosis. PLoS ONE 2013, 8, e84175. [Google Scholar] [CrossRef]
- Manago, A.; Leanza, L.; Carraretto, L.; Sassi, N.; Grancara, S.; Quintana-Cabrera, R.; Trimarco, V.; Toninello, A.; Scorrano, L.; Trentin, L.; et al. Early effects of the antineoplastic agent salinomycin on mitochondrial function. Cell Death Dis. 2015, 6, e1930. [Google Scholar] [CrossRef]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Zdanowicz, A.; Grosicka-Maciag, E. The Interplay between Autophagy and Mitochondria in Cancer. Int. J. Mol. Sci. 2024, 25, 9143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, C.; Feng, C.; Yan, C.; Yu, Y.; Chen, Z.; Guo, C.; Wang, X. Role of mitochondrial reactive oxygen species in homeostasis regulation. Redox Rep. 2022, 27, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Casanova, A.; Wevers, A.; Navarro-Ledesma, S.; Pruimboom, L. Mitochondria: It is all about energy. Front. Physiol. 2023, 14, 1114231. [Google Scholar] [CrossRef]
- Li, J.; Min, Y. Pre-clinical evidence that salinomycin is active against retinoblastoma via inducing mitochondrial dysfunction, oxidative damage and AMPK activation. J. Bioenerg. Biomembr. 2021, 53, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hao, Y.; Li, Y.; Zheng, Y.; Dai, J.; Zhong, F.; Wei, W.; Fang, Z. Salinomycin induces autophagic cell death in salinomycin-sensitive melanoma cells through inhibition of autophagic flux. Sci. Rep. 2020, 10, 18515. [Google Scholar] [CrossRef]
- Xipell, E.; Gonzalez-Huarriz, M.; Martinez de Irujo, J.J.; Garcia-Garzon, A.; Lang, F.F.; Jiang, H.; Fueyo, J.; Gomez-Manzano, C.; Alonso, M.M. Salinomycin induced ROS results in abortive autophagy and leads to regulated necrosis in glioblastoma. Oncotarget 2016, 7, 30626–30641. [Google Scholar] [CrossRef]
- Yu, S.N.; Kim, S.H.; Kim, K.Y.; Ji, J.H.; Seo, Y.K.; Yu, H.S.; Ahn, S.C. Salinomycin induces endoplasmic reticulum stress-mediated autophagy and apoptosis through generation of reactive oxygen species in human glioma U87MG cells. Oncol. Rep. 2017, 37, 3321–3328. [Google Scholar] [CrossRef]
- Jedrzejczyk, M.; Sulik, M.; Mielczarek-Puta, M.; Lim, G.Y.; Podsiad, M.; Hoser, J.; Bednarczyk, P.; Struga, M.; Huczynski, A. Anticancer activity of salinomycin quaternary phosphonium salts. Eur. J. Med. Chem. 2025, 282, 117055. [Google Scholar] [CrossRef]
- Korshunov, S.S.; Skulachev, V.P.; Starkov, A.A. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997, 416, 15–18. [Google Scholar] [CrossRef]
- Suski, J.M.; Lebiedzinska, M.; Bonora, M.; Pinton, P.; Duszynski, J.; Wieckowski, M.R. Relation between mitochondrial membrane potential and ROS formation. Methods Mol. Biol. 2012, 810, 183–205. [Google Scholar] [CrossRef]
- Salaroglio, I.C.; Belisario, D.C.; Akman, M.; La Vecchia, S.; Godel, M.; Anobile, D.P.; Ortone, G.; Digiovanni, S.; Fontana, S.; Costamagna, C.; et al. Mitochondrial ROS drive resistance to chemotherapy and immune-killing in hypoxic non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2022, 41, 243. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Hsu, S.C.; Ann, D.K.; Yen, Y.; Kung, H.J. Arginine Signaling and Cancer Metabolism. Cancers 2021, 13, 3541. [Google Scholar] [CrossRef]
- Qiu, F.; Chen, Y.R.; Liu, X.; Chu, C.Y.; Shen, L.J.; Xu, J.; Gaur, S.; Forman, H.J.; Zhang, H.; Zheng, S.; et al. Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Sci. Signal. 2014, 7, ra31. [Google Scholar] [CrossRef]
- Cheng, C.T.; Qi, Y.; Wang, Y.C.; Chi, K.K.; Chung, Y.; Ouyang, C.; Chen, Y.R.; Oh, M.E.; Sheng, X.; Tang, Y.; et al. Arginine starvation kills tumor cells through aspartate exhaustion and mitochondrial dysfunction. Commun. Biol. 2018, 1, 178. [Google Scholar] [CrossRef]
- Kremer, J.C.; Prudner, B.C.; Lange, S.E.S.; Bean, G.R.; Schultze, M.B.; Brashears, C.B.; Radyk, M.D.; Redlich, N.; Tzeng, S.C.; Kami, K.; et al. Arginine Deprivation Inhibits the Warburg Effect and Upregulates Glutamine Anaplerosis and Serine Biosynthesis in ASS1-Deficient Cancers. Cell Rep. 2017, 18, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Brashears, C.B.; Barlin, M.; Ehrhardt, W.R.; Rathore, R.; Schultze, M.; Tzeng, S.C.; Van Tine, B.A.; Held, J.M. Systems level profiling of arginine starvation reveals MYC and ERK adaptive metabolic reprogramming. Cell Death Dis. 2020, 11, 662. [Google Scholar] [CrossRef]
- Ji, J.X.; Cochrane, D.R.; Tessier-Cloutier, B.; Chen, S.Y.; Ho, G.; Pathak, K.V.; Alcazar, I.N.; Farnell, D.; Leung, S.; Cheng, A.; et al. Arginine Depletion Therapy with ADI-PEG20 Limits Tumor Growth in Argininosuccinate Synthase-Deficient Ovarian Cancer, Including Small-Cell Carcinoma of the Ovary, Hypercalcemic Type. Clin. Cancer Res. 2020, 26, 4402–4413. [Google Scholar] [CrossRef] [PubMed]
- Bean, G.R.; Kremer, J.C.; Prudner, B.C.; Schenone, A.D.; Yao, J.C.; Schultze, M.B.; Chen, D.Y.; Tanas, M.R.; Adkins, D.R.; Bomalaski, J.; et al. A metabolic synthetic lethal strategy with arginine deprivation and chloroquine leads to cell death in ASS1-deficient sarcomas. Cell Death Dis. 2016, 7, e2406. [Google Scholar] [CrossRef]
- Savaraj, N.; Wu, C.; Kuo, M.T.; You, M.; Wangpaichitr, M.; Robles, C.; Spector, S.; Feun, L. The relationship of arginine deprivation, argininosuccinate synthetase and cell death in melanoma. Drug Target Insights 2007, 2, 119–128. [Google Scholar] [CrossRef]
- Delage, B.; Luong, P.; Maharaj, L.; O’Riain, C.; Syed, N.; Crook, T.; Hatzimichael, E.; Papoudou-Bai, A.; Mitchell, T.J.; Whittaker, S.J.; et al. Promoter methylation of argininosuccinate synthetase-1 sensitises lymphomas to arginine deiminase treatment, autophagy and caspase-dependent apoptosis. Cell Death Dis. 2012, 3, e342. [Google Scholar] [CrossRef]
- Fultang, L.; Vardon, A.; De Santo, C.; Mussai, F. Molecular basis and current strategies of therapeutic arginine depletion for cancer. Int. J. Cancer 2016, 139, 501–509. [Google Scholar] [CrossRef] [PubMed]



| Step | Type | Method | Equation |
|---|---|---|---|
| 1 | Missing Value Imputation | Half-minimum | |
| 2 | Data Transformation | Log10 | |
| 3 | Data Centering | Min–max centering | |
| 4 | Data Normalization | Pareto |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zdanowicz, A.; Ilchenko, O.; Ciechanowicz, A.; Chi, H.; Struga, M.; Pyrzynska, B. Low-Dose Salinomycin Alters Mitochondrial Function and Reprograms Global Metabolism in Burkitt Lymphoma. Int. J. Mol. Sci. 2025, 26, 5125. https://doi.org/10.3390/ijms26115125
Zdanowicz A, Ilchenko O, Ciechanowicz A, Chi H, Struga M, Pyrzynska B. Low-Dose Salinomycin Alters Mitochondrial Function and Reprograms Global Metabolism in Burkitt Lymphoma. International Journal of Molecular Sciences. 2025; 26(11):5125. https://doi.org/10.3390/ijms26115125
Chicago/Turabian StyleZdanowicz, Aleksandra, Oleksandr Ilchenko, Andrzej Ciechanowicz, Haoyu Chi, Marta Struga, and Beata Pyrzynska. 2025. "Low-Dose Salinomycin Alters Mitochondrial Function and Reprograms Global Metabolism in Burkitt Lymphoma" International Journal of Molecular Sciences 26, no. 11: 5125. https://doi.org/10.3390/ijms26115125
APA StyleZdanowicz, A., Ilchenko, O., Ciechanowicz, A., Chi, H., Struga, M., & Pyrzynska, B. (2025). Low-Dose Salinomycin Alters Mitochondrial Function and Reprograms Global Metabolism in Burkitt Lymphoma. International Journal of Molecular Sciences, 26(11), 5125. https://doi.org/10.3390/ijms26115125






