Integrative Analysis of Immune- and Metabolism-Related Genes Identifies Robust Prognostic Signature and PYCR1 as a Carcinogenic Regulator in Clear Cell Renal Cell Carcinoma
Abstract
1. Introduction
2. Results
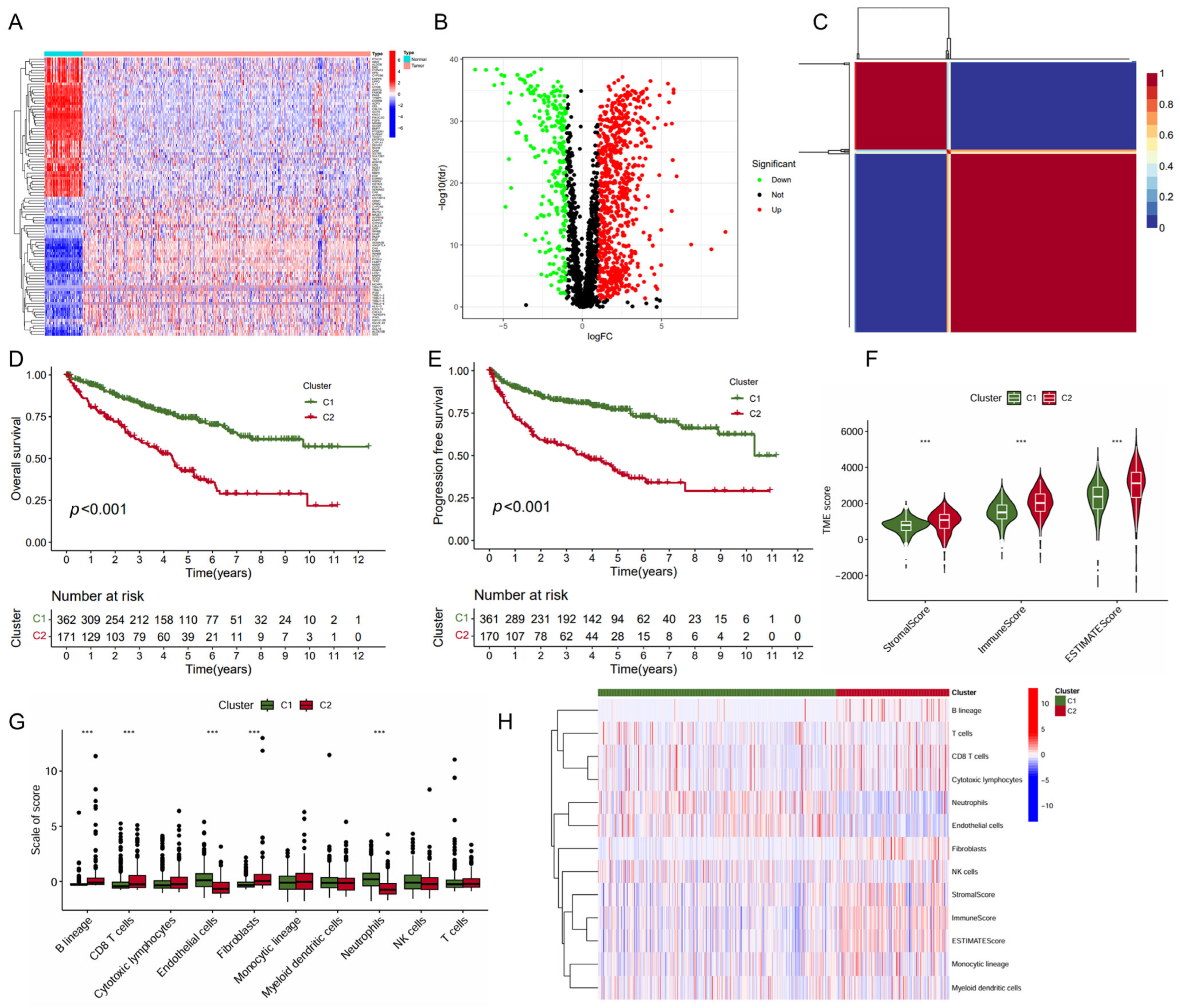
2.1. Identification of ccRCC Subtypes with Prognosis Differences Based on Immune- and Metabolism-Related Genes
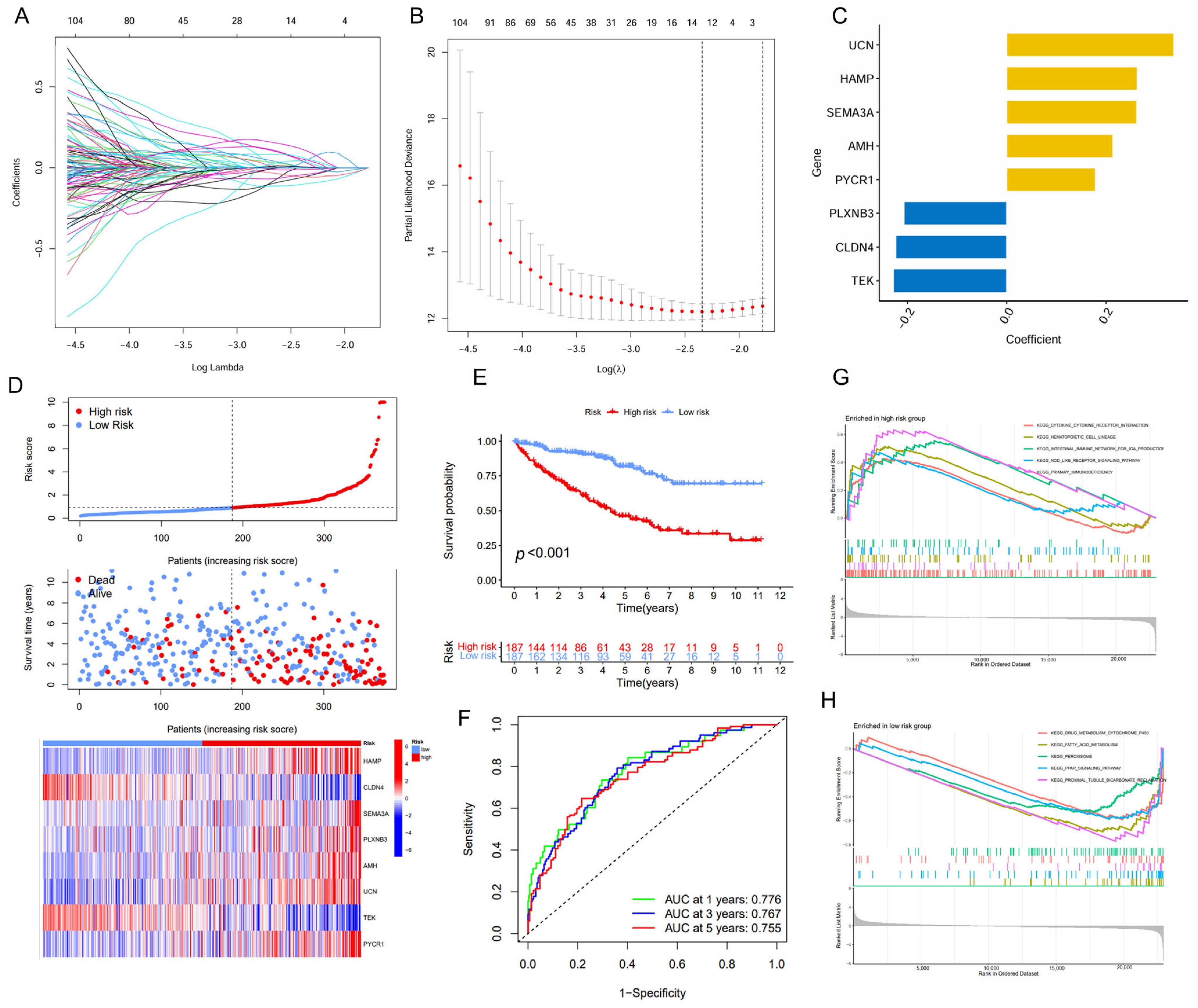
2.2. Construction of a Prognostic Signature with Immune- and Metabolism-Related Genes
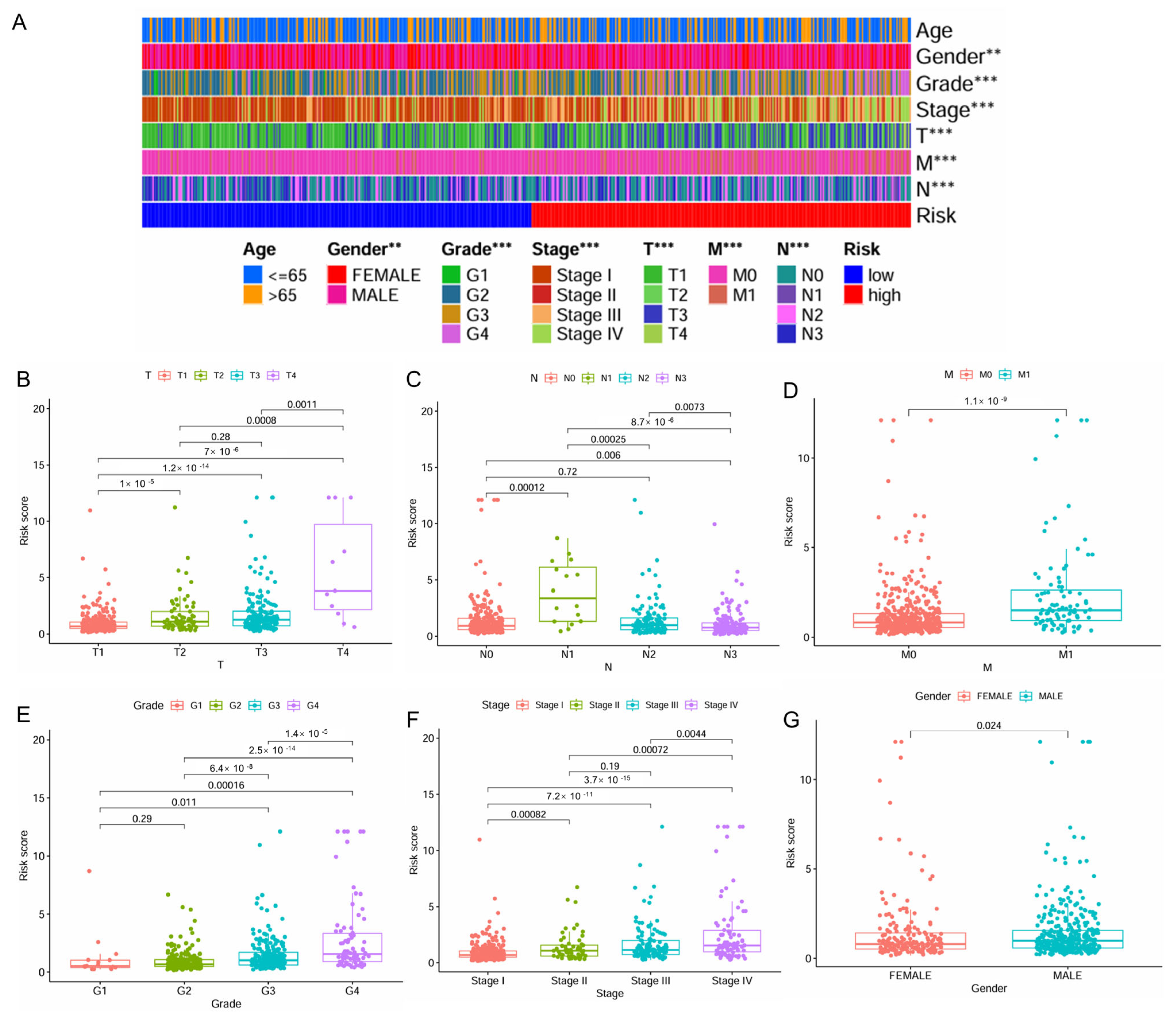
2.3. IMRG Score Correlates with Patient Clinicopathologic Characters
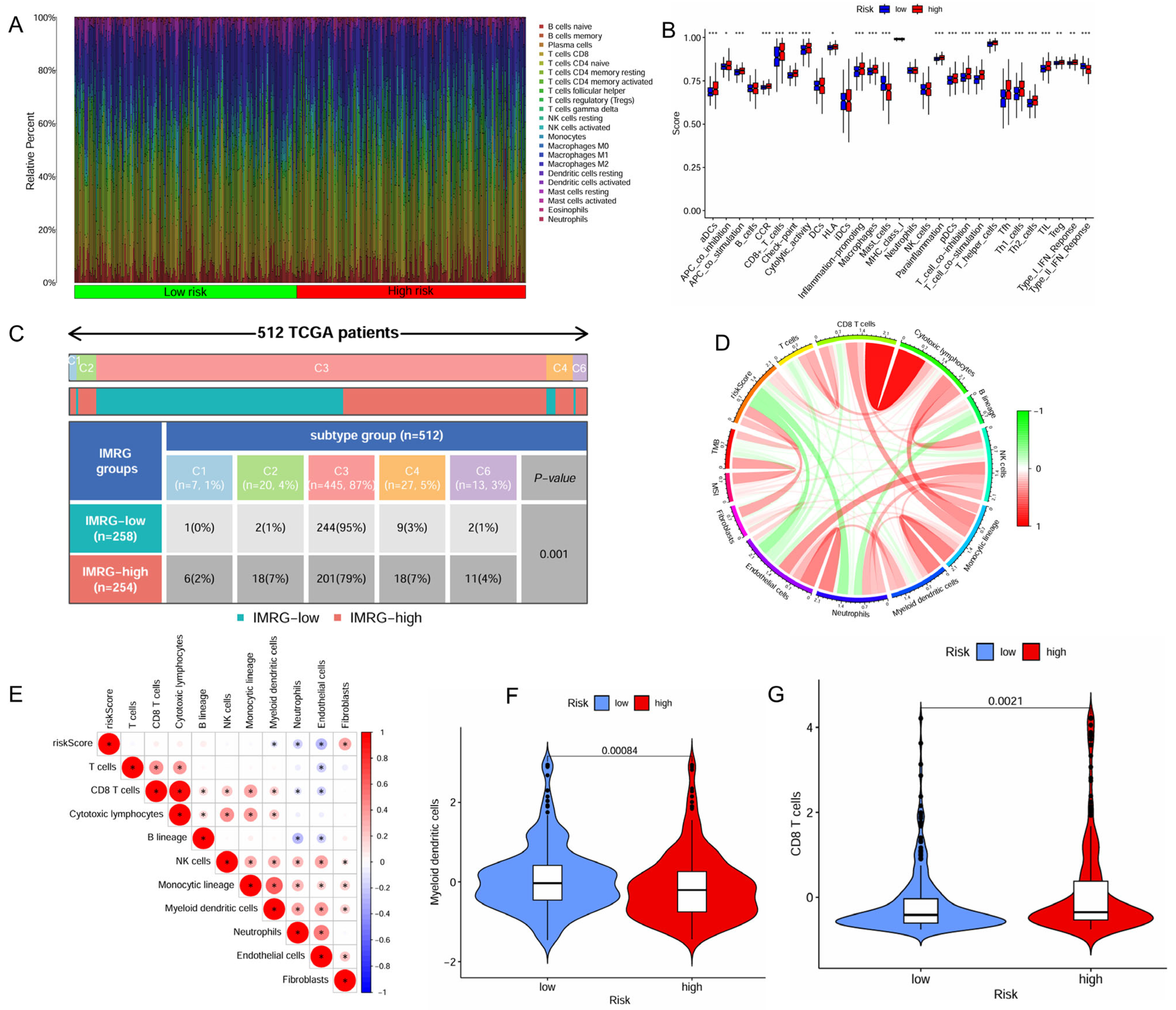
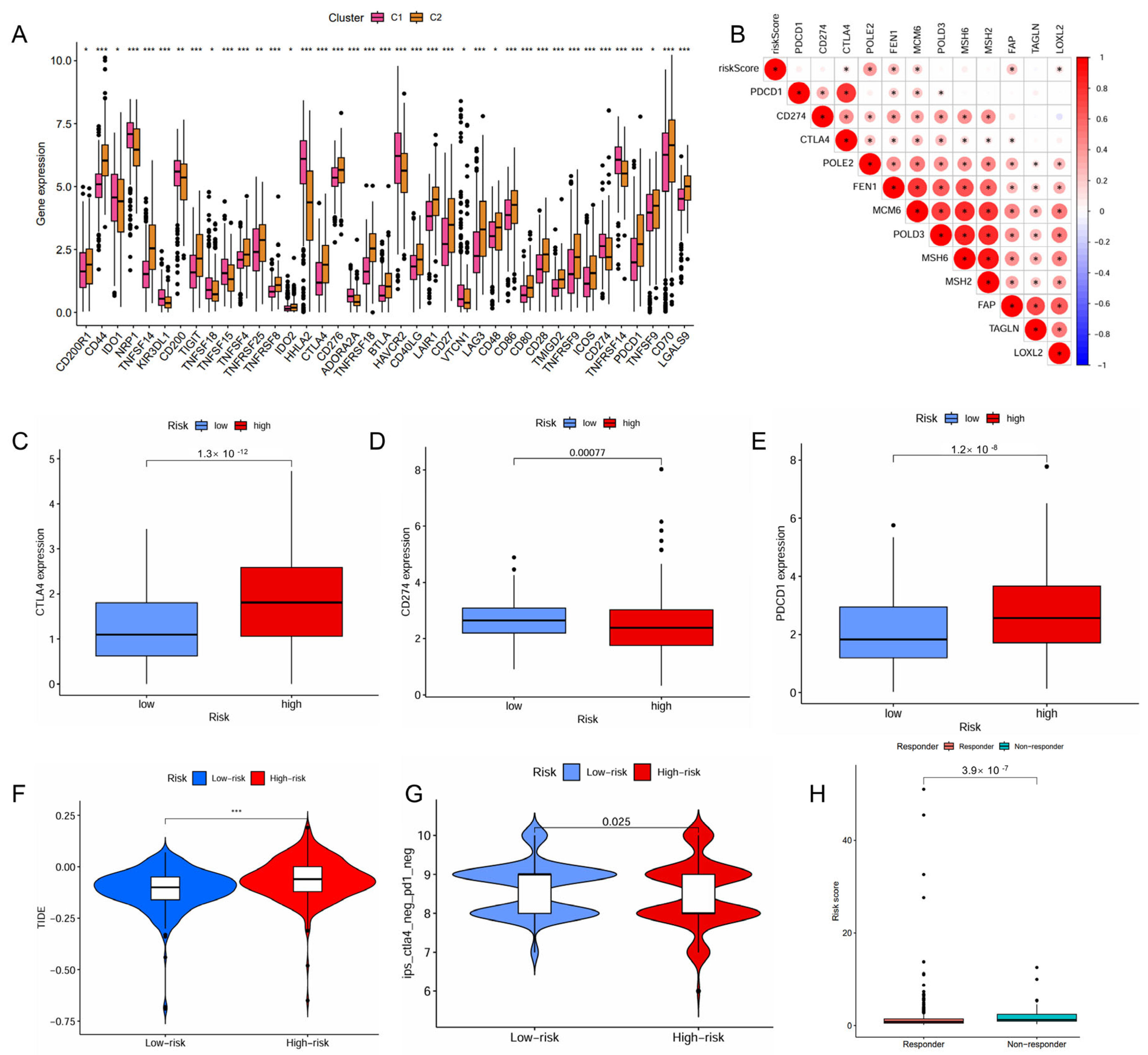
2.4. IMRG Score Correlates with Immune Microenvironments and Responses to Immunotherapy
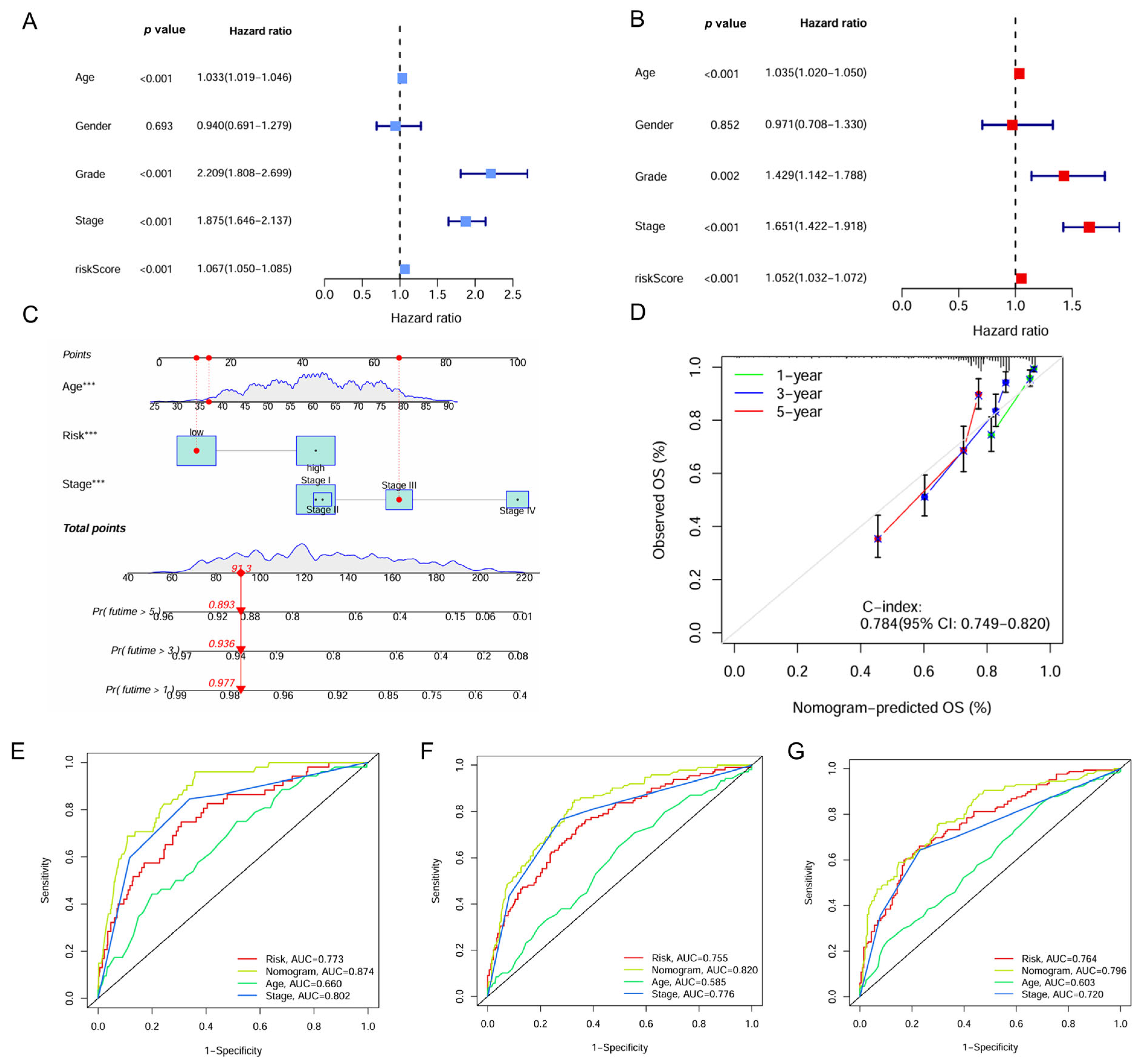
2.5. Construction and Evaluation of the IMRG-Based Nomogram
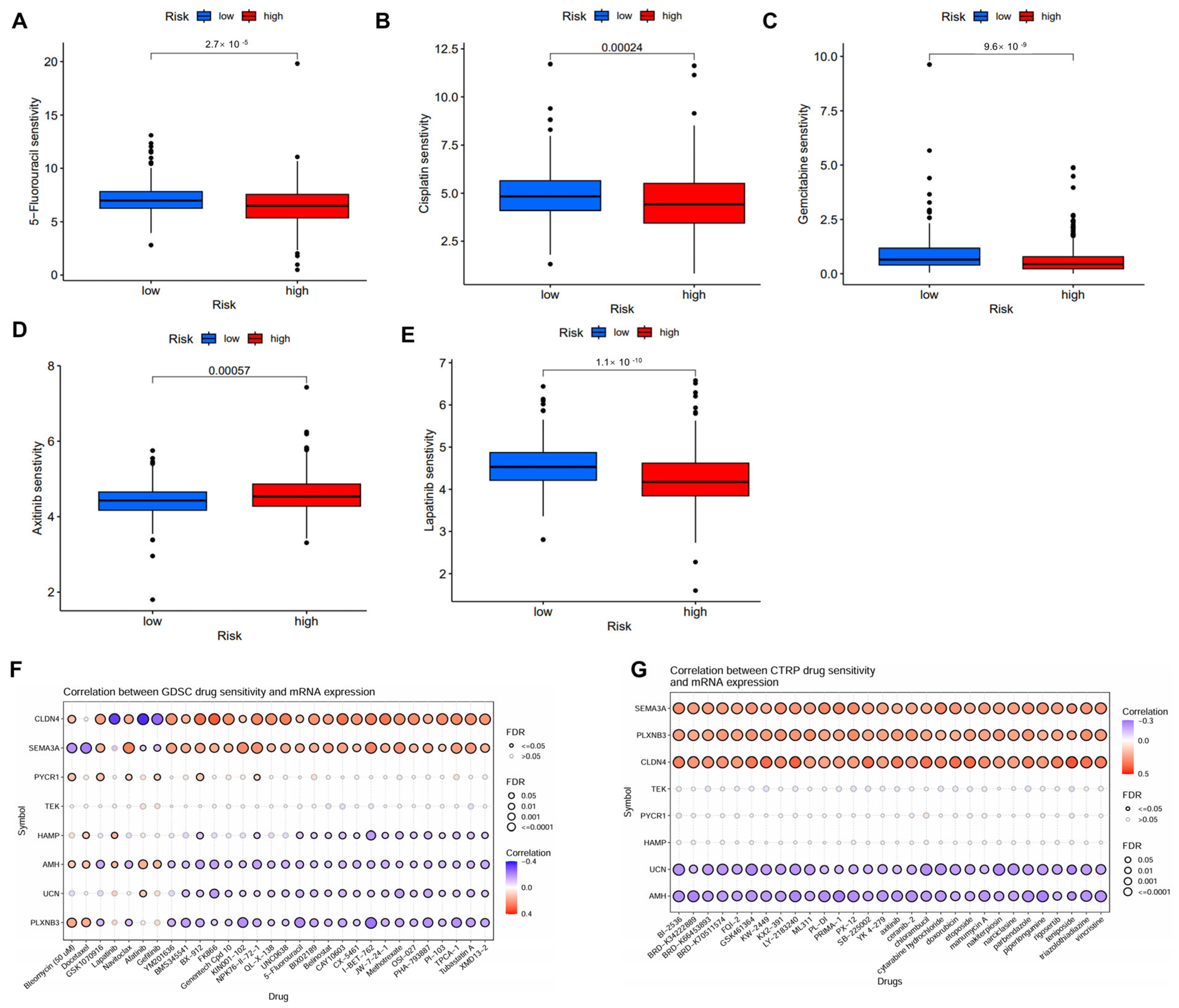
2.6. Correlation Between IMRG and Drug Sensitivity
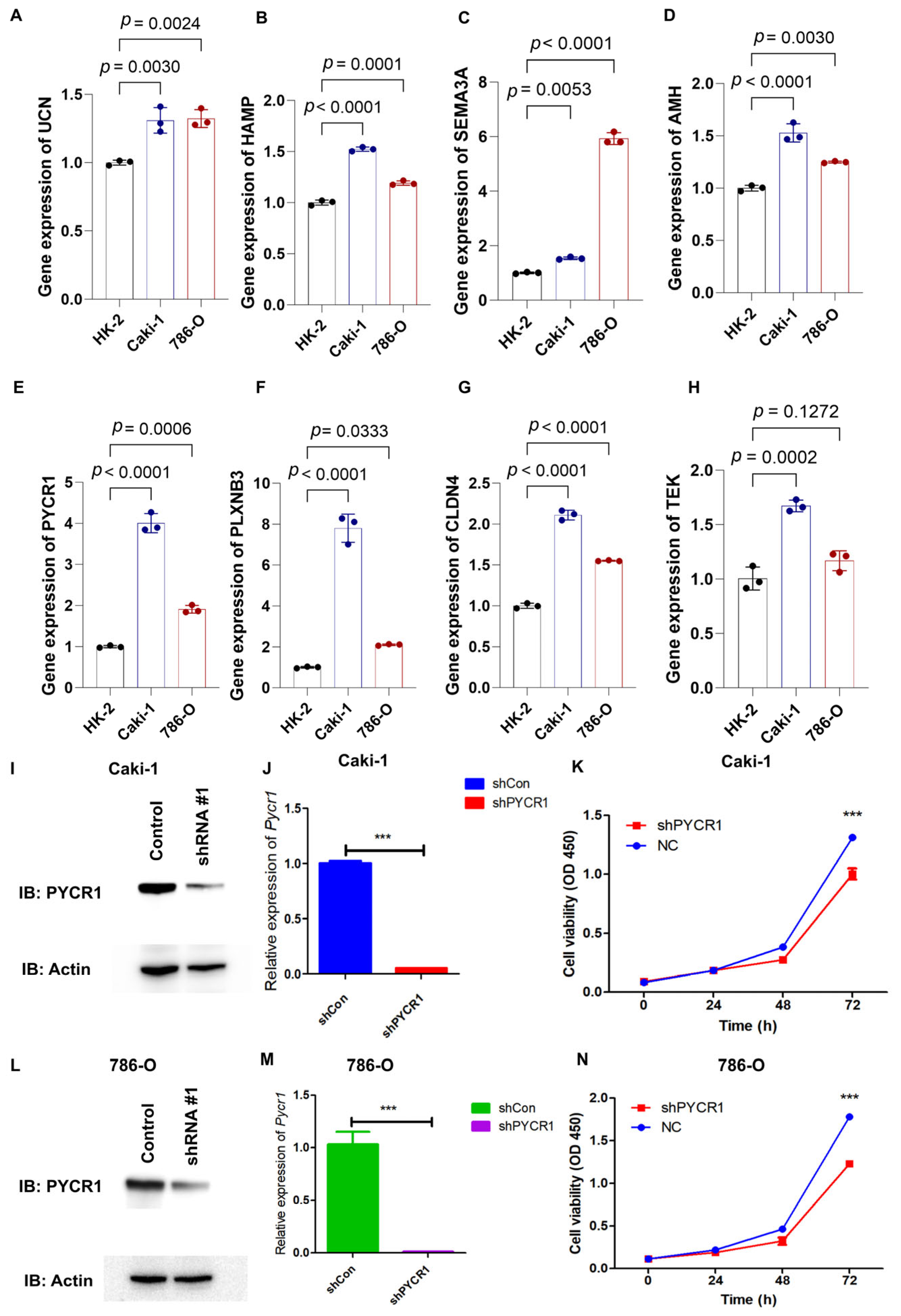
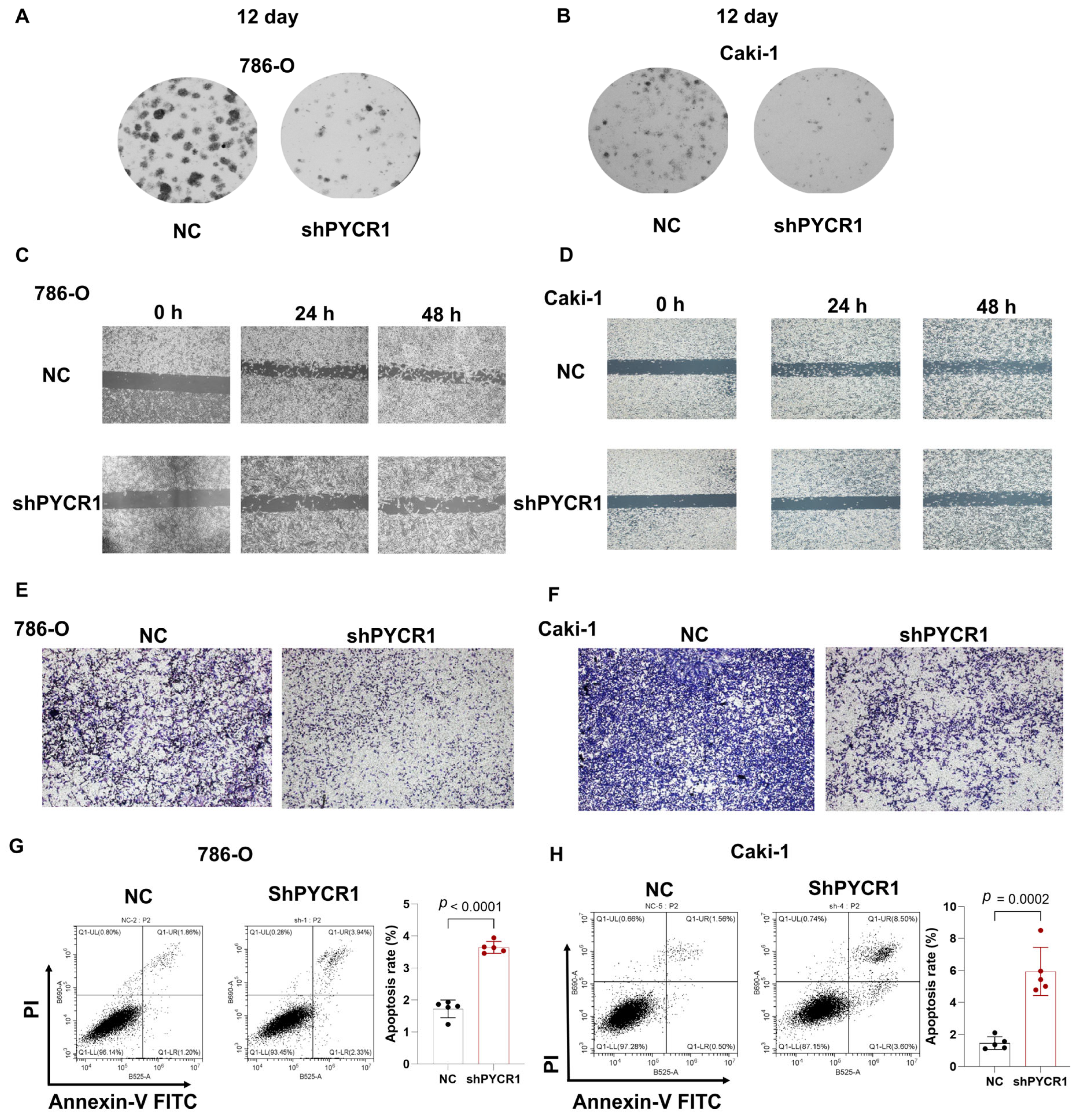
2.7. Knockdown of PYCR1 Inhibits the Malignant Behaviors in ccRCC
3. Discussion
4. Materials and Methods
4.1. Acquisition and Processing of Datasets
4.2. Collection of Metabolism and Immune-Related Gene Sets
4.3. Construction and Validation of IMRGs Signature
4.4. Gene Set Enrichment Analysis (GSEA)
4.5. Evaluation of the Tumor Microenvironment and Response to Immunotherapy
4.6. Comparisons of Drug Sensitivity
4.7. Cell Lines and Culture
4.8. RNA Isolation and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.9. Construction and Infection of Lentivirus Plasmid
4.10. Protein Extraction and Western Blot
4.11. Cell Growth and Colony Formation Assays
4.12. Wound Healing Assay
4.13. Transwell Assays
4.14. Cell Apoptosis by Flow Cytometry
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roerden, M.; Spranger, S. Cancer immune evasion, immunoediting and intratumour heterogeneity. Nat. Rev. Immunol. 2025, 25, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Han, Y.; Wu, N.; Yi, P.; Tang, L.; Pan, Q.; Rao, S.; et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 2021, 20, 28. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Kawase, K.; Nishi, T.; Watanabe, T.; Takenaga, K.; Inozume, T.; Ishino, T.; Aki, S.; Lin, J.; Kawashima, S.; et al. Immune evasion through mitochondrial transfer in the tumour microenvironment. Nature 2025, 638, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Xu, H.; Yuan, S.; Li, X.; Chen, X.; Fan, X.; Gao, X.; Zhang, L.; Sun, S.; Zhu, X. LncRNA mediated metabolic reprogramming: The chief culprits of solid tumor malignant progression: An update review. Nutr. Metab. 2024, 21, 89. [Google Scholar] [CrossRef]
- Leone, R.D.; Zhao, L.; Englert, J.M.; Sun, I.M.; Oh, M.H.; Sun, I.H.; Arwood, M.L.; Bettencourt, I.A.; Patel, C.H.; Wen, J.; et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 2019, 366, 1013–1021. [Google Scholar] [CrossRef]
- Xie, Y.; Guan, S.; Li, Z.; Cai, G.; Liu, Y.; Li, G.; Huang, P.; Lin, M. Identification of a metabolic-immune signature associated with prognosis in colon cancer and exploration of potential predictive efficacy of immunotherapy response. Clin. Exp. Med. 2025, 25, 46. [Google Scholar] [CrossRef]
- Kao, K.C.; Vilbois, S.; Tsai, C.H.; Ho, P.C. Metabolic communication in the tumour-immune microenvironment. Nat. Cell Biol. 2022, 24, 1574–1583. [Google Scholar] [CrossRef]
- Lyu, F.; Zhong, Y.; He, Q.; Xiao, W.; Zhang, X. Identification and validation of prognostic biomarkers in ccRCC: Immune-stromal score and survival prediction. BMC Cancer 2025, 25, 148. [Google Scholar] [CrossRef]
- Huang, T.; Peng, Y.; Liu, R.; Ma, B.; Chen, J.; Wei, W.; Zhong, W.; Liu, Y.; Guo, S.; Han, H.; et al. Prognostic significance of immune evasion-related genes in clear cell renal cell carcinoma immunotherapy. Int. Immunopharmacol. 2024, 142, 113106. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, P.; Gao, J.; Li, X.; Wei, C.; Liu, W.; He, Q.; Zhang, Y. Integrated bulk and single-cell transcriptome data identify clinically relevant cell populations in clear cell renal cell carcinoma. Genes Dis. 2024, 11, 42–45. [Google Scholar] [CrossRef]
- Hu, J.; Wang, S.G.; Hou, Y.; Chen, Z.; Liu, L.; Li, R.; Li, N.; Zhou, L.; Yang, Y.; Wang, L.; et al. Multi-omic profiling of clear cell renal cell carcinoma identifies metabolic reprogramming associated with disease progression. Nat. Genet. 2024, 56, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ferdinand, J.R.; Loudon, K.W.; Bowyer, G.S.; Laidlaw, S.; Muyas, F.; Mamanova, L.; Neves, J.B.; Bolt, L.; Fasouli, E.S.; et al. Mapping single-cell transcriptomes in the intra-tumoral and associated territories of kidney cancer. Cancer Cell 2022, 40, 1583–1599.e1510. [Google Scholar] [CrossRef] [PubMed]
- Golkaram, M.; Kuo, F.; Gupta, S.; Carlo, M.I.; Salmans, M.L.; Vijayaraghavan, R.; Tang, C.; Makarov, V.; Rappold, P.; Blum, K.A.; et al. Spatiotemporal evolution of the clear cell renal cell carcinoma microenvironment links intra-tumoral heterogeneity to immune escape. Genome Med. 2022, 14, 143. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Street, K.; Burke, K.P.; Cookmeyer, D.L.; Denize, T.; Pedersen, C.B.; Gohil, S.H.; Schindler, N.; Pomerance, L.; Hirsch, L.; et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell 2021, 39, 632–648.e638. [Google Scholar] [CrossRef]
- Kay, E.J.; Paterson, K.; Riera-Domingo, C.; Sumpton, D.; Dabritz, J.H.M.; Tardito, S.; Boldrini, C.; Hernandez-Fernaud, J.R.; Athineos, D.; Dhayade, S.; et al. Cancer-associated fibroblasts require proline synthesis by PYCR1 for the deposition of pro-tumorigenic extracellular matrix. Nat. Metab. 2022, 4, 693–710. [Google Scholar] [CrossRef]
- Wang, H.; Xu, M.; Zhang, T.; Pan, J.; Li, C.; Pan, B.; Zhou, L.; Huang, Y.; Gao, C.; He, M.; et al. PYCR1 promotes liver cancer cell growth and metastasis by regulating IRS1 expression through lactylation modification. Clin. Transl. Med. 2024, 14, e70045. [Google Scholar] [CrossRef]
- Chen, H.; Chen, Q.; Chen, J.; Mao, Y.; Duan, L.; Ye, D.; Cheng, W.; Chen, J.; Gao, X.; Lin, R.; et al. Deciphering the Effects of the PYCR Family on Cell Function, Prognostic Value, Immune Infiltration in ccRCC and Pan-Cancer. Int. J. Mol. Sci. 2024, 25, 8096. [Google Scholar] [CrossRef]
- D’Aniello, C.; Patriarca, E.J.; Phang, J.M.; Minchiotti, G. Proline Metabolism in Tumor Growth and Metastatic Progression. Front Oncol. 2020, 10, 776. [Google Scholar] [CrossRef]
- Ding, Z.; Ericksen, R.E.; Lee, Q.Y.; Han, W. Reprogramming of mitochondrial proline metabolism promotes liver tumorigenesis. Amino Acids 2021, 53, 1807–1815. [Google Scholar] [CrossRef]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef]
- Rees, M.G.; Seashore-Ludlow, B.; Cheah, J.H.; Adams, D.J.; Price, E.V.; Gill, S.; Javaid, S.; Coletti, M.E.; Jones, V.L.; Bodycombe, N.E.; et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016, 12, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wang, Z.; Wang, J.; Xu, Y.; Kong, W.; Zhang, J. Development of a novel gene signature to predict prognosis and response to PD-1 blockade in clear cell renal cell carcinoma. Oncoimmunology 2021, 10, 1933332. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yan, Z.; Liu, C.; Yang, R.; Zheng, Q.; Jian, J.; Wang, M.; Wang, L.; Weng, X.; Chen, Z.; et al. Integrated RNA sequencing analysis and machine learning identifies a metabolism-related prognostic signature in clear cell renal cell carcinoma. Sci. Rep. 2025, 15, 1691. [Google Scholar] [CrossRef]
- Zhang, H.; Li, S.; Wang, D.; Liu, S.; Xiao, T.; Gu, W.; Yang, H.; Wang, H.; Yang, M.; Chen, P. Metabolic reprogramming and immune evasion: The interplay in the tumor microenvironment. Biomark. Res. 2024, 12, 96. [Google Scholar] [CrossRef]
- Zhang, L.; Romero, P. Metabolic Control of CD8+ T Cell Fate Decisions and Antitumor Immunity. Trends Mol. Med. 2018, 24, 30–48. [Google Scholar] [CrossRef]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Roh, W.; Chen, P.L.; Reuben, A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Gopalakrishnan, V.; Wang, F.; Cooper, Z.A.; Reddy, S.M.; et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017, 9, eaah3560. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Wei, X.; Ye, J.; Pei, Y.; Wang, C.; Yang, H.; Tian, J.; Si, G.; Ma, Y.; Wang, K.; Liu, G. Extracellular vesicles from colorectal cancer cells promote metastasis via the NOD1 signalling pathway. J. Extracell Vesicles 2022, 11, e12264. [Google Scholar] [CrossRef]
- Gu, J.; Zhang, X.; Peng, Z.; Peng, Z.; Liao, Z. A novel immune-related gene signature for predicting immunotherapy outcomes and survival in clear cell renal cell carcinoma. Sci. Rep. 2023, 13, 18922. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wu, J.; Liu, S.; Sun, Y.; Liu, G.; Zhang, L.; Yu, Q.; Xu, J.; Meng, L. Immune-related risk prognostic model for clear cell renal cell carcinoma: Implications for immunotherapy. Medicine 2023, 102, e34786. [Google Scholar] [CrossRef]
- Li, W.; Meng, X.; Yuan, H.; Xiao, W.; Zhang, X. A Novel Immune-Related ceRNA Network and Relative Potential Therapeutic Drug Prediction in ccRCC. Front. Genet. 2021, 12, 755706. [Google Scholar] [CrossRef]
- Wan, B.; Liu, B.; Huang, Y.; Yu, G.; Lv, C. Prognostic value of immune-related genes in clear cell renal cell carcinoma. Aging 2019, 11, 11474–11489. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, X.; Zhu, S.; Hu, B.; Deng, Z.; Feng, H.; Liu, B.; Luan, Y.; Liu, Z.; Wang, S.; et al. Prediction of clear cell renal cell carcinoma prognosis based on an immunogenomic landscape analysis. Heliyon 2024, 10, e36156. [Google Scholar] [CrossRef]
- Barnkob, M.B.; Michaels, Y.S.; Andre, V.; Macklin, P.S.; Gileadi, U.; Valvo, S.; Rei, M.; Kulicke, C.; Chen, J.L.; Jain, V.; et al. Semmaphorin 3 A causes immune suppression by inducing cytoskeletal paralysis in tumour-specific CD8+ T cells. Nat. Commun. 2024, 15, 3173. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, X.; Wang, S.; Wang, Y.; Ji, C.; Yao, L.; Song, N. PYCR1 regulates glutamine metabolism to construct an immunosuppressive microenvironment for the progression of clear cell renal cell carcinoma. Am. J. Cancer Res. 2022, 12, 3780–3798. [Google Scholar]
- Yu, Z.; Zhan, Y.; Guo, Y.; He, D. Better prediction of clinical outcome in clear cell renal cell carcinoma based on a 6 metabolism-related gene signature. Sci. Rep. 2023, 13, 11490. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, X.; Wang, E.; Yang, Y.; Hu, L.; Xu, H.; Zhang, B. PYCR1 promotes the malignant progression of lung cancer through the JAK-STAT3 signaling pathway via PRODH-dependent glutamine synthesize. Transl. Oncol. 2023, 32, 101667. [Google Scholar] [CrossRef]
- Zhou, B.; Mai, Z.; Ye, Y.; Song, Y.; Zhang, M.; Yang, X.; Xia, W.; Qiu, X. The role of PYCR1 in inhibiting 5-fluorouracil-induced ferroptosis and apoptosis through SLC25A10 in colorectal cancer. Hum. Cell 2022, 35, 1900–1911. [Google Scholar] [CrossRef]
- Oudaert, I.; Satilmis, H.; Vlummens, P.; De Brouwer, W.; Maes, A.; Hose, D.; De Bruyne, E.; Ghesquiere, B.; Vanderkerken, K.; De Veirman, K.; et al. Pyrroline-5-Carboxylate Reductase 1: A novel target for sensitizing multiple myeloma cells to bortezomib by inhibition of PRAS40-mediated protein synthesis. J. Exp. Clin. Cancer Res. 2022, 41, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Yang, Y.; Deng, X.; Chao, N.; Chen, Z.; Ye, Y.; Zhang, W.; Liu, W.; Zhao, S. High CD8+ tumor-infiltrating lymphocytes indicate severe exhaustion and poor prognosis in angioimmunoblastic T-cell lymphoma. Front. Immunol. 2023, 14, 1228004. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Pu, D.; Ren, J.; Liu, M.; Zhang, Z.; Liu, Z.; Li, J. CD8+ T-cell exhaustion: Impediment to triple-negative breast cancer (TNBC) immunotherapy. Biochim. Biophys. Acta Rev. Cancer 2024, 1879, 189193. [Google Scholar] [CrossRef] [PubMed]
- Buttner, F.; Winter, S.; Rausch, S.; Hennenlotter, J.; Kruck, S.; Stenzl, A.; Scharpf, M.; Fend, F.; Agaimy, A.; Hartmann, A.; et al. Clinical utility of the S3-score for molecular prediction of outcome in non-metastatic and metastatic clear cell renal cell carcinoma. BMC Med. 2018, 16, 108. [Google Scholar] [CrossRef]
- Serie, D.J.; Joseph, R.W.; Cheville, J.C.; Ho, T.H.; Parasramka, M.; Hilton, T.; Thompson, R.H.; Leibovich, B.C.; Parker, A.S.; Eckel-Passow, J.E. Clear Cell Type A and B Molecular Subtypes in Metastatic Clear Cell Renal Cell Carcinoma: Tumor Heterogeneity and Aggressiveness. Eur. Urol. 2017, 71, 979–985. [Google Scholar] [CrossRef]
- Rini, B.; Goddard, A.; Knezevic, D.; Maddala, T.; Zhou, M.; Aydin, H.; Campbell, S.; Elson, P.; Koscielny, S.; Lopatin, M.; et al. A 16-gene assay to predict recurrence after surgery in localised renal cell carcinoma: Development and validation studies. Lancet Oncol. 2015, 16, 676–685. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, Y.; Senbabaoglu, Y.; Ciriello, G.; Yang, L.; Reznik, E.; Shuch, B.; Micevic, G.; De Velasco, G.; Shinbrot, E.; et al. Multilevel Genomics-Based Taxonomy of Renal Cell Carcinoma. Cell Rep. 2016, 14, 2476–2489. [Google Scholar] [CrossRef]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326e315. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Geeleher, P.; Cox, N.J.; Huang, R.S. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014, 15, R47. [Google Scholar] [CrossRef]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef]
- Chi, C.; Ye, Y.; Chen, B.; Huang, H. Bipartite graph-based approach for clustering of cell lines by gene expression-drug response associations. Bioinformatics 2021, 37, 2617–2626. [Google Scholar] [CrossRef]
| Gene Name | Full Name | Functions | Immunity or Metabolism Gene |
|---|---|---|---|
| UCN | Urocortin | Activation of cAMP-dependent PKA and peptide hormone metabolism | Immunity |
| HAMP | Hepcidin antimicrobial peptide | Hfe effect on hepcidin production and TAR syndrome | Immunity |
| SEMA3A | Semaphorin 3A | Apoptotic pathways in synovial fibroblasts and GPCR pathway | Immunity |
| AMH | Anti-mullerian hormone | Mammalian disorder of sexual development and signaling by TGFB family members | Immunity |
| PYCR1 | Pyrroline-5-Carboxylate Reductase 1 | Glutamate and glutamine metabolism and superpathway of L-citrulline metabolism | Metabolism |
| PLXNB3 | Plexin B3 | Nervous system development and semaphorin interactions | Immunity |
| CLDN4 | Claudin 4 | Blood–brain barrier and immune cell transmigration: VCAM-1/CD106 signaling and cell junction organization | Immunity |
| TEK | TEK Receptor Tyrosine Kinase | GPCR pathway and ERK signaling | Immunity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, G.; Ding, J.; Ma, J.; Jiang, Y.; Wang, Y.; Wang, S.; Li, N. Integrative Analysis of Immune- and Metabolism-Related Genes Identifies Robust Prognostic Signature and PYCR1 as a Carcinogenic Regulator in Clear Cell Renal Cell Carcinoma. Int. J. Mol. Sci. 2025, 26, 4953. https://doi.org/10.3390/ijms26104953
Zhao G, Ding J, Ma J, Jiang Y, Wang Y, Wang S, Li N. Integrative Analysis of Immune- and Metabolism-Related Genes Identifies Robust Prognostic Signature and PYCR1 as a Carcinogenic Regulator in Clear Cell Renal Cell Carcinoma. International Journal of Molecular Sciences. 2025; 26(10):4953. https://doi.org/10.3390/ijms26104953
Chicago/Turabian StyleZhao, Guo, Jiatong Ding, Jiaxiu Ma, Yale Jiang, Yuning Wang, Shuhang Wang, and Ning Li. 2025. "Integrative Analysis of Immune- and Metabolism-Related Genes Identifies Robust Prognostic Signature and PYCR1 as a Carcinogenic Regulator in Clear Cell Renal Cell Carcinoma" International Journal of Molecular Sciences 26, no. 10: 4953. https://doi.org/10.3390/ijms26104953
APA StyleZhao, G., Ding, J., Ma, J., Jiang, Y., Wang, Y., Wang, S., & Li, N. (2025). Integrative Analysis of Immune- and Metabolism-Related Genes Identifies Robust Prognostic Signature and PYCR1 as a Carcinogenic Regulator in Clear Cell Renal Cell Carcinoma. International Journal of Molecular Sciences, 26(10), 4953. https://doi.org/10.3390/ijms26104953








