Noninvasive Biomarkers of Human Embryo Developmental Potential
Abstract
1. Introduction
2. Noninvasive Biomarkers: History, Current State, and Strengths and Limitations
2.1. Morphology and Kinetics
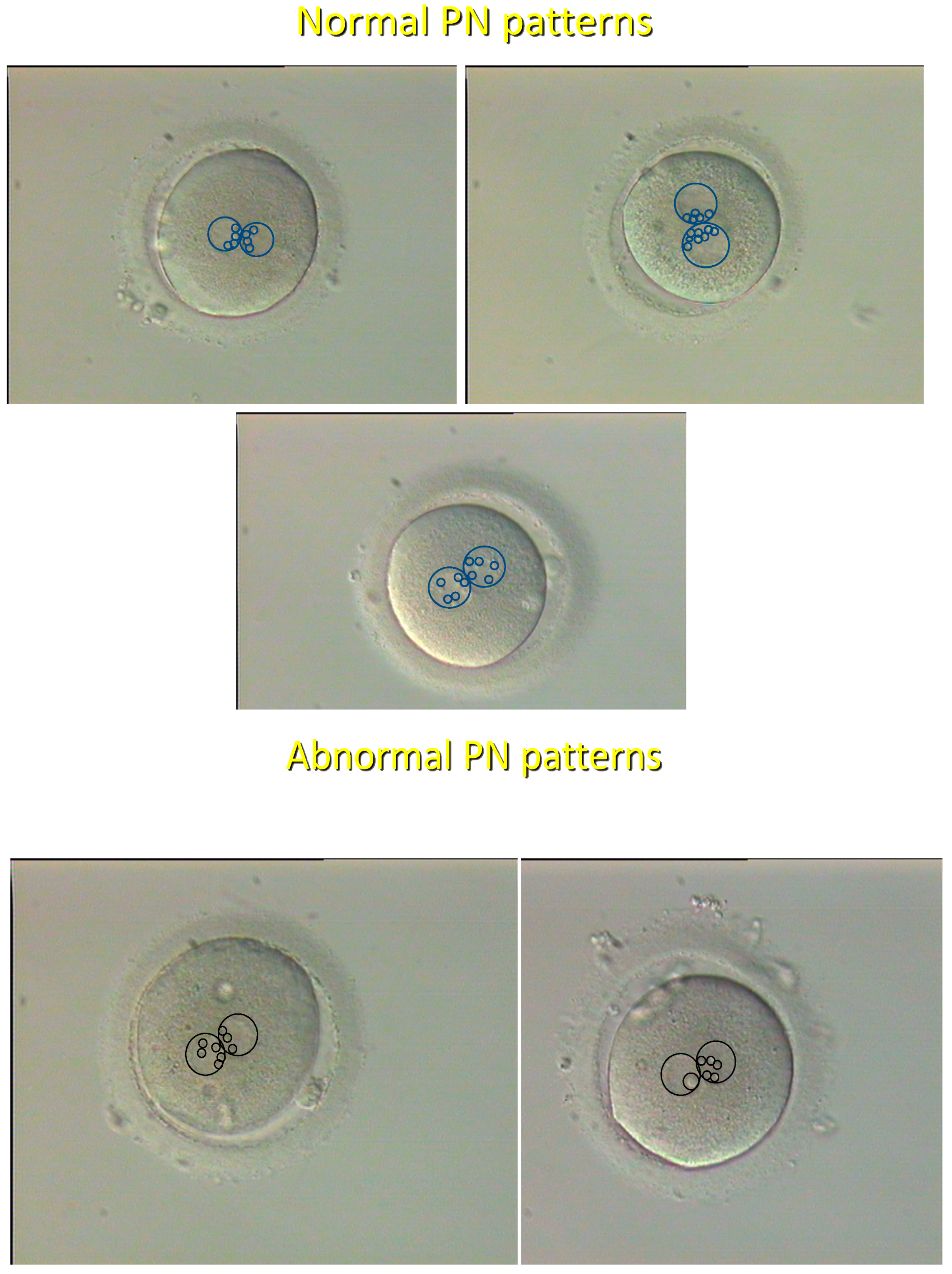
2.1.1. History
2.1.2. Current Status
| Marker | Reference Outcome | Predictive Capacity | Techniques | References |
|---|---|---|---|---|
| Pronuclear | Ongoing development | Very high | Single microscopic | [8] |
| morphology | Chromosomal ploidy | Very high | observation | [20] |
| Cleavage speed and regularity | Pregnancy | Low | Repeated microscopic observation | [21,22,23] |
| Proportion of MNB | Developmental arrest and pregnancy failure | High | Repeated microscopic observation | [24] |
| Noninvasive continuous recording | Developmental arrest and pregnancy failureChromosomal ploidy | Debated High with 3D setting (holotomography)High | Time-lapse photography | [25,26,27,28,29] [30,31,32] |
2.1.3. Strengths and Limitations
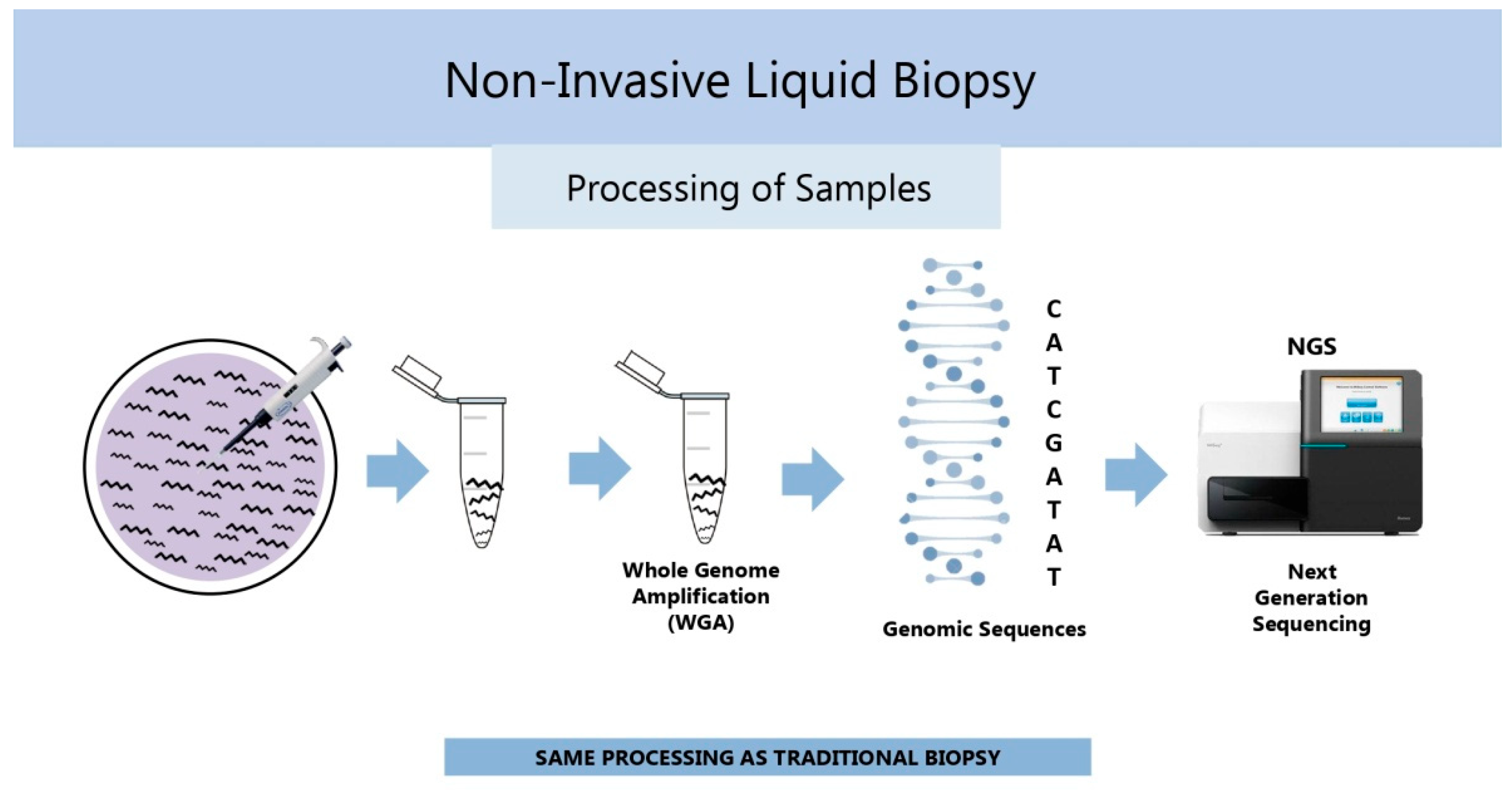
2.2. Chromosomal Ploidy Status (Genomics)
2.2.1. History
2.2.2. Current Status
2.2.3. Strengths and Limitations
2.3. Gene Activity
2.3.1. Transcription (Transcriptomics)
History
Current Status
Strengths and Limitations
2.3.2. Translation (Proteomics and Secretomics)
History
| Marker | Reference Outcome | Predictive Capacity | Techniques | References |
|---|---|---|---|---|
| Transcriptome miRNAs piRNAs | Embryo viability and pregnancy | Very high | Massive-parallel sequencing, qRT-PCR | [56,57,58,59,60,61,62,63,64,65,66,67] |
| Proteome | Embryo viability, ploidy, and pregnancy | Very high | Historical: ELISA, Western blotting | [70] |
| Current: Mass spectrometry | [71,72,73,74,75,76,77] |
Current Status
Strengths and Limitations
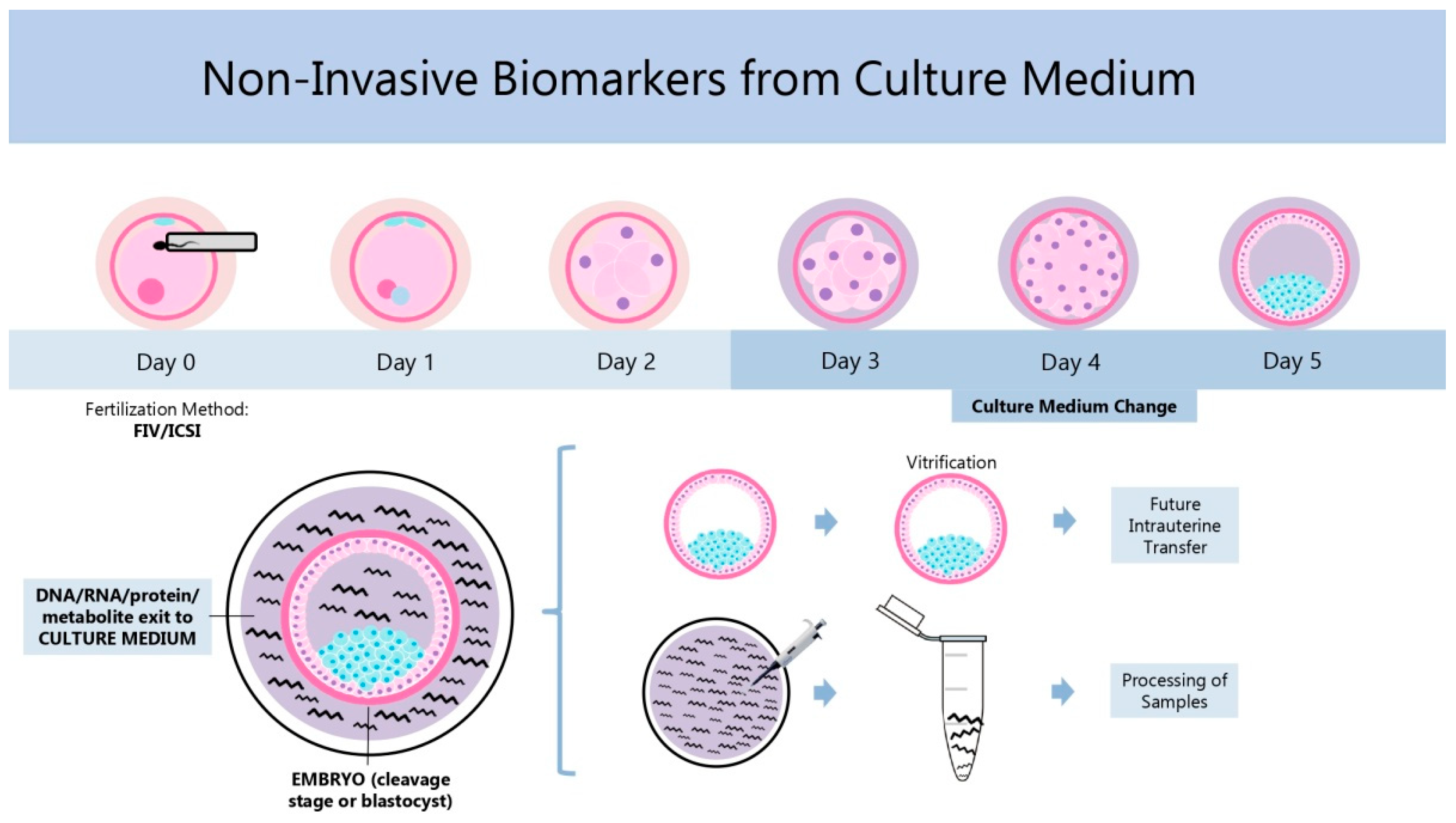
2.4. Substrate Uptake and Secretion (Metabolomics)
2.4.1. History
2.4.2. Current Status
2.4.3. Strengths and Limitations
2.5. Extracellular Vesicles
2.5.1. History
2.5.2. Current Status
2.5.3. Strengths and Limitations
2.6. Combined Use of Different Biomarkers
3. Conclusions
Funding
Conflicts of Interest
References
- Steptoe, P.C.; Edwards, R.G. Birth after reimplantation of a human embryo. Lancet 1978, 2, 366. [Google Scholar] [CrossRef]
- Tesarik, J. Assisted reproduction: New challenges and future prospects. In 40 Years After In Vitro Fertilisation; Tesarik, J., Ed.; Cambridge Scholars Publishing: Newcastle upon Tyne, UK, 2019; pp. 269–286. [Google Scholar]
- Bashiri, A.; Halper, K.I.; Orvieto, R. Recurrent implantation failure-update overview on etiology, diagnosis, treatment and future directions. Reprod. Biol. Endocrinol. 2018, 16, 121. [Google Scholar] [CrossRef] [PubMed]
- Tesarik, J. Complementarity between early embryogenesis and uterine receptivity: Toward integrative approach to female infertility management. Editorial to the Special Issue “Molecular Mechanisms of Human Oogenesis and Early Embryogenesis”. Int. J. Mol. Sci. 2023, 24, 1557. [Google Scholar] [CrossRef]
- Rødgaard, T.; Heegaard, P.M.; Callesen, H. Non-invasive assessment of in-vitro embryo quality to improve transfer success. Reprod. Biomed. Online 2015, 31, 585–592. [Google Scholar] [CrossRef]
- Ziebe, S.; Petersen, K.; Lindenberg, S.; Andersen, A.G.; Gabrielsen, A.; Andersen, A.N. Embryo morphology or cleavage stage: How to select the best embryos for transfer after in-vitro fertilization. Hum. Reprod. 1997, 12, 1545–1549. [Google Scholar] [CrossRef]
- Scott, L.A.; Smith, S. The successful use of pronuclear embryo transfers the day following oocyte retrieval. Hum. Reprod. 1998, 13, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Tesarik, J.; Greco, E. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear stage morphology. Hum. Reprod. 1999, 14, 1318–1323. [Google Scholar] [CrossRef]
- Gardner, D.K.; Lane, M.; Stevens, J.; Schlenker, T.; Schoolcraft, W.B. Blastocyst score affects implantation and pregnancy outcome: Towards a single blastocyst transfer. Fertil. Steril. 2000, 73, 1155–1158. [Google Scholar] [CrossRef]
- Lundin, K.; Bergh, C.; Hardarson, T. Early embryo cleavage is a strong indicator of embryo quality in human IVF. Hum. Reprod. 2001, 16, 2652–2657. [Google Scholar] [CrossRef]
- Fenwick, J.; Platteau, P.; Murdoch, A.P.; Herbert, M. Time from insemination to first cleavage predicts developmental competence of human preimplantation embryos in vitro. Hum. Reprod. 2002, 17, 407–412. [Google Scholar] [CrossRef]
- Scott, L. Pronuclear scoring as a predictor of embryo development. Reprod. Biomed. Online 2003, 6, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.F.; Raburn, D.J.; Couchman, G.M.; Price, T.M.; Jamison, M.G.; Walmer, D.K. Relationship between pre-embryo pronuclear morphology (zygote score) and standard day 2 or 3 embryo morphology with regard to assisted reproductive technique outcomes. Fertil. Steril. 2005, 84, 900–909. [Google Scholar] [CrossRef]
- Chen, C.; Kattera, S. Comparison of pronuclear zygote morphology and early cleavage status of zygotes as additional criteria in the selection of day 3 embryos: A randomized study. Fertil. Steril. 2006, 85, 347–352. [Google Scholar] [CrossRef]
- Weitzman, V.N.; Schnee-Riesz, J.; Benadiva, C.; Nulsen, J.; Siano, L.; Maier, D. Predictive value of embryo grading for embryos with known outcomes. Fertil. Steril. 2010, 93, 658–662. [Google Scholar] [CrossRef]
- Lemmen, J.G.; Agerholm, I.; Ziebe, S. Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI-fertilized oocytes. Reprod. Biomed. Online 2008, 17, 385–391. [Google Scholar] [CrossRef]
- Gardner, D.K.; Balaban, B. Assessment of human embryo development using morphological criteria in an era of time-lapse, algorithms and ‘OMICS’: Is looking good still important? Mol. Hum. Reprod. 2016, 22, 704–718. [Google Scholar] [CrossRef]
- Cummins, J.M.; Breen, T.M.; Harrison, K.L.; Shaw, J.M.; Wilson, L.M.; Hennessey, J.F. A formula for scoring human embryo growth rates in in vitro fertilization: Its value in predicting pregnancy and in comparison with visual estimates of embryo quality. J. Vitr. Fert. Embryo Transf. 1986, 3, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Racowsky, C.; Vernon, M.; Mayer, J.; Ball, G.D.; Behr, B.; Pomeroy, K.O.; Wininger, D.; Gibbons, W.; Conaghan, J.; Stern, J.E. Standardization of grading embryo morphology. J. Assist. Reprod. Genet. 2010, 27, 437–439. [Google Scholar] [CrossRef] [PubMed]
- Balaban, B.; Yakin, K.; Urman, B.; Isiklar, A.; Tesarik, J. Pronuclear morphology predicts embryo development and chromosome constitution. Reprod. Biomed. Online 2004, 8, 695–700. [Google Scholar] [CrossRef]
- Prados, F.J.; Debrock, S.; Lemmen, J.G.; Agerholm, I. The cleavage stage embryo. Hum. Reprod. 2012, 27 (Suppl. S1), i50–i71. [Google Scholar] [CrossRef]
- Majumdar, G.; Majumdar, A.; Verma, I.C.; Upadhyaya, K.C. Relationship Between Morphology, Euploidy and Implantation Potential of Cleavage and Blastocyst Stage Embryos. J. Hum. Reprod. Sci. 2017, 10, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Paternot, G.; Wetzels, A.M.; Thonon, F.; Vansteenbrugge, A.; Willemen, D.; Devroe, J.; Debrock, S.; D’Hooghe, T.M.; Spiessens, C. Intra- and interobserver analysis in the morphological assessment of early stage embryos during an IVF procedure: A multicentre study. Reprod. Biol. Endocrinol. 2011, 9, 127. [Google Scholar] [CrossRef]
- Barberet, J.; Bruno, C.; Valot, E.; Antunes-Nunes, C.; Jonval, L.; Chammas, J.; Choux, C.; Ginod, P.; Sagot, P.; Soudry-Faure, A.; et al. Can novel early non-invasive biomarkers of embryo quality be identified with time-lapse imaging to predict live birth? Hum. Reprod. 2019, 34, 1439–1449. [Google Scholar] [CrossRef]
- Yoneyama, M.; Ito, A.; Katagiri, Y.; Oigawa, S.; Nagao, K.; Nakata, M. Blastocyst re-expansion rate immediately after warming is a strong dynamic indicator of embryo quality. Reprod. Biomed. Online, 2025; in press. [Google Scholar] [CrossRef]
- Lee, C.; Kim, G.; Shin, C.; Lee, S.; Kim, J.Y.; Choi, K.H.; Do, J.; Park, J.; Do, J.; Kim, J.H.; et al. Noninvasive time-lapse 3D subcellular analysis of embryo development for machine learning-enabled prediction of blastocyst formation. bioRxiv 2024. [Google Scholar] [CrossRef]
- Illingworth, P.J.; Venetis, C.; Gardner, D.K.; Nelson, S.M.; Berntsen, J.; Larman, M.G.; Agresta, F.; Ahitan, S.; Ahlström, A.; Cattrall, F.; et al. Deep learning versus manual morphology-based embryo selection in IVF: A randomized, double-blind noninferiority trial. Nat. Med. 2024, 30, 3114–3120. [Google Scholar] [CrossRef]
- Sacks, G.C.; Mozes, H.; Ronn, R.; Elder-Geva, T.; Schonberger, O.; Ben-Ami, I.; Srebnik, N. Time-Lapse Incubation for Embryo Culture-Morphokinetics and Environmental Stability May Not Be Enough: Results from a Pilot Randomized Controlled Trial. J. Clin. Med. 2024, 13, 1701. [Google Scholar] [CrossRef]
- Porokh, V.; Kyjovská, D.; Martonová, M.; Klenková, T.; Otevřel, P.; Kloudová, S.; Holubcová, Z. Zygotic spindle orientation defines cleavage pattern and nuclear status of human embryos. Nat. Commun. 2024, 15, 6369. [Google Scholar] [CrossRef]
- Danardono, G.B.; Handayani, N.; Louis, C.M.; Polim, A.A.; Sirait, B.; Periastiningrum, G.; Afadlal, S.; Boediono, A.; Sini, I. Embryo ploidy status classification through computer-assisted morphology assessment. AJOG Glob. Rep. 2023, 3, 100209. [Google Scholar] [CrossRef]
- Rajendran, S.; Brendel, M.; Barnes, J.; Zhan, Q.; Malmsten, J.E.; Zisimopoulos, P.; Sigaras, A.; Ofori-Atta, K.; Meseguer, M.; Miller, K.A.; et al. Automatic ploidy prediction and quality assessment of human blastocysts using time-lapse imaging. Nat. Commun. 2024, 15, 7756. [Google Scholar] [CrossRef]
- Serrano-Novillo, C.; Uroz, L.; Márquez, C. Novel time-lapse parameters correlate with embryo ploidy and suggest an improvement in non-invasive embryo selection. J. Clin. Med. 2023, 12, 2983. [Google Scholar] [CrossRef] [PubMed]
- Hassold, T.; Hunt, P. To ERR (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001, 2, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Leem, J.; Gowett, M.; Bolarinwa, S.; Mogessie, B. On the origin of mitosis-derived human embryo aneuploidy. Nat. Commun. 2024, 15, 10391. [Google Scholar] [CrossRef]
- Nagaoka, S.I.; Hassold, T.J.; Hunt, P.A. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012, 13, 493–504. [Google Scholar] [CrossRef] [PubMed]
- McCoy, R.C. Mosaicism in preimplantation human embryos: When chromosomal abnormalities are the norm. Trends Genet. 2017, 33, 448–463. [Google Scholar] [CrossRef]
- Munné, S.; Lee, A.; Rosenwaks, Z.; Grifo, J.; Cohen, J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum. Reprod. 1993, 8, 2185–2191. [Google Scholar] [CrossRef] [PubMed]
- Dokras, A.; Sargent, I.L.; Ross, C.; Gardner, R.L.; Barlow, D.H. Trophectoderm biopsy in human blastocysts. Hum. Reprod. 1990, 5, 821–825. [Google Scholar] [CrossRef]
- Burks, C.; Van Heertum, K.; Weinerman, R. The technological advances in embryo selection and genetic testing: A look back at the evolution of aneuploidy screening and the prospects of non-invasive PGT. Reprod. Med. 2021, 2, 26–34. [Google Scholar] [CrossRef]
- Bellver, J.; Bosch, E.; Espinós, J.J.; Fabregues, F.; Fontes, J.; García-Velasco, J.; Llácer, J.; Requena, A.; Checa, M.A.; Spanish Infertility SWOT Group (SISG). Second-generation preimplantation genetic testing for aneuploidy in assisted reproduction: A SWOT analysis. Reprod. Biomed. Online 2019, 39, 905–915. [Google Scholar] [CrossRef]
- Cimadomo, D.; Capalbo, A.; Ubaldi, F.M.; Scarica, C.; Palagiano, A.; Canipari, R.; Rienzi, L. The impact of biopsy on human embryo developmental potential during preimplantation genetic diagnosis. Biomed. Res. Int. 2016, 2016, 7193075. [Google Scholar] [CrossRef]
- Xiong, S.; Liu, W.; Wang, J.; Liu, J.; Gao, Y.; Wu, L.; Zhu, J.; Hao, X.; Li, J.; Liu, D.; et al. Trophectoderm biopsy protocols may impact the rate of mosaic blastocysts in cycles with pre-implantation genetic testing for aneuploidy. J. Assist. Reprod. Genet. 2021, 38, 1153–1162. [Google Scholar] [CrossRef]
- Skinner, B.M.; Viotti, M.; International Registry of Mosaic Embryo Transfers (IRMET); Griffin, G.K.; Ellis, P.J.I. Explaining the counter-intuitive effectiveness of trophectoderm biopsy for PGT-A using computational modelling. Elife 2024, 13, RP94506. [Google Scholar] [CrossRef]
- Xu, J.; Fang, R.; Chen, L.; Chen, D.; Xiao, J.P.; Yang, W.; Wang, H.; Song, X.; Ma, T.; Bo, S.; et al. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc. Natl. Acad. Sci. USA 2016, 113, 11907–11912. [Google Scholar] [CrossRef] [PubMed]
- Shitara, A.; Takahashi, K.; Goto, M.; Takahashi, H.; Iwasawa, T.; Onodera, Y.; Makino, K.; Miura, H.; Shirasawa, H.; Sato, W.; et al. Cell-free DNA in spent culture medium effectively reflects the chromosomal status of embryos following culturing beyond implantation compared to trophectoderm biopsy. PLoS ONE 2021, 16, e0246438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, Y.; Wang, X.; Li, Q.; Tan, J.; Liang, B.; Gao, M.; Wu, J.; Ling, X.; Liu, J.; et al. Preimplantation genetic testing for structural rearrangements by genome-wide SNP genotyping and haplotype analysis: A prospective multicenter clinical study. EBioMedicine 2025, 111, 105514. [Google Scholar] [CrossRef]
- Chang, X.; Yao, Y.; Zou, Y.; Lv, Y.; Qiao, Y.; Wang, X.; Zhao, S.; Li, J.; Zhao, D.; Lu, S.; et al. Minimizing the impact of maternal contamination in embryo ploidy assessment using spent culture medium in couples with chromosomal rearrangements couples: Algorithms development and validation. Reprod. Biomed. Online, 2025; in press. [Google Scholar] [CrossRef]
- Barnes, J.; Brendel, M.; Gao, V.R.; Rajendran, S.; Kim, J.; Li, Q.; Malmsten, J.E.; Sierra, J.T.; Zisimopoulos, P.; Sigaras, A.; et al. A non-invasive artificial intelligence approach for the prediction of human blastocyst ploidy: A retrospective model development and validation study. Lancet Digit. Health 2023, 5, e28–e40. [Google Scholar] [CrossRef]
- Tesarik, J. Involvement of oocyte-coded message in cell differentiation control of early human embryos. Development 1989, 105, 317–322. [Google Scholar] [CrossRef]
- Tesarik, J. Control of Maternal-to-Zygotic Transition in Human Embryos and Other Animal Species (Especially Mouse): Similarities and Differences. Int. J. Mol. Sci. 2022, 23, 8562. [Google Scholar] [CrossRef]
- Tesarik, J.; Kopecny, V. Nucleic acid synthesis and development of human male pronucleus. J. Reprod. Fertil. 1989, 86, 549–558. [Google Scholar] [CrossRef]
- Tesarik, J.; Kopecny, V. Assembly of the nucleolar precursor bodies in human male pronuclei is correlated with an early RNA synthetic activity. Exp. Cell Res. 1990, 191, 153–156. [Google Scholar] [CrossRef]
- Asami, M.; Lam, B.Y.H.; Ma, M.K.; Rainbow, K.; Braun, S.; VerMilyea, M.D.; Yeo, G.S.H.; Perry, A.C.F. Human embryonic genome activation initiates at the one-cell stage. Cell Stem. Cell 2022, 29, 209–216.e4. [Google Scholar] [CrossRef]
- Tesarik, J.; Kopecny, V.; Plachot, M.; Mandelbaum, J. Activation of nucleolar and extranucleolar RNA synthesis and changes in the ribosomal content of human embryos developing in vitro. J. Reprod. Fertil. 1986, 78, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Tesarik, J.; Kopecny, V.; Plachot, M.; Mandelbaum, J. Ultrastructural and autoradiographic observations on multinucleated blastomeres of human cleaving embryos obtained by in-vitro fertilization. Hum. Reprod. 1987, 2, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Toporcerová, S.; Špaková, I.; Šoltys, K.; Klepcová, Z.; Kľoc, M.; Bohošová, J.; Trachtová, K.; Peterová, L.; Mičková, H.; Urdzík, P.; et al. Small Non-Coding RNAs as New Biomarkers to Evaluate the Quality of the Embryo in the IVF Process. Biomolecules 2022, 12, 1687. [Google Scholar] [CrossRef]
- Huang, W.; Chen, A.C.H.; Ng, E.H.Y.; Yeung, W.S.B.; Lee, Y.L. Non-coding RNAs as biomarkers for embryo quality and pregnancy outcomes: A systematic review and meta-analysis. Int. J. Mol. Sci. 2023, 24, 5751. [Google Scholar] [CrossRef]
- Kolanska, K.; Zerbib, E.; Dabi, Y.; Chabbert-Buffet, N.; Mathieu-d’Argent, E.; Favier, A.; Ferrier, C.; Touboul, C.; Hamamah, S.; Daraï, E. Narrative review on biofluid ncRNAs expressions in conditions associated with couple infertility. Reprod. Biomed. Online, 2025; in press. [Google Scholar] [CrossRef]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ action through miRNA editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Cuman, C.; Van Sinderen, M.; Gantier, M.P.; Rainczuk, K.; Sorby, K.; Rombauts, L.; Osianlis, T.; Dimitriadis, E. Human blastocyst secreted microRNA regulate endometrial epithelial cell adhesion. EBioMedicine 2015, 2, 1528–1535. [Google Scholar] [CrossRef]
- Rosenbluth, E.M.; Shelton, D.N.; Wells, L.M.; Sparks, A.E.; Van Voorhis, B.J. Human embryos secrete microRNAs into culture media--a potential biomarker for implantation. Fertil. Steril. 2014, 101, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Capalbo, A.; Ubaldi, F.M.; Cimadomo, D.; Noli, L.; Khalaf, Y.; Farcomeni, A.; Ilic, D.; Rienzi, L. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil. Steril. 2016, 105, e1–e3. [Google Scholar] [CrossRef]
- Azzahra, T.B.; Febri, R.R.; Iffanolida, P.A.; Mutia, K.; Wiweko, B. The correlation of chronological age and micro ribonucleic acid-135b expression in spent culture media of in vitro fertilisation patient. J. Hum. Reprod. Sci. 2022, 15, 78–81. [Google Scholar] [CrossRef]
- Mutia, K.; Wiweko, B.; Abinawanto, A.; Dwiranti, A.; Bowolaksono, A. microRNAs as a biomarker to predict embryo quality assessment in in vitro fertilization. Int. J. Fertil. Steril. 2023, 17, 85–91. [Google Scholar] [CrossRef]
- Shen, L.; Zeng, H.; Fu, Y.; Ma, W.; Guo, X.; Luo, G.; Hua, R.; Wang, X.; Shi, X.; Wu, B.; et al. Specific plasma microRNA profiles could be potential non-invasive biomarkers for biochemical pregnancy loss following embryo transfer. BMC Pregnancy Childbirth 2024, 24, 351. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, L.; Zhu, Y.; Jiang, W. Characterization of microRNAs in spent culture medium associated with human embryo quality and development. Ann. Transl. Med. 2021, 9, 1648. [Google Scholar] [CrossRef]
- Jin, J.; Ma, J.; Wang, X.; Hong, F.; Zhang, Y.; Zhou, F.; Wan, C.; Zou, Y.; Yang, J.; Lu, S.; et al. Multi-omics PGT: Re-evaluation of euploid blastocysts for implantation potential based on RNA sequencing. Hum. Reprod. 2024, 39, 2861–2872. [Google Scholar] [CrossRef]
- Braude, P.; Bolton, V.; Moore, S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 1988, 332, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Tesarik, J.; Kopecny, V.; Plachot, M.; Mandelbaum, J. Early morphological signs of embryonic genome expression in human preimplantation development as revealed by quantitative electron microscopy. Dev. Biol. 1988, 128, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Ferrick, L.; Lee, Y.S.L.; Gardner, D.K. Reducing time to pregnancy and facilitating the birth of healthy children through functional analysis of embryo physiology. Biol. Reprod. 2019, 101, 1124–1139. [Google Scholar] [CrossRef]
- Nilsson, T.; Mann, M.; Aebersold, R.; Yates, J.R., 3rd; Bairoch, A.; Bergeron, J.J. Mass spectrometry in high-throughput proteomics: Ready for the big time. Nat. Methods 2010, 7, 681–685. [Google Scholar] [CrossRef]
- Pais, R.J.; Sharara, F.; Zmuidinaite, R.; Butler, S.; Keshavarz, S.; Iles, R. Bioinformatic identification of euploid and aneuploid embryo secretome signatures in IVF culture media based on MALDI-ToF mass spectrometry. J. Assist. Reprod. Genet. 2020, 37, 2189–2198. [Google Scholar] [CrossRef]
- Iles, R.K.; Sharara, F.I.; Zmuidinaite, R.; Abdo, G.; Keshavarz, S.; Butler, S.A. Secretome profile selection of optimal IVF embryos by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Assist. Reprod. Genet. 2019, 36, 1153–1160. [Google Scholar] [CrossRef]
- Butler, S.A.; Luttoo, J.; Freire, M.O.; Abban, T.K.; Borrelli, P.T.; Iles, R.K. Human chorionic gonadotropin (hCG) in the secretome of cultured embryos: Hyperglycosylated hCG and hCG-free beta subunit are potential markers for infertility management and treatment. Reprod. Sci. 2013, 20, 1038–1045. [Google Scholar] [CrossRef]
- Dominguez, F.; Meseguer, M.; Aparicio-Ruiz, B.; Piqueras, P.; Quiñonero, A.; Simón, C. New strategy for diagnosing embryo implantation potential by combining proteomics and time-lapse technologies. Fertil. Steril. 2015, 104, 908–914. [Google Scholar] [CrossRef]
- Díaz, R.R.; Zamora, R.B.; Sánchez, R.V.; Pérez, J.G.; Bethencourt, J.C.A. Embryo sHLA-G secretion is related to pregnancy rate. Zygote 2019, 27, 78–81. [Google Scholar] [CrossRef]
- Bouvier, S.; Paulmyer-Lacroix, O.; Molinari, N.; Bertaud, A.; Paci, M.; Leroyer, A.; Robert, S.; Dignat George, F.; Blot-Chabaud, M.; Bardin, N. Soluble CD146, an innovative and non-invasive biomarker of embryo selection for in vitro fertilization. PLoS ONE 2017, 12, e0173724. [Google Scholar] [CrossRef]
- Cortezzi, S.S.; Garcia, J.S.; Ferreira, C.R.; Braga, D.P.; Figueira, R.C.; Iaconelli, A., Jr.; Souza, G.H.; Borges, E., Jr.; Eberlin, M.N. Secretome of the preimplantation human embryo by bottom-up label-free proteomics. Anal. Bioanal. Chem. 2011, 401, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Leese, H.J.; Barton, A.M. Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J. Reprod. Fertil. 1984, 72, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Leese, H.J.; Conaghan, J.; Martin, K.L.; Hardy, K. Early human embryo metabolism. Bioessays 1993, 15, 259–264. [Google Scholar] [CrossRef]
- Mahmoud, A.I. Metabolic switches during development and regeneration. Development 2023, 150, dev202008. [Google Scholar] [CrossRef]
- Houghton, F.D.; Hawkhead, J.A.; Humpherson, P.G.; Hogg, J.E.; Balen, A.H.; Rutherford, A.J.; Leese, H.J. Non-invasive amino acid turnover predicts human embryo developmental capacity. Hum. Reprod. 2002, 17, 999–1005. [Google Scholar] [CrossRef]
- Gardner, D.K.; Lane, M.; Stevens, J.; Schoolcraft, W.B. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil. Steril. 2001, 76, 1175–1180. [Google Scholar] [CrossRef]
- Huo, P.; Zhu, Y.; Liang, C.; Yao, J.; Le, J.; Qin, L.; Lei, X.; Zhang, S. Non-invasive amino acid profiling of embryo culture medium using HPLC correlates with embryo implantation potential in women undergoing in vitro fertilization. Front. Physiol. 2020, 11, 405. [Google Scholar] [CrossRef]
- Eldarov, C.; Gamisonia, A.; Chagovets, V.; Ibragimova, L.; Yarigina, S.; Smolnikova, V.; Kalinina, E.; Makarova, N.; Zgoda, V.; Sukhikh, G.; et al. LC-MS analysis revealed the significantly different metabolic profiles in spent culture media of human embryos with distinct morphology, karyotype and implantation outcomes. Int. J. Mol. Sci. 2022, 23, 2706. [Google Scholar] [CrossRef] [PubMed]
- Nami, S.; Govahi, A.; Najjar, N.; Ghasemi, S.; Rezaei, F.; Amjadi, F.; Taheripak, G. Metabolomic profiling of embryo culture media in patients with repeated implantation failure during assisted reproductive technology cycles. Clin. Exp. Reprod. Med. 2024, 51, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Z.P.; Sakkas, D.; Behr, B. Symposium: Innovative techniques in human embryo viability assessment. Non-invasive assessment of embryo viability by metabolomic profiling of culture media (‘metabolomics’). Reprod. Biomed. Online 2008, 17, 502–507. [Google Scholar] [CrossRef]
- Zhao, Q.; Yin, T.; Peng, J.; Zou, Y.; Yang, J.; Shen, A.; Hu, J. Noninvasive metabolomic profiling of human embryo culture media using a simple spectroscopy adjunct to morphology for embryo assessment in in vitro fertilization (IVF). Int. J. Mol. Sci. 2013, 14, 6556–6570. [Google Scholar] [CrossRef]
- Baştu, E.; Parlatan, U.; Başar, G.; Yumru, H.; Bavili, N.; Sağ, F.; Bulgurcuoğlu, S.; Buyru, F. Spectroscopic analysis of embryo culture media for predicting reproductive potential in patients undergoing in vitro fertilization. Turk. J. Obstet. Gynecol. 2017, 14, 145–150. [Google Scholar] [CrossRef]
- Vergouw, C.G.; Kieslinger, D.C.; Kostelijk, E.H.; Botros, L.L.; Schats, R.; Hompes, P.G.; Sakkas, D.; Lambalk, C.B. Day 3 embryo selection by metabolomic profiling of culture medium with near-infrared spectroscopy as an adjunct to morphology: A randomized controlled trial. Hum. Reprod. 2012, 27, 2304–2311. [Google Scholar] [CrossRef]
- Rinaudo, P.; Shen, S.; Hua, J.; Qian, S.; Prabhu, U.; Garcia, E.; Cedars, M.; Sukumaran, D.; Szyperski, T.; Andrews, C. (1)H NMR based profiling of spent culture media cannot predict success of implantation for day 3 human embryos. J. Assist. Reprod. Genet. 2012, 29, 1435–1442. [Google Scholar] [CrossRef]
- Meng, H.; Huang, S.; Diao, F.; Gao, C.; Zhang, J.; Kong, L.; Gao, Y.; Jiang, C.; Qin, L.; Chen, Y.; et al. Rapid and non-invasive diagnostic techniques for embryonic developmental potential: A metabolomic analysis based on Raman spectroscopy to identify the pregnancy outcomes of IVF-ET. Front. Cell Dev. Biol. 2023, 11, 1164757. [Google Scholar] [CrossRef]
- Zagers, M.S.; Laverde, M.; Goddijn, M.; de Groot, J.J.; Schrauwen, F.A.P.; Vaz, F.M.; Mastenbroek, S. The composition of commercially available human embryo culture media. Hum. Reprod. 2025, 40, 30–40. [Google Scholar] [CrossRef]
- Cuhadar, S.; Koseoglu, M.; Atay, A.; Dirican, A. The effect of storage time and freeze-thaw cycles on the stability of serum samples. Biochem. Med. 2013, 23, 70–777. [Google Scholar] [CrossRef]
- Veraguas, D.; Aguilera, C.; Henriquez, C.; Velasquez, A.E.; Melo-Baez, B.; Silva-Ibañez, P.; Castro, F.O.; Rodriguez-Alvarez, L. Evaluation of extracellular vesicles and gDNA from culture medium as a possible indicator of developmental competence in human embryos. Zygote 2021, 29, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Pavani, K.C.; Meese, T.; Pascottini, O.B.; Guan, X.; Lin, X.; Peelman, L.; Hamacher, J.; Van Nieuwerburgh, F.; Deforce, D.; Boel, A.; et al. Hatching is modulated by microRNA-378a-3p derived from extracellular vesicles secreted by blastocysts. Proc. Natl. Acad. Sci. USA 2022, 119, e2122708119. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liang, J.; Qin, T.; Zhang, Y.; Chen, X.; Wang, Z. The role of extracellular vesicles in embryo implantation. Front. Endocrinol. 2022, 13, 809596. [Google Scholar] [CrossRef] [PubMed]
- Abu-Halima, M.; Häusler, S.; Backes, C.; Fehlmann, T.; Staib, C.; Nestel, S.; Nazarenko, I.; Meese, E.; Keller, A. Micro-ribonucleic acids and extracellular vesicles repertoire in the spent culture media is altered in women undergoing In Vitro Fertilization. Sci. Rep. 2017, 7, 13525. [Google Scholar] [CrossRef]
- Pallinger, E.; Bognar, Z.; Bodis, J.; Csabai, T.; Farkas, N.; Godony, K.; Varnagy, A.; Buzas, E.; Szekeres-Bartho, J. A simple and rapid flow cytometry-based assay to identify a competent embryo prior to embryo transfer. Sci. Rep. 2017, 7, 39927. [Google Scholar] [CrossRef]
- Fernandez, E.I.; Ferreira, A.S.; Cecílio, M.H.M.; Chéles, D.S.; de Souza, R.C.M.; Nogueira, M.F.G.; Rocha, J.C. Artificial intelligence in the IVF laboratory: Overview through the application of different types of algorithms for the classification of reproductive data. J. Assist. Reprod. Genet. 2020, 37, 2359–2376. [Google Scholar] [CrossRef]
- Zhao, C.; Plaza Reyes, A.; Schell, J.P.; Weltner, J.; Ortega, N.M.; Zheng, Y.; Björklund, Å.K.; Baqué-Vidal, L.; Sokka, J.; Trokovic, R.; et al. A comprehensive human embryo reference tool using single-cell RNA-sequencing data. Nat. Methods 2025, 22, 193–206. [Google Scholar] [CrossRef]
- Kieslinger, D.C.; Lambalk, C.B.; Vergouw, C.G. The inconvenient reality of AI-assisted embryo selection in IVF. Nat. Med. 2024, 30, 3059–3060. [Google Scholar] [CrossRef]
| Marker | Reference Outcome | Predictive Capacity | Techniques | References |
|---|---|---|---|---|
| PGT-A | Whole-embryo ploidy and pregnancy | Intermediate and questionable | FISH, PCR, CGH, NGS | [37,38,39,40,41,42,43] |
| NICS (niPGT-A) | Whole-embryo ploidy and pregnancy | Very high but needing confirmation by RCTs | NGS after WGA | [44,45,46,47] |
| Marker | Reference Outcome | Predictive Capacity | Techniques | References |
|---|---|---|---|---|
| Carbohydrate profile | Embryo viability and pregnancy | Doubtful for NIR and NMR spectroscopies | Historical: TLC, ELISA Current: HPLC and Raman, NIR, or NMR spectroscopies | [83,84,85,86,87,88,89,90,91,92] |
| Amino acid profile | Embryo viability and pregnancy | High for HPLC and Raman spectroscopy |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tesarik, J. Noninvasive Biomarkers of Human Embryo Developmental Potential. Int. J. Mol. Sci. 2025, 26, 4928. https://doi.org/10.3390/ijms26104928
Tesarik J. Noninvasive Biomarkers of Human Embryo Developmental Potential. International Journal of Molecular Sciences. 2025; 26(10):4928. https://doi.org/10.3390/ijms26104928
Chicago/Turabian StyleTesarik, Jan. 2025. "Noninvasive Biomarkers of Human Embryo Developmental Potential" International Journal of Molecular Sciences 26, no. 10: 4928. https://doi.org/10.3390/ijms26104928
APA StyleTesarik, J. (2025). Noninvasive Biomarkers of Human Embryo Developmental Potential. International Journal of Molecular Sciences, 26(10), 4928. https://doi.org/10.3390/ijms26104928







