Mass Spectrometric ITEM-FOUR Analysis Reveals Coding Single-Nucleotide Polymorphisms in Human Cardiac Troponin T That Evade Detection by Sandwich ELISAs Which Use Monoclonal Antibodies M7 and M11.7 from the Elecsys Troponin T® Assay
Abstract
1. Introduction
2. Results
2.1. Characterization of Antibodies and Epitope Peptides
2.2. Binding Strength Analysis by ITEM-FOUR Mass Spectrometry
2.3. Binding Motif Deduction
3. Discussion
4. Materials and Methods
4.1. Preparation of Solutions with Peptides, Antibodies, and Antibody–Peptide Complexes
4.2. Protein and Peptide Concentration Determination
4.3. SDS-PAGE Analysis of the hcTnT Antigen
4.4. Western Blot Analysis of the Anti-hcTnT Antibodies
4.5. Mass Calibration of Mass Spectrometry Instruments
4.6. Preparation of Nano-ESI-MS Emitters, Filling and Mounting
4.7. Q-ToF 2 Instrument Settings and Data Acquisition
4.8. Synapt G2S Instrument Settings and Data Acquisition
4.9. ITEM-FOUR Spectral Data Analysis
4.10. ITEM-FOUR Calculations of Apparent Kinetic and Apparent Thermodynamic Values
4.11. Molecular Modeling of Protein and Peptide Structures
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| Å | Ångström |
| ACC | American College of Cardiology |
| anti-TNFα | anti-tumor necrosis factor alpha |
| BSA | bovine serum albumin |
| cSNP | coding single-nucleotide polymorphism |
| cTnT | cardiac troponin T |
| DCM | dilative cardiomyopathy |
| dbSNP | single-nucleotide polymorphism database |
| ELISA | enzyme-linked immunosorbent assay |
| ESC | European Society for Cardiology |
| ESI | electrospray ionization |
| HCM | hypertrophic cardiomyopathy |
| hcTNT | human cardiac troponin T |
| hs | high-sensitivity |
| ITEM | intact transition epitope mapping |
| ITEM FOUR | intact transition epitope mapping—force differences between original and unusual residues |
| kDa | kiloDalton |
| MI | myocardial infarction |
| m/z | mass-to-charge ratio |
| nano-ESI-MS | nano-electrospray ionization–mass spectrometry |
| PBS | phosphate-buffered saline |
| POC | point of care |
| PRIDE | PRoteomics IDEntification |
| PVDF | polyvinylidene fluoride |
| Q-ToF | quadrupole time of flight |
| RCM | restrictive cardiomyopathy |
| rpm | revolutions per minute |
| RIPA | radioimmunoprecipitation assay |
| SDC | sodium deoxycholate |
| SDS | sodium dodecyl sulfate |
| SDS-PAGE | sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| SNP | single-nucleotide polymorphism |
| tif | tagged image file |
| Tn | troponin |
| TnI | troponin I |
| TNNT2 | troponin T gene |
| TnT | troponin T |
| TRIS | tris(hydroxymethyl)aminomethane |
| ΔCV | voltage difference in collision cell |
| 3D | three-dimensional |
References
- Dwivedi, S.; Purohit, P.; Misra, R.; Pareek, P.; Goel, A.; Khattri, S.; Pant, K.K.; Misra, S.; Sharma, P. Diseases and Molecular Diagnostics: A Step Closer to Precision Medicine. Indian J. Clin. Biochem. 2017, 32, 374–398. [Google Scholar] [CrossRef] [PubMed]
- Jeremias, A. The utility of troponin measurement to detect myocardial infarction: Review of the current findings. Vasc. Health Risk Manag. 2010, 6, 691–699. [Google Scholar] [CrossRef]
- Mingels, A.M.; Mills, N.L.; Mueller, C. Cardiac troponin T and I: Back to basics. Eur. Heart J. Acute Cardiovasc. Care 2023, 12, 631–632. [Google Scholar] [CrossRef]
- Mohammed, A.A.; Januzzi, J.L., Jr. Clinical applications of highly sensitive troponin assays. Cardiol. Rev. 2010, 18, 12–19. [Google Scholar] [CrossRef]
- Mair, J.; Hammarsten, O. Potential analytical interferences in cardiac troponin immunoassays. J. Lab. Precis. Med. 2023, 8, 12. [Google Scholar] [CrossRef]
- von Meyer, A.; Albert, G.; Kunzelmann, S.; Rank, C.; Zerback, R.; Imdahl, R. Evaluating the performance of an updated high-sensitivity troponin T assay with increased tolerance to biotin. Clin. Chem. Lab. Med. 2021, 59, 591–597. [Google Scholar] [CrossRef]
- Lippi, G.; Cervellin, G. Genetic polymorphisms of human cardiac troponins as an unrecognized challenge for diagnosing myocardial injury. Int. J. Cardiol. 2014, 171, 467–470. [Google Scholar] [CrossRef]
- Antontseva, E.V.; Degtyareva, A.O.; Korbolina, E.E.; Damarov, I.S.; Merkulova, T.I. Human-genome single nucleotide polymorphisms affecting transcription factor binding and their role in pathogenesis. Vavilov J. Genet. Breed. 2023, 27, 662–675. [Google Scholar] [CrossRef]
- Manolio, T.A.; Brooks, L.D.; Collins, F.S. A HapMap harvest of insights into the genetics of common disease. J. Clin. Investig. 2008, 118, 1590–1605. [Google Scholar] [CrossRef]
- Alwi, Z.B. The Use of SNPs in Pharmacogenomics Studies. Malays. J. Med. Sci. 2005, 12, 4–12. [Google Scholar]
- Li, X.; Luo, R.; Gu, H.; Deng, Y.; Xu, X.; Wu, X.; Hua, W. Cardiac troponin T (TNNT2) mutations in chinese dilated cardiomyopathy patients. Biomed. Res. Int. 2014, 2014, 907360. [Google Scholar] [CrossRef]
- Myers, M.C.; Wang, S.; Zhong, Y.; Maruyama, S.; Bueno, C.; Bastien, A.; Fazeli, M.S.; Golchin, N. Prevalence of Genetically Associated Dilated Cardiomyopathy: A Systematic Literature Review and Meta-Analysis. Cardiol. Res. 2024, 15, 233–245. [Google Scholar] [CrossRef]
- Glavaški, M.; Velicki, L.; Vučinić, N. Hypertrophic Cardiomyopathy: Genetic Foundations, Outcomes, Interconnections, and Their Modifiers. Medicina 2023, 59, 1424. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, R.L.; Hollander, J.E.; Peacock, W.F.; Limkakeng, A.T.; Breitenbeck, N.; Rivers, E.J.; Ziegler, A.; Laimighofer, M.; deFilippi, C. The 99th percentile upper reference limit for the 5th generation cardiac troponin T assay in the United States. Clin. Chim. Acta 2020, 504, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Macht, M.; Fiedler, W.; Kürzinger, K.; Przybylski, M. Mass spectrometric mapping of protein epitope structures of myocardial infarct markers myoglobin and troponin T. Biochemistry 1996, 35, 15633–15639. [Google Scholar] [CrossRef]
- Schmid, J.; Liesinger, L.; Birner-Gruenberger, R.; Stojakovic, T.; Scharnagl, H.; Dieplinger, B.; Asslaber, M.; Radl, R.; Beer, M.; Polacin, M.; et al. Elevated Cardiac Troponin T in Patients With Skeletal Myopathies. J. Am. Coll. Cardiol. 2018, 71, 1540–1549. [Google Scholar] [CrossRef]
- Macht, M.; Marquardt, A.; Deininger, S.O.; Damoc, E.; Kohlmann, M.; Przybylski, M. “Affinity-proteomics”: Direct protein identification from biological material using mass spectrometric epitope mapping. Anal. Bioanal. Chem. 2004, 378, 1102–1111. [Google Scholar] [CrossRef]
- Röwer, C.; Ortmann, C.; Neamtu, A.; El-Kased, R.F.; Glocker, M.O. Intact Transition Epitope Mapping-Force Differences between Original and Unusual Residues (ITEM-FOUR). Biomolecules 2023, 13, 187. [Google Scholar] [CrossRef]
- Yefremova, Y.; Danquah, B.D.; Opuni, K.F.; El-Kased, R.; Koy, C.; Glocker, M.O. Mass spectrometric characterization of protein structures and protein complexes in condensed and gas phase. Eur. J. Mass Spectrom. 2017, 23, 445–459. [Google Scholar] [CrossRef]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F.; Whitehouse, C.M. Electrospray Ionization for Mass Spectrometry of Large Biomolecules. Science 1989, 246, 64–71. [Google Scholar] [CrossRef]
- Alexandrov, M.L.; Gall, L.N.; Krasnov, N.V.; Nikolaev, V.I.; Pavlenko, V.A.; Shkurov, V.A. Extraction of ions from solutions under atmospheric pressure as a method for mass spectrometric analysis of bioorganic compounds. Rapid Commun. Mass Spectrom. 2008, 22, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Danquah, B.D.; Opuni, K.F.M.; Roewer, C.; Koy, C.; Glocker, M.O. Mass Spectrometric Analysis of Antibody-Epitope Peptide Complex Dissociation: Theoretical Concept and Practical Procedure of Binding Strength Characterization. Molecules 2020, 25, 4776. [Google Scholar] [CrossRef] [PubMed]
- Glish, G.L.; Vachet, R.W. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug Discov. 2003, 2, 140–150. [Google Scholar] [CrossRef]
- Morris, C.B.; Poland, J.C.; May, J.C.; McLean, J.A. Fundamentals of Ion Mobility-Mass Spectrometry for the Analysis of Biomolecules. Meth Mol. Biol. 2020, 2084, 1–31. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, Z.; Li, S.; Rong, X.; Bu, J.; Liu, Q.; Ouyang, Z. Differentiating enantiomers by directional rotation of ions in a mass spectrometer. Science 2024, 383, 612–618. [Google Scholar] [CrossRef]
- Nibbering, N.M.M. The role of mass spectrometric methods in ionic reaction mechanistic studies. Int. J. Mass Spectrom. 2000, 200, 27–42. [Google Scholar] [CrossRef]
- Mehmood, S.; Allison, T.M.; Robinson, C.V. Mass spectrometry of protein complexes: From origins to applications. Annu. Rev. Phys. Chem. 2015, 66, 453–474. [Google Scholar] [CrossRef]
- Christ, P.; Rutzinger, S.; Seidel, W.; Uchaikin, S.; Pro, F.; Koy, C.; Glocker, M.O. High detection sensitivity achieved with cryogenic detectors in combination with matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Eur. J. Mass Spectrom. 2004, 10, 469–476. [Google Scholar] [CrossRef]
- Strauß, M.; Shayeghi, A.; Mauser, M.F.X.; Geyer, P.; Kostersitz, T.; Salapa, J.; Dobrovolskiy, O.; Daly, S.; Commandeur, J.; Hua, Y.; et al. Highly sensitive single-molecule detection of macromolecule ion beams. Sci. Adv. 2023, 9, eadj2801. [Google Scholar] [CrossRef]
- Scherf, M.; Danquah, B.D.; Koy, C.; Lorenz, P.; Steinbeck, F.; Neamtu, A.; Thiesen, H.J.; Glocker, M.O. Epitope Fine Mapping by Mass Spectrometry: Investigations of Immune Complexes Consisting of Monoclonal Anti-HpTGEKP Antibody and Zinc Finger Protein Linker Phospho-Hexapeptides. ChemBioChem 2022, 23, e202200390. [Google Scholar] [CrossRef]
- Puniya, B.L.; Verma, M.; Damiani, C.; Bakr, S.; Dräger, A. Perspectives on computational modeling of biological systems and the significance of the SysMod community. Bioinf. Adv. 2024, 4, vbae090. [Google Scholar] [CrossRef]
- Hager-Braun, C.; Tomer, K.B. Determination of protein-derived epitopes by mass spectrometry. Expert Rev. Proteom. 2005, 2, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Opuni, K.F.; Ruß, M.; Geens, R.; De Vocht, L.; Van Wielendaele, P.; Debuy, C.; Sterckx, Y.G.-J.; Glocker, M.O. Mass spectrometry-complemented molecular modeling predicts the interaction interface for a camelid single-domain antibody targeting the Plasmodium falciparum circumsporozoite protein’s C-terminal domain. Comput. Struct. Biotechnol. J. 2024, 23, 3300–3314. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, N.D.; Mayrose, I.; Halperin, D.; Yekutieli, D.; Gershoni, J.M.; Pupko, T. Computational characterization of B-cell epitopes. Mol. Immunol. 2008, 45, 3477–3489. [Google Scholar] [CrossRef] [PubMed]
- Al-Majdoub, M.; Koy, C.; Lorenz, P.; Thiesen, H.J.; Glocker, M.O. Mass spectrometric and peptide chip characterization of an assembled epitope: Analysis of a polyclonal antibody model serum directed against the Sjøgren/systemic lupus erythematosus autoantigen TRIM21. J. Mass Spectrom. 2013, 48, 651–659. [Google Scholar] [CrossRef]
- Saul, F.A.; Alzari, P.M. Crystallographic Studies of Antigen-Antibody Interactions. In Epitope Mapping Protocols; Morris, G.E., Ed.; Humana Press: Totowa, NJ, USA, 1996; pp. 11–23. [Google Scholar]
- Al-Majdoub, M.; Opuni, K.; Koy, C.; Glocker, M. Facile fabrication and instant application of miniaturized antibody-decorated affinity columns for higher-order structure and functional characterization of TRIM21 epitope peptides. Anal. Chem. 2013, 85, 10479–10487. [Google Scholar] [CrossRef]
- Finkelstein, A.V.; Badretdinov, A.Y.; Ptitsyn, O.B. Physical reasons for secondary structure stability: Alpha-helices in short peptides. Proteins 1991, 10, 287–299. [Google Scholar] [CrossRef]
- Opuni, K.F.; Al-Majdoub, M.; Yefremova, Y.; El-Kased, R.F.; Koy, C.; Glocker, M.O. Mass spectrometric epitope mapping. Mass Spectrom. Rev. 2018, 37, 229–241. [Google Scholar] [CrossRef]
- Yu, W.-Y.; Watts, R.; Karr, R.W. Identification of amino acids in HLA-DPw4bβ and -DR5β1 chains that are involved in antibody binding epitodes using site-directed mutagenesis and DNA-mediated gene transfer. Hum. Immunol. 1990, 27, 122–135. [Google Scholar] [CrossRef]
- Wiedemann, P.; Giehl, K.; Almo, S.C.; Fedorov, A.A.; Girvin, M.; Steinberger, P.; Rüdiger, M.; Ortner, M.; Sippl, M.; Dolecek, C.; et al. Molecular and Structural Analysis of a Continuous Birch Profilin Epitope Defined by a Monoclonal Antibody*. J. Biol. Chem. 1996, 271, 29915–29921. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, X.; Tian, Y.; Li, F.; Zhao, X.; Liu, J.; Yao, C.; Zhang, Y. Point mutation in CD19 facilitates immune escape of B cell lymphoma from CAR-T cell therapy. J. Immunother. Cancer 2020, 8, e001150. [Google Scholar] [CrossRef]
- Cheng, H.D.; Grimm, S.K.; Gilman, M.S.; Gwom, L.C.; Sok, D.; Sundling, C.; Donofrio, G.; Karlsson Hedestam, G.B.; Bonsignori, M.; Haynes, B.F.; et al. Fine epitope signature of antibody neutralization breadth at the HIV-1 envelope CD4-binding site. JCI Insight 2018, 3, e97018. [Google Scholar] [CrossRef] [PubMed]
- Hoffmüller, U.; Knaute, T.; Hahn, M.; Höhne, W.; Schneider-Mergener, J.; Kramer, A. Evolutionary transition pathways for changing peptide ligand specificity and structure. Embo J. 2000, 19, 4866–4874. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yefremova, Y.; Opuni, K.F.-M.; Danquah, B.D.; Thiesen, H.-J.; Glocker, M.O. Intact Transition Epitope Mapping (ITEM). J. Am. Soc. Mass Spectrom. 2017, 28, 1612–1622. [Google Scholar] [CrossRef] [PubMed]
- Richard, P.; Charron, P.; Carrier, L.; Ledeuil, C.L.; Cheav, T.; Pichereau, C.; Benaiche, A.; Isnard, R.; Dubourg, O.; Burban, M.; et al. Hypertrophic Cardiomyopathy. Circulation 2003, 107, 2227–2232. [Google Scholar] [CrossRef]
- Huang, S.; Li, J.; Li, Q.; Wang, Q.; Zhou, X.; Chen, J.; Chen, X.; Bellou, A.; Zhuang, J.; Lei, L. Cardiomyopathy: Pathogenesis and therapeutic interventions. MedComm 2024, 5, e772. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Luo, R.; Gu, H.; Zhang, C.; Zhang, Y.; Hui, R.; Wu, X.; Hua, W. TNNT2 gene polymorphisms are associated with susceptibility to idiopathic dilated cardiomyopathy in the Han Chinese population. Biomed Res. Int. 2013, 2013, 201372. [Google Scholar] [CrossRef]
- Rani, D.S.; Dhandapany, P.S.; Nallari, P.; Narasimhan, C.; Thangaraj, K. A novel arginine to tryptophan (R144W) mutation in troponin T (cTnT) gene in an indian multigenerational family with dilated cardiomyopathy (FDCM). PLoS ONE 2014, 9, e101451. [Google Scholar] [CrossRef]
- Arbelo, E.; Protonotarios, A.; Gimeno, J.R.; Arbustini, E.; Barriales-Villa, R.; Basso, C.; Bezzina, C.R.; Biagini, E.; Blom, N.A.; de Boer, R.A.; et al. 2023 ESC Guidelines for the Management of Cardiomyopathies, ESC Clinical Practice Guidelines; European Society of Cardiology, 2023; Available online: https://www.escardio.org/Guidelines/Clinical-Practice-Guidelines/Cardiomyopathy-Guidelines (accessed on 30 November 2023).
- Ommen, S.R.; Ho, C.Y.; Asif, I.M.; Balaji, S.; Burke, M.A.; Day, S.M.; Dearani, J.A.; Epps, K.C.; Evanovich, L.; Ferrari, V.A.; et al. 2024 AHA/ACC/AMSSM/HRS/PACES/SCMR Guideline for the Management of Hypertrophic Cardiomyopathy: A Report of the American Heart Association/American College of Cardiology Joint Committee on Clinical Practice Guidelines. Circulation 2024, 149, e1239–e1311. [Google Scholar] [CrossRef]
- Kockum, I.; Huang, J.; Stridh, P. Overview of Genotyping Technologies and Methods. Curr. Protoc. 2023, 3, e727. [Google Scholar] [CrossRef]
- Grant, P.; Langlois, S.; Lynd, L.D.; Austin, J.C.; Elliott, A.M. Out-of-pocket and private pay in clinical genetic testing: A scoping review. Clin. Genet. 2021, 100, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, A.P. Collaborative Paradigm of Preventive, Personalized, and Precision Medicine With Point-of-Care Technologies. IEEE J. Transl. Eng. Health Med. 2016, 4, 2800908. [Google Scholar] [CrossRef]
- Koy, C.; Opuni, K.F.M.; Danquah, B.D.; Neamtu, A.; Glocker, M.O. Mass Spectrometric and Bio-Computational Binding Strength Analysis of Multiply Charged RNAse S Gas-Phase Complexes Obtained by Electrospray Ionization from Varying In-Solution Equilibrium Conditions. Int. J. Mol. Sci. 2021, 22, 10183. [Google Scholar] [CrossRef] [PubMed]
- Danquah, B.D.; Yefremova, Y.; Opuni, K.F.; Röwer, C.; Koy, C.; Glocker, M.O. Intact Transition Epitope Mapping—Thermodynamic Weak-force Order (ITEM-TWO). J. Proteom. 2019, 212, 103572. [Google Scholar] [CrossRef]
- Opuni, K.F.M.; Koy, C.; Russ, M.; Reepmeyer, M.; Danquah, B.D.; Weresow, M.; Alef, A.; Lorenz, P.; Thiesen, H.J.; Glocker, M.O. ITEM-THREE analysis of a monoclonal anti-malaria antibody reveals its assembled epitope on the pfMSP119 antigen. J. Biol. Chem. 2020, 295, 14987–14997. [Google Scholar] [CrossRef]
- Heitner, J.C.; Koy, C.; Kreutzer, M.; Gerber, B.; Reimer, T.; Glocker, M.O. Differentiation of HELLP patients from healthy pregnant women by proteome analysis–on the way towards a clinical marker set. J. Chromatogr. B Anal. Technol. Biomed Life Sci. 2006, 840, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Röwer, C.; George, C.; Reimer, T.; Stengel, B.; Radtke, A.; Gerber, B.; Glocker, M.O. Distinct Ezrin Truncations Differentiate Metastases in Sentinel Lymph Nodes from Unaffected Lymph Node Tissues, from Primary Breast Tumors, and from Healthy Glandular Breast Tissues. Transl. Oncol. 2017, 11, 1–10. [Google Scholar] [CrossRef]
- Kang, D.-H.; Gho, Y.-S.; Suh, M.-K.; Kang, C.-H. Highly sensitive and fast protein detection with coomassie brilliant blue in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Bull. Korean Chem. Soc. 2002, 23, 1511–1512. [Google Scholar] [CrossRef]
- Lorenz, P.; Koczan, D.; Thiesen, H.-J. Transcriptional Repression Mediated by the KRAB Domain of the Human C2H2 Zinc Finger Protein Kox1/ZNF10 Does Not Require Histone Deacetylation. Biol. Chem. 2001, 382, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Kyhse-Andersen, J. Electroblotting of multiple gels: A simple apparatus without buffer tank for rapid transfer of proteins from polycrylamide to nitrocellulose. J. Biochem. Biophys. Methods 1984, 10, 203–209. [Google Scholar] [CrossRef]
- Eickner, T. Anreicherung und Charakterisierung von TRIM28-Interaktionspartnern mit Massenspektrometrischen Methoden. Ph.D. Thesis, Proteom-Zentrum Rostock, Rostock, Germany, 2012. [Google Scholar]
- Born, N.; Thiesen, H.-J.; Lorenz, P. The B-Subdomain of the Xenopus laevis XFIN KRAB-AB Domain Is Responsible for Its Weaker Transcriptional Repressor Activity Compared to Human ZNF10/Kox1. PLoS ONE 2014, 9, e87609. [Google Scholar] [CrossRef] [PubMed]
- Al Chiblak, M.; Steinbeck, F.; Thiesen, H.-J.; Lorenz, P. DUF3669, a “domain of unknown function” within ZNF746 and ZNF777, oligomerizes and contributes to transcriptional repression. BMC Mol. Cell Biol. 2019, 20, 60. [Google Scholar] [CrossRef]
- Scherf, M.; Koy, C.; Röwer, C.; Neamtu, A.; Glocker, M.O. Characterization of Phosphorylation-Dependent Antibody Binding to Cancer-Mutated Linkers of C2H2 Zinc Finger Proteins by Intact Transition Epitope Mapping-Thermodynamic Weak-Force Order Analysis. J. Am. Soc. Mass Spectrom. 2023, 34, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Danquah, B.D.; Röwer, C.; Opuni, K.M.; El-Kased, R.; Frommholz, D.; Illges, H.; Koy, C.; Glocker, M.O. Intact Transition Epitope Mapping–Targeted High-Energy Rupture of Extracted Epitopes (ITEM-THREE)*[S]. Mol. Cell. Proteom. 2019, 18, 1543–1555. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Bannister, D.; Popovic, B.; Sridharan, S.; Giannotta, F.; Filée, P.; Yilmaz, N.; Minter, R. Epitope mapping and key amino acid identification of anti-CD22 immunotoxin CAT-8015 using hybrid β-lactamase display. Protein Eng. Des. Sel. 2010, 24, 351–360. [Google Scholar] [CrossRef][Green Version]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef] [PubMed]
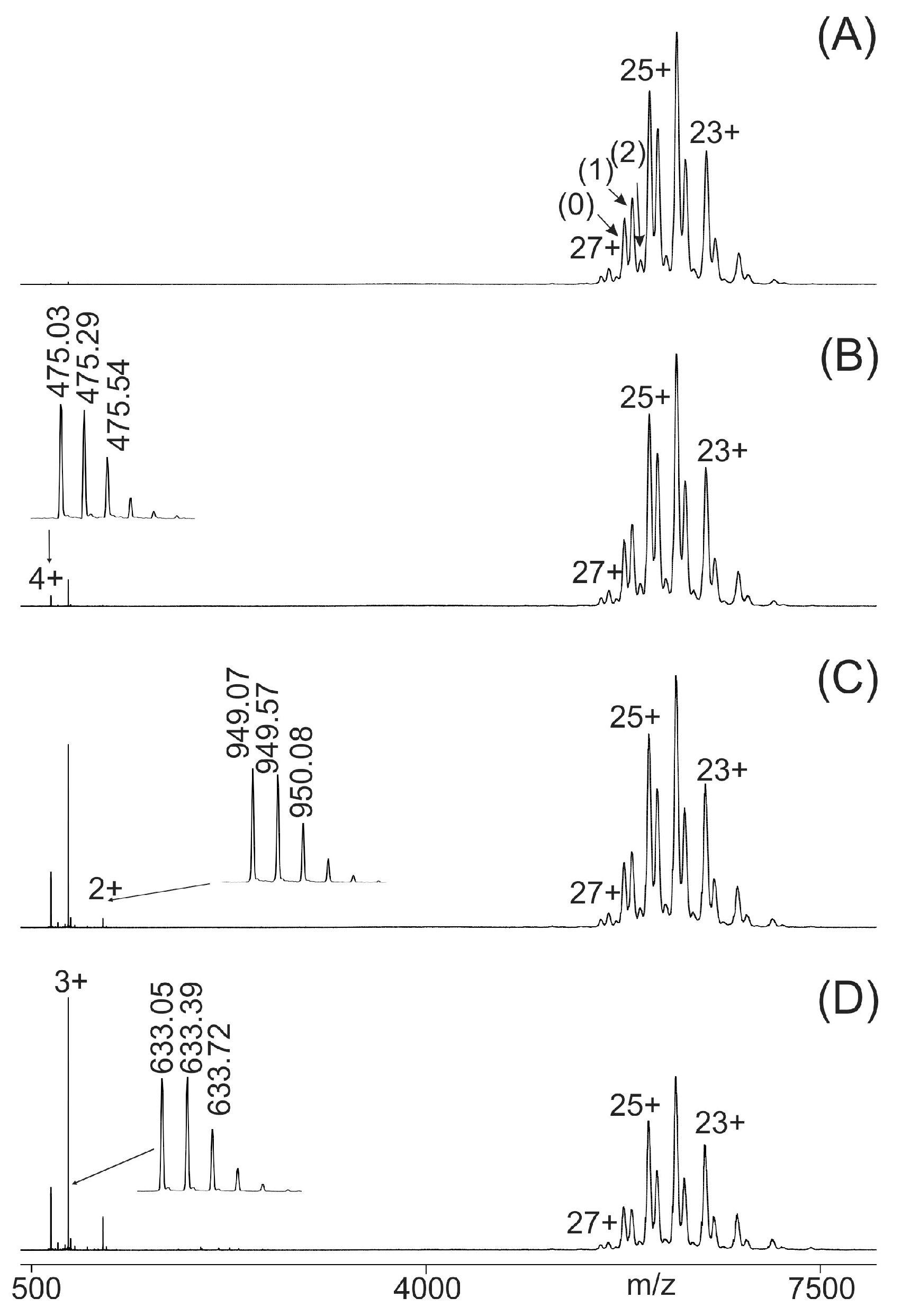
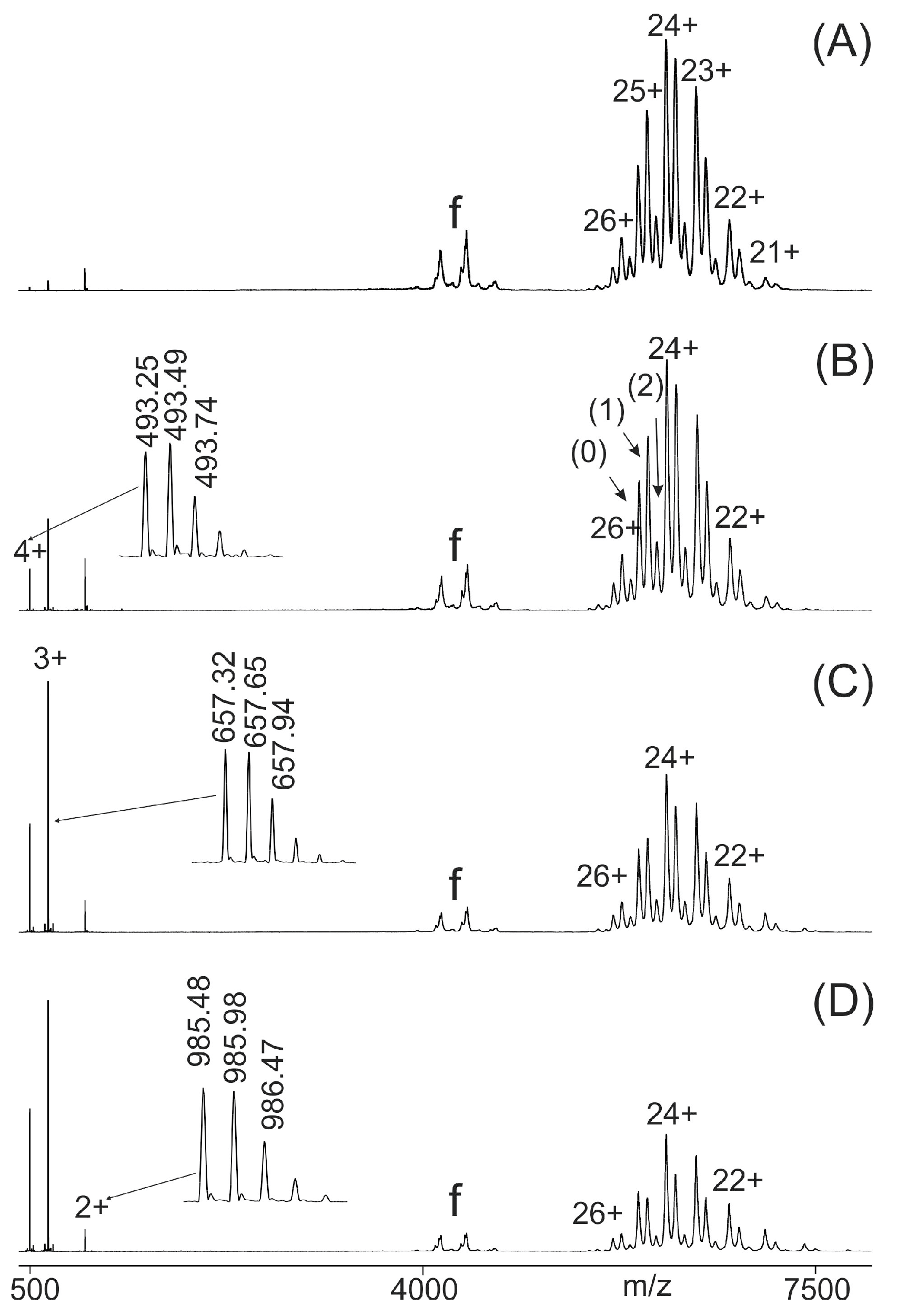
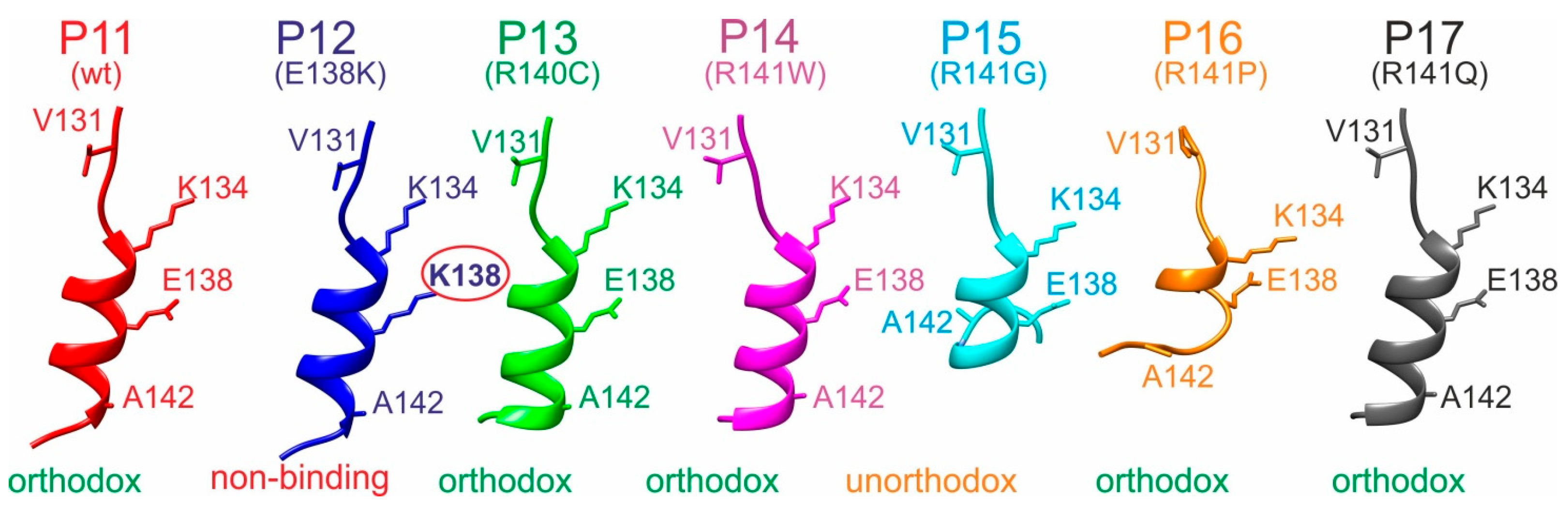


| Peptide No. (wt or SAP) (a,b) | Amino Acid Sequence (wt or SAP) (a,b) | SNP Entry (c) | SNP (c) | Cardiomyopathy Association (c) | Atom No. | Mr (Mono) (d) | MM (Exp.) (e) |
|---|---|---|---|---|---|---|---|
| 11 (wt) | LVSLKDRIERRRAER | n.a. | n.a. | n.a. | 278 | 1896.14 | 1896.91 |
| 12 (E138K) | LVSLKDRIKRRRAER | rs 730881100 | G > A | hypertrophic | 283 | 1896.27 | 1896.44 |
| 13 (R140C) | LVSLKDRIERCRAER | rs 397516463 | C > T | hypertrophic/familial restrictive | 264 | 1842.03 | 1842.09 |
| 14 (R141W) | LVSLKDRIERRWAER | rs 74315380 | C > T | dilated | 279 | 1926.11 | 1926.36 |
| 15 (R141G) | LVSLKDRIERRGAER | rs 74315380 | C > G | not provided | 262 | 1797.07 | 1797.19 |
| 16 (R141P) | LVSLKDRIERRPAER | rs 397516464 | G > C | dilated | 269 | 1837.08 | 1837.43 |
| 17 (R141Q) | LVSLKDRIERRQAER | rs 397516464 | G > A | dilated | 272 | 1868.09 | 1868.18 |
| 21 (wt) | AEQQRIRNEREKERQ | n.a. | n.a. | n.a. | 274 | 1969.04 | 1969.67 |
| 22 (E146Q) | AQQQRIRNEREKERQ | rs 371142225 | G > C | dilated | 275 | 1968.07 | 1968.78 |
| 23 (E146K) | AKQQRIRNEREKERQ | rs 371142225 | G > A | dilated | 279 | 1968.10 | 1969.02 |
| 24 (Q148R) | AEQRRIRNEREKERQ | rs 730880232 | A > G | not provided | 280 | 1997.08 | 1997.43 |
| 25 (R149C) | AEQQCIRNEREKERQ | rs 397516465 | C > T | familial dilated | 261 | 1914.94 | 1915.46 |
| 26 (R149S) | AEQQSIRNEREKERQ | rs 397516465 | C > A | not provided | 262 | 1899.96 | 1899.56 |
| 27 (R149H) | AEQQHIRNEREKERQ | rs 397516466 | G > A | dilated | 268 | 1950.00 | 1951.24 |
| 28 (R151W) | AEQQRIWNEREKERQ | rs 74315379 | C > T | dilated | 275 | 1999.03 | 2000.02 |
| 29 (R151Q) | AEQQRIQNEREKERQ | rs 730881101 | G > A | familial restrictive | 268 | 1941.00 | 1941.99 |
| 30 (R154W) | AEQQRIRNEWEKERQ | rs 483352832 | C > T | dilated | 275 | 1999.03 | 2000.95 |
| 31 (R154Q) | AEQQRIRNEQEKERQ | rs 745632066 | G > A | familial restrictive | 268 | 1941.00 | 1941.90 |
| 32 (E155K) | AEQQRIRNERKKERQ | rs 984218824 | G > A | familial restrictive | 279 | 1968.10 | 1969.03 |
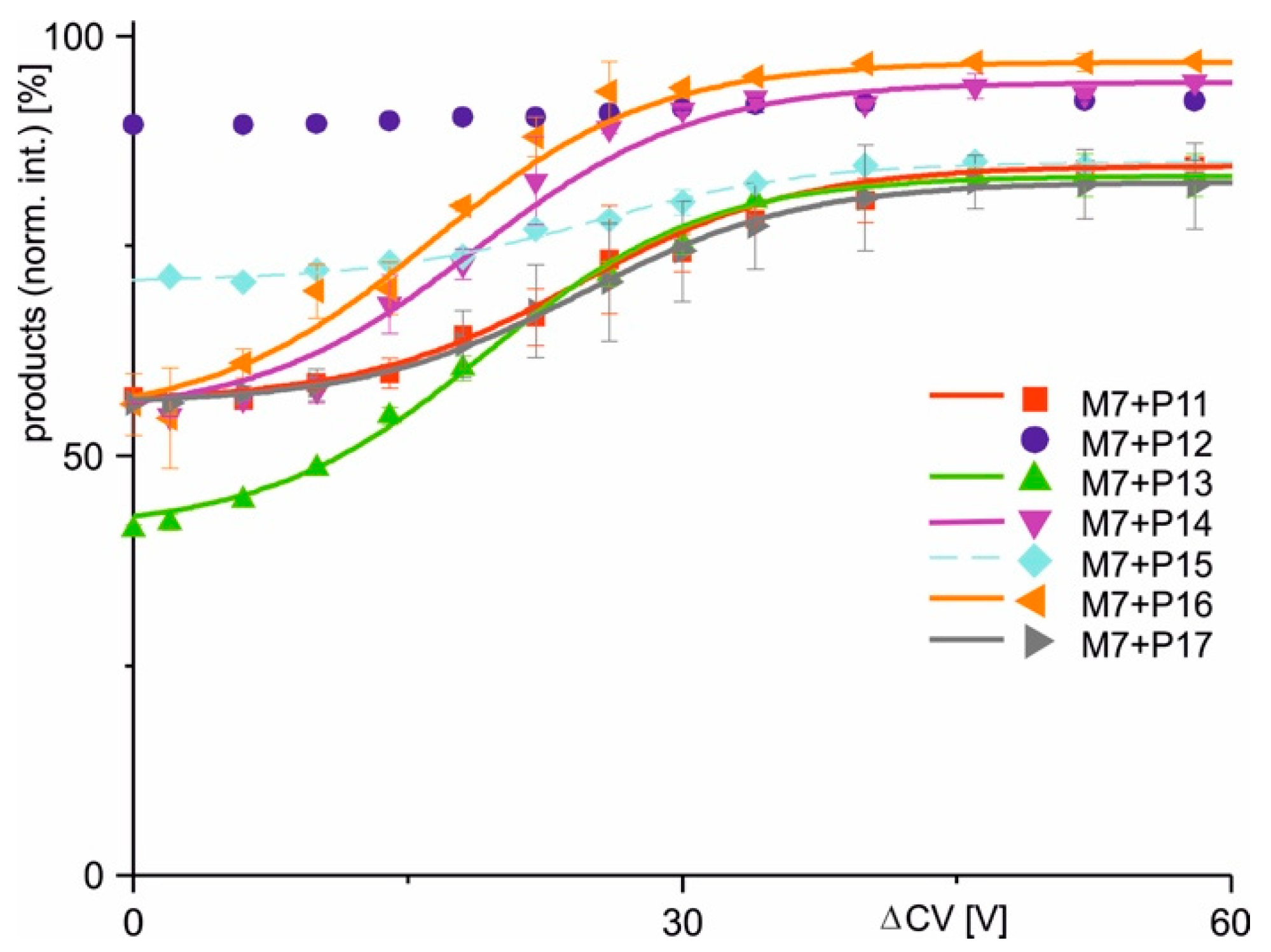
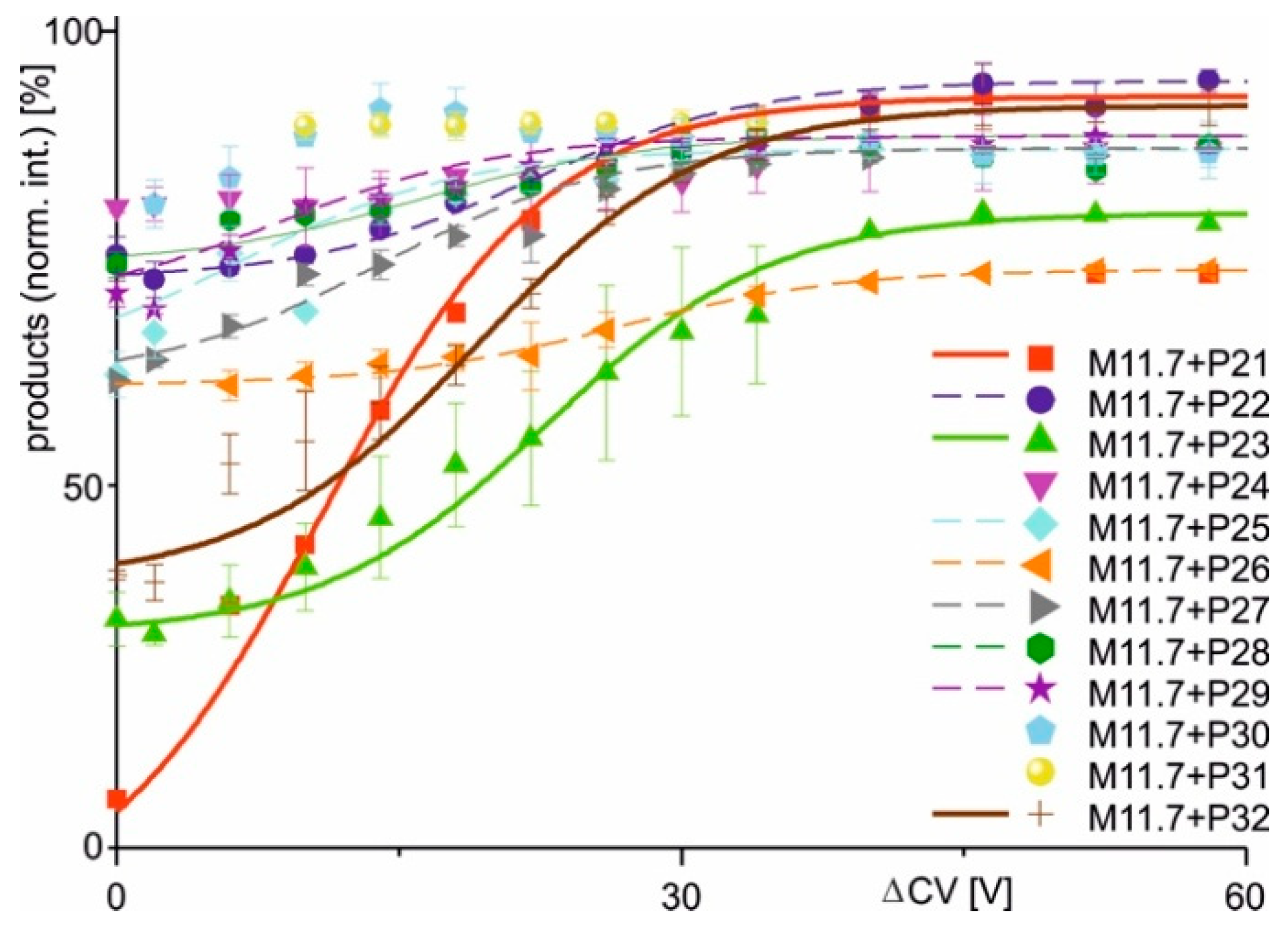
| Complex (a) | Peptide Sequence | Mean Charge ± SD. (b,c) | Initial (%) (b,c,d) | Final (%) (b,c,e) | Δ (% pts) | ΔCV50 (V) (b) | dx (V) (b) | Slope (%/V) (b) | R2 (b,c) |
|---|---|---|---|---|---|---|---|---|---|
| M7 + P11 | LVSLKDIERRRAER | 24.1 ± 0.23 | 56.43 | 84.36 | 27.93 | 25.27 | 6.69 | 1.04 | 0.995 |
| M7 + P12 | LVSLKDIKRRRAER | 24.5 ± 0.67 | 89.38 | 92.28 | 2.90 | n.a. | n.a. | n.a. | n.a. |
| M7 + P13 | LVSLKDIERCRAER | 24.3 ± 0.29 | 40.36 | 83.40 | 43.04 | 23.30 | 6.55 | 1.64 | 0.999 |
| M7 + P14 | LVSLKDIERRWAER | 24.1 ± 0.22 | 54.37 | 93.86 | 39.49 | 18.02 | 4.33 | 2.28 | 0.997 |
| M7 + P15 | LVSLKDIERRGAER | 24.1 ± 0.01 | 70.69 | 84.98 | 14.29 | 24.89 | 6.09 | 0.59 | 0.995 |
| M7 + P16 | LVSLKDIERRPAER | 24.2 ± 0.15 | 55.34 | 96.90 | 41.56 | 15.83 | 5.05 | 2.06 | 0.993 |
| M7 + P17 | LVSLKDIERRQAER | 24.0 ± 0.01 | 56.18 | 82.69 | 26.51 | 24.61 | 6.46 | 1.03 | 0.999 |
| M11.7 + P21 | AEQQRIRNEREKERQ | 23.6 ± 0.11 | 56.91 | 96.29 | 39.38 | 12.77 | 5.77 | 1.71 | 0.996 |
| M11.7 + P22 | AQQQRIRNEREKERQ | 23.7 ± 0.10 | 87.08 | 97.12 | 10.04 | 25.18 | 7.70 | 0.33 | 0.970 |
| M11.7 + P23 | AKQQRIRNEREKERQ | 23.3 ± 0.01 | 67.00 | 89.77 | 22.77 | 22.47 | 7.29 | 0.78 | 0.994 |
| M11.7 + P24 | AEQRRIRNEREKERQ | 23.3 ± 0.06 | 90.25 | 93.57 | 3.32 | n.a. | n.a. | n.a. | n.a. |
| M11.7 + P25 | AEQQCIRNEREKERQ | 23.2 ± 0.17 | 80.75 | 93.33 | 12.58 | 10.06 | 6.00 | 0.52 | 0.962 |
| M11.7 + P26 | AEQQSIRNEREKERQ | 22.9 ± 0.00 | 80.48 | 86.48 | 6.00 | 26.67 | 6.49 | 0.25 | 0.996 |
| M11.7 + P27 | AEQQHIRNEREKERQ | 23.1 ± 0.07 | 80.22 | 93.49 | 13.27 | 14.04 | 7.60 | 0.44 | 0.987 |
| M11.7 + P28 | AEQQRIWNEREKERQ | 23.4 ± 0.01 | 86.91 | 93.90 | 6.99 | 13.80 | 6.90 | 0.25 | 0.955 |
| M11.7 + P29 | AEQQRIQNEREKERQ | 23.2 ± 0.15 | 84.39 | 93.92 | 9.53 | 9.96 | 5.17 | 0.46 | 0.981 |
| M11.7 + P30 | AEQQRIRNEWEKERQ | 23.5 ± 0.09 | 90.48 | 94.03 | 3.55 | n.a. | n.a. | n.a. | n.a. |
| M11.7 + P31 | AEQQRIRNEQEKERQ | 23.2 ± 0.02 | 93.60 | 93.39 | −0.21 | n.a. | n.a. | n.a. | n.a. |
| M11.7 + P32 | AEQQRIRNERKKERQ | 23.1 ± 0.06 | 69.09 | 96.03 | 26.94 | 17.63 | 7.32 | 0.92 | 0.992 |
| Complex (a) | Peptide Sequence | (b) [1/s] | (b,c) [Ø] | (b) [kJ/mol] | (b) [kJ/mol] | (b) [kJ/mol] | Binding Type |
|---|---|---|---|---|---|---|---|
| M7 + P11 | LVSLKDIERRRAER | 3.69∙1012 | 0.59 | 1.28 | 40.25 | 38.96 | orthodox |
| M7 + P12 | LVSLKDIKRRRAER | n.a. | n.a. | n.a. | n.a. | n.a. | non-binding |
| M7 + P13 | LVSLKDIERCRAER | 1.72∙1012 | 0.28 | 3.17 | 55.16 | 51.99 | orthodox |
| M7 + P14 | LVSLKDIERRWAER | 1.75∙1012 | 0.28 | 3.13 | 92.09 | 88.96 | orthodox |
| M7 + P15 | LVSLKDIERRGAER | n.a. | n.a. | n.a. | n.a. | n.a. | unorthodox |
| M7 + P16 | LVSLKDIERRPAER | 2.95∙1012 | 0.48 | 1.84 | 85.61 | 83.77 | orthodox |
| M7 + P17 | LVSLKDIERRQAER | 3.82∙1012 | 0.62 | 1.14 | 38.06 | 36.92 | orthodox |
| M11.7 + P21 | AEQQRIRNEREKERQ | 5.70∙1012 | 0.92 | 0.21 | 72.53 | 72.32 | orthodox |
| M11.7 + P22 | AQQQRIRNEREKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | unorthodox |
| M11.7 + P23 | AKQQRIRNEREKERQ | 7.33∙1012 | 1.18 | −0.42 | 37.41 | 37.81 | orthodox |
| M11.7 + P24 | AEQRRIRNEREKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | non-binding |
| M11.7 + P25 | AEQQCIRNEREKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | unorthodox |
| M11.7 + P26 | AEQQSIRNEREKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | unorthodox |
| M11.7 + P27 | AEQQHIRNEREKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | unorthodox |
| M11.7 + P28 | AEQQRIWNEREKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | unorthodox |
| M11.7 + P29 | AEQQRIQNEREKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | unorthodox |
| M11.7 + P30 | AEQQRIRNEWEKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | non-binding |
| M11.7 + P31 | AEQQRIRNEQEKERQ | n.a. | n.a. | n.a. | n.a. | n.a. | non-binding |
| M11.7 + P32 | AEQQRIRNERKKERQ | 8.85∙1012 | 1.43 | −0.88 | 50.35 | 51.23 | orthodox |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kormann, K.; Ruß, M.; Röwer, C.; Koy, C.; Glocker, M.O. Mass Spectrometric ITEM-FOUR Analysis Reveals Coding Single-Nucleotide Polymorphisms in Human Cardiac Troponin T That Evade Detection by Sandwich ELISAs Which Use Monoclonal Antibodies M7 and M11.7 from the Elecsys Troponin T® Assay. Int. J. Mol. Sci. 2025, 26, 4892. https://doi.org/10.3390/ijms26104892
Kormann K, Ruß M, Röwer C, Koy C, Glocker MO. Mass Spectrometric ITEM-FOUR Analysis Reveals Coding Single-Nucleotide Polymorphisms in Human Cardiac Troponin T That Evade Detection by Sandwich ELISAs Which Use Monoclonal Antibodies M7 and M11.7 from the Elecsys Troponin T® Assay. International Journal of Molecular Sciences. 2025; 26(10):4892. https://doi.org/10.3390/ijms26104892
Chicago/Turabian StyleKormann, Kristjan, Manuela Ruß, Claudia Röwer, Cornelia Koy, and Michael O. Glocker. 2025. "Mass Spectrometric ITEM-FOUR Analysis Reveals Coding Single-Nucleotide Polymorphisms in Human Cardiac Troponin T That Evade Detection by Sandwich ELISAs Which Use Monoclonal Antibodies M7 and M11.7 from the Elecsys Troponin T® Assay" International Journal of Molecular Sciences 26, no. 10: 4892. https://doi.org/10.3390/ijms26104892
APA StyleKormann, K., Ruß, M., Röwer, C., Koy, C., & Glocker, M. O. (2025). Mass Spectrometric ITEM-FOUR Analysis Reveals Coding Single-Nucleotide Polymorphisms in Human Cardiac Troponin T That Evade Detection by Sandwich ELISAs Which Use Monoclonal Antibodies M7 and M11.7 from the Elecsys Troponin T® Assay. International Journal of Molecular Sciences, 26(10), 4892. https://doi.org/10.3390/ijms26104892






