Patterns of Heartwood Formation and Its Key Response Signaling Molecules in Dalbergia odorifera T. Chen
Abstract
1. Introduction
2. Results
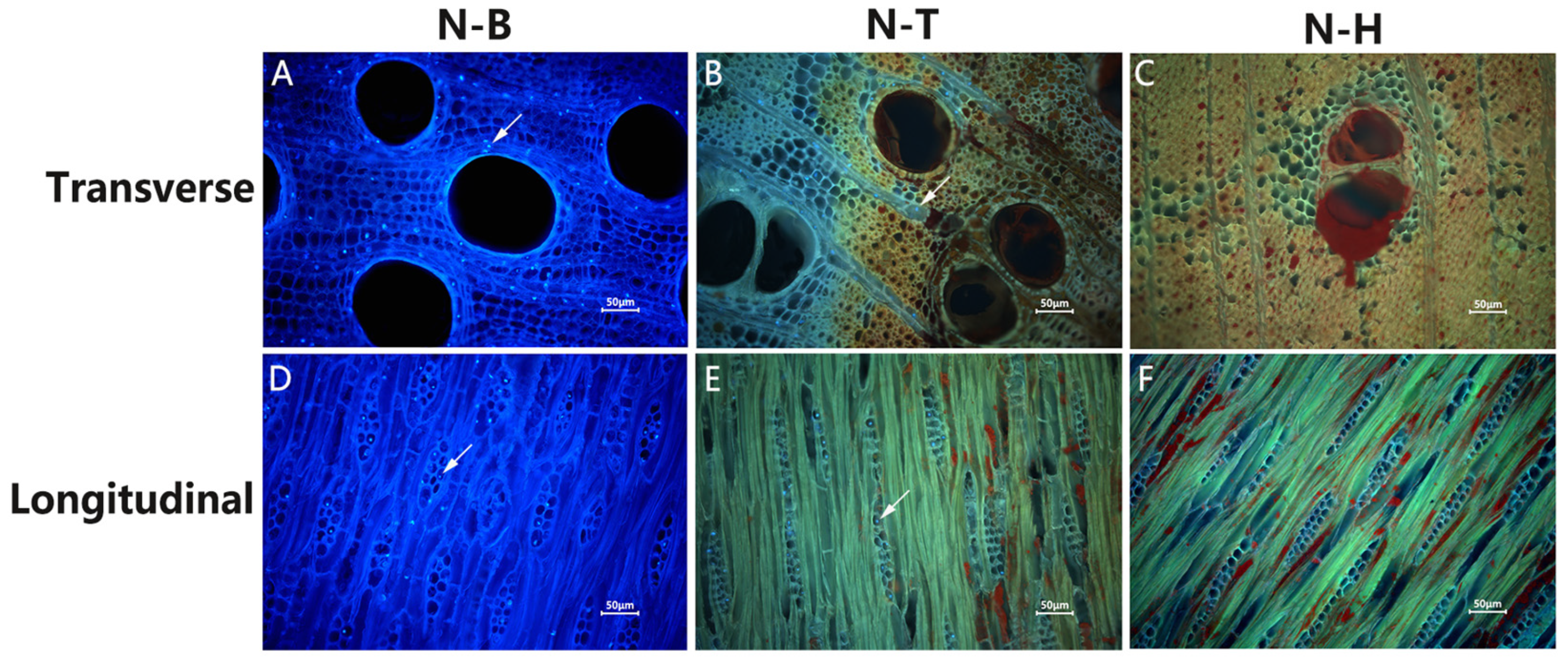
2.1. Observation of Fluorescence Staining of the Nuclei of D. odorifera Cells
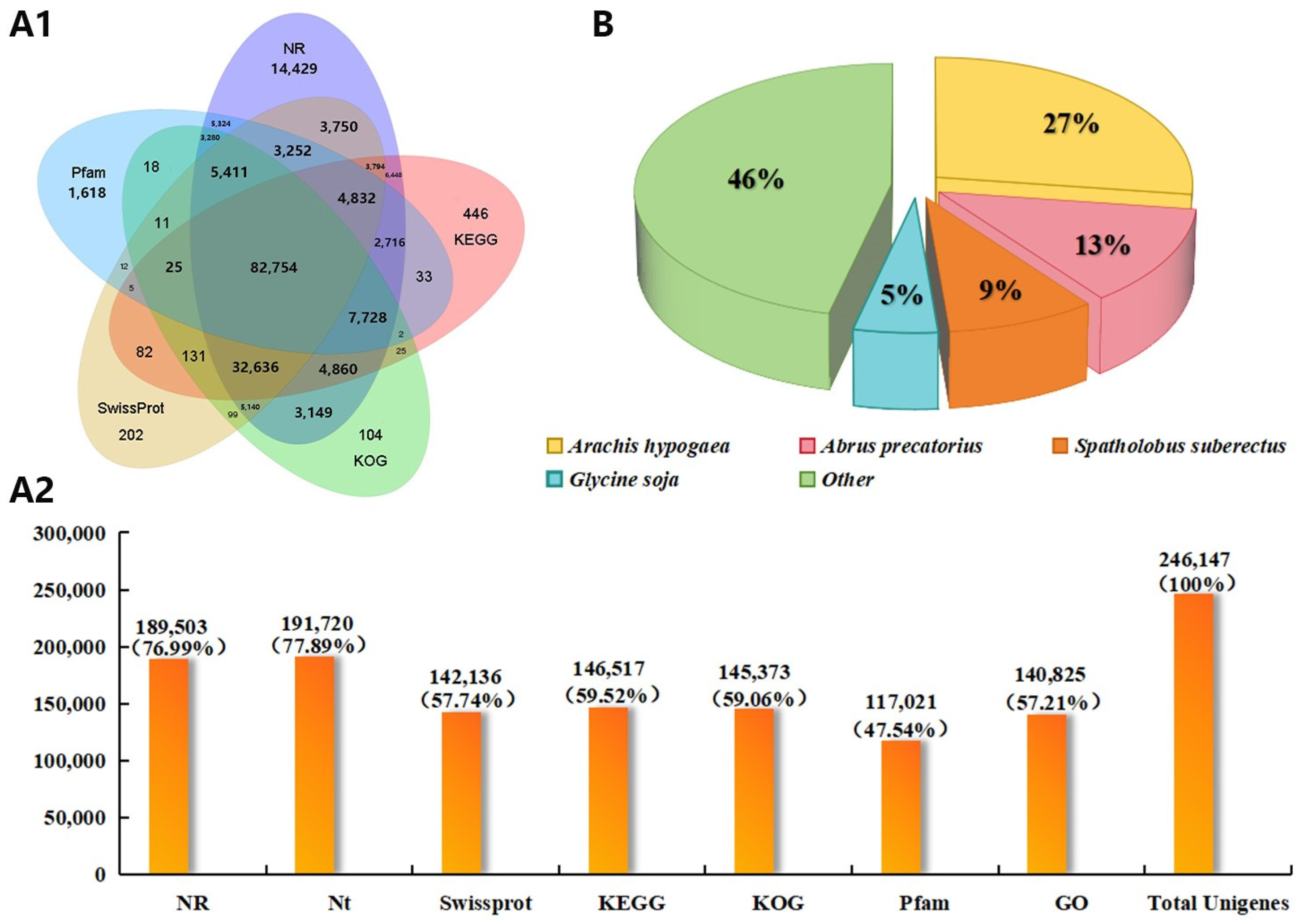
2.2. Transcriptome Sequencing and Quality Control Analysis of D. odorifera
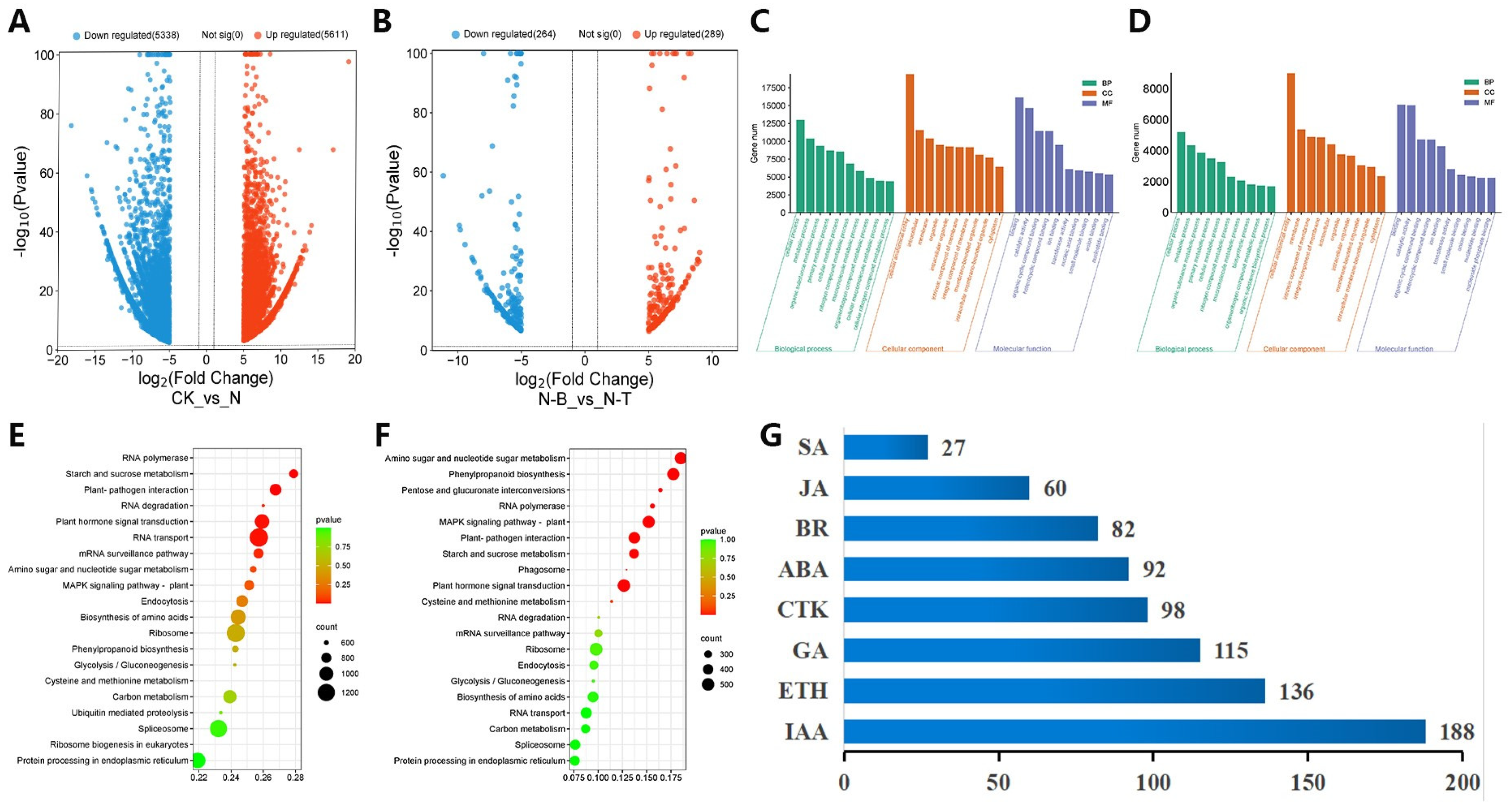
2.3. Differentially Expressed Genes Analysis
2.4. Enrichment Analysis of Differentially Expressed Genes
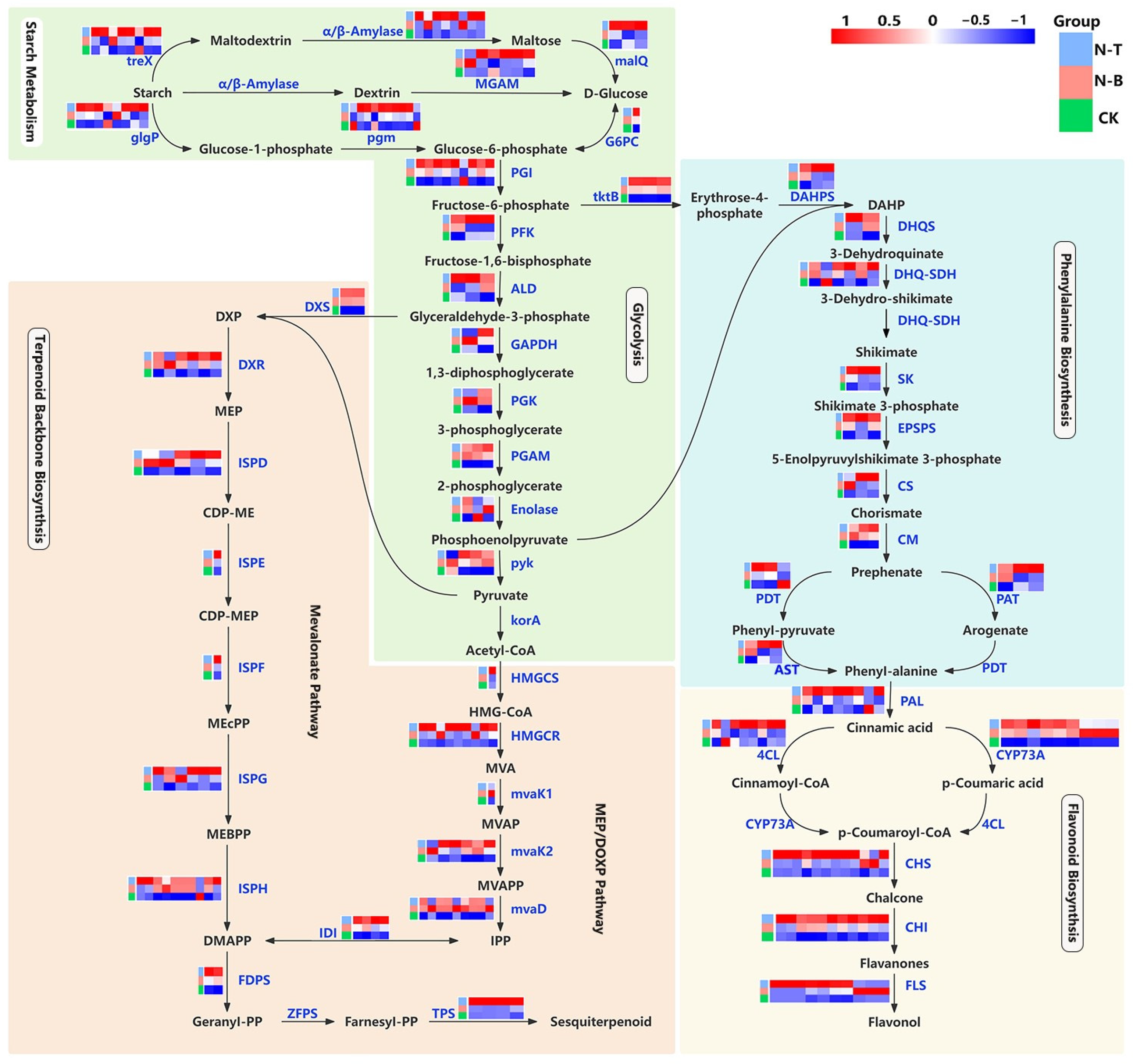
2.5. Expression Pattern Analysis of Differentially Expressed Genes in Primary and Secondary Metabolic Pathways
2.6. Expression Pattern Analysis of DEGs of Plant Hormone Synthesis and Signal Transduction Pathways
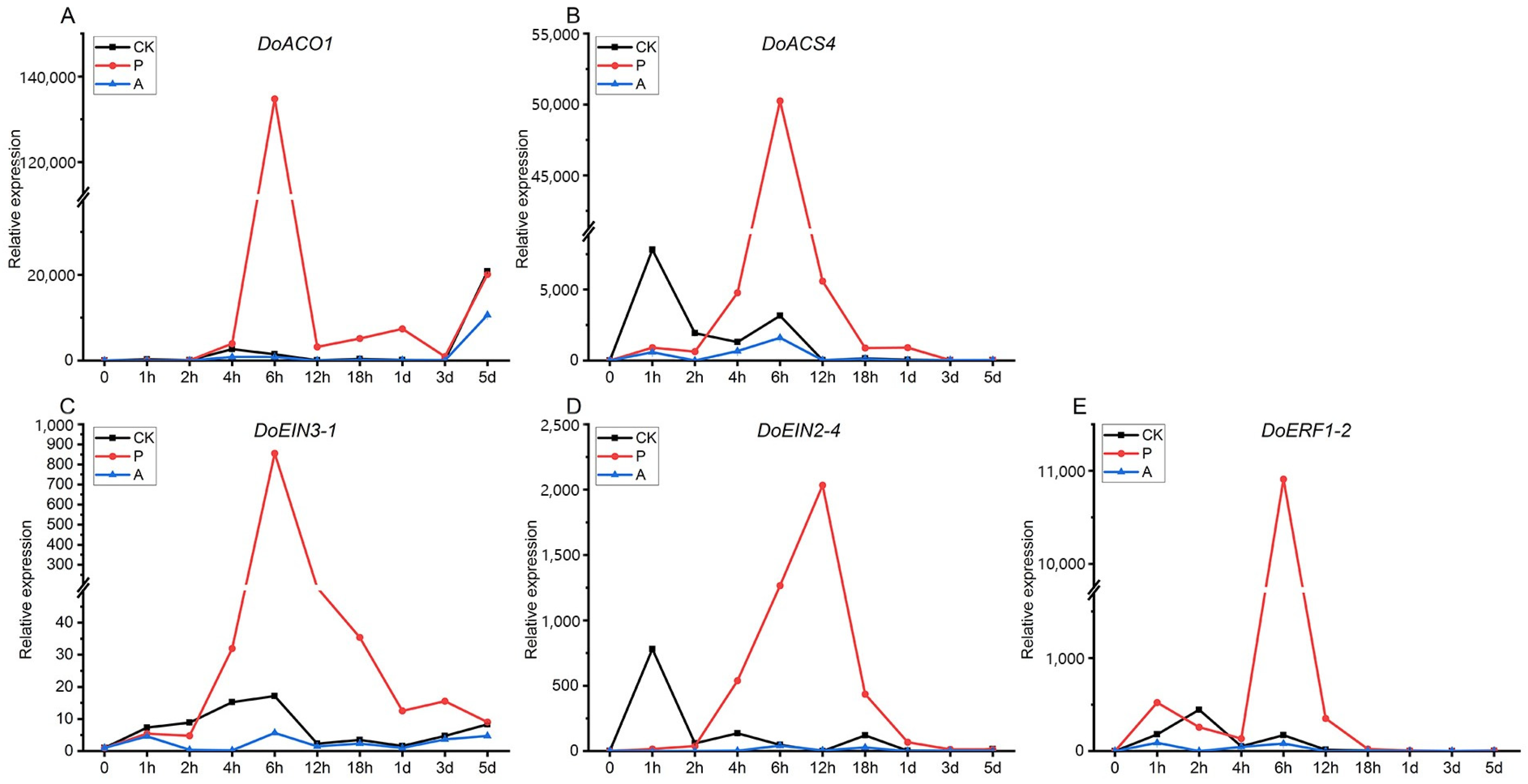
2.7. Expression Analysis of Genes Related to Ethylene Synthesis and Transduction
2.8. Expression of Secondary Metabolism-Related Genes Validation
2.9. Analysis of Changes in Ethylene Concentration and Trans-Nerolidol Content
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Observation of Nuclear Staining in Different Compartments of Xylem Cells
4.3. Iso-Seq Library RNA Preparation and Sequencing
4.4. RNA-Seq Data Generation and Analysis
4.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.6. PEG, AVG Treatment of D. odorifera Suspension Cells, and Expression Validation
4.7. Detection of Ethylene and Trans-Nerolidol Content
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, D.; Chen, Y.; Li, W.; Li, Q.; Lu, M.; Zhou, G.; Chai, G. Vascular Cambium: The Source of Wood Formation. Front. Plant Sci. 2021, 12, 1765. [Google Scholar] [CrossRef] [PubMed]
- Celedon, J.M.; Bohlmann, J. An extended model of heartwood secondary metabolism informed by functional genomics. Tree Physiol. 2018, 38, 311–319. [Google Scholar] [CrossRef]
- Saha Tchinda, J.-B.; Ndikontar, M.K.; Belinga, A.D.F.; Mounguengui, S.; Njankouo, J.M.; Durmaçay, S.; Gerardin, P. Inhibition of fungi with wood extractives and natural durability of five Cameroonian wood species. Ind. Crop. Prod. 2018, 123, 183–191. [Google Scholar] [CrossRef]
- Taylor, A.M.; Gartner, B.L.; Morrell, J.J. Heartwood formation and natural durability—A review. Wood Fiber Sci. 2002, 34, 587–611. [Google Scholar]
- Cao, S.; Deng, H.; Zhao, Y.; Zhang, Z.; Tian, Y.; Sun, Y.; Li, Y.; Zheng, H. Metabolite Profiling and Transcriptome Analysis Unveil the Mechanisms of Red-HeartChinese Fir [Cunninghamia lanceolata (Lamb.) Hook] Heartwood Coloration. Front. Plant Sci. 2022, 13, 854716. [Google Scholar] [CrossRef]
- Cui, Z.; Li, X.; Xu, D.; Yang, Z.; Zhang, N.; Liu, X.; Hong, Z. Physiological changes during heartwood formation induced by plant growth regulators in Dalbergia odorifera (Leguminosae). IAWA J. 2021, 42, 217–234. [Google Scholar] [CrossRef]
- Chen, S.Y.; Yen, P.L.; Chang, T.C.; Chang, S.T.; Huang, S.K.; Yeh, T.F. Distribution of living ray parenchyma cells and major bioactive compounds during the heartwood formation of Taiwania cryptomerioides hayata. J. Wood Chem. Technol. 2018, 38, 84–95. [Google Scholar] [CrossRef]
- Spicer, R.; Holbrook, N.M. Parenchyma cell respiration and survival in secondary xylem: Does metabolic activity decline with cell age? Plant Cell Environ. 2007, 30, 934–943. [Google Scholar] [CrossRef]
- Magel, E.; Jay-Allemand, C.; Ziegler, H. Formation of heartwood substances in the stemwood of Robinia pseudoacacia L. II. Distribution of nonstructural carbohydrates and wood extractives across the trunk. Trees 1994, 8, 165–171. [Google Scholar] [CrossRef]
- Burtin, P.; Jay-Allemand, C.; Charpentier, J.-P.; Janin, G. Natural wood colouring process in Juglans sp. (J. nigra, J. regia and hybrid J. nigra 23 × J. regia) depends on native phenolic compounds accumulated in the transition zone between sapwood and heartwood. Trees 1998, 12, 258–264. [Google Scholar] [CrossRef]
- Celedon, J.M.; Chiang, A.; Yuen, M.M.S.; Diaz-Chavez, M.L.; Madilao, L.L.; Finnegan, P.M.; Barbour, E.L.; Bohlmann, J. Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J. 2016, 86, 289–299. [Google Scholar] [CrossRef]
- Hong, Z.; Li, J.; Liu, X.; Lian, J.; Zhang, N.; Yang, Z.; Niu, Y.; Cui, Z.; Xu, D. The chromosome-level draft genome of Dalbergia odorifera. Gigascience 2020, 9, giaa084. [Google Scholar] [CrossRef]
- Yu, X.; Wang, W.; Yang, M. Antioxidant activities of compounds isolated from Dalbergia odorifera T. Chen and their inhibition effects on the decrease of glutathione level of rat lens induced by UV irradiation. Food Chem. 2007, 104, 715–720. [Google Scholar] [CrossRef]
- Meng, H.; Xia, X.Y.; Ma, G.X.; Yang, Y.; Liu, Y.; Wei, J.; Chen, D. Chemical constituents and anti-inflammatory activity from heartwood of Dalbergia odorifera. Nat. Prod. Res. Dev. 2018, 30, 800. [Google Scholar]
- Zan, N.; Lu, Z.; Wang, X.; Wang, R.; Liang, N.; Huo, H.; Zhao, Y.; Song, Y.; Tu, P.; Zheng, J.; et al. Anti-inflammatory flavonoid derivatives from the heartwood of Dalbergia odorifera T. Chen. Nat. Prod. Res. 2022, 37, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Wang, Y. Bioactive sesquiterpenes isolated from the essential oil of Dalbergia odorifera T. Chen. Fitoterapia 2010, 81, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Liu, Y.Y.; Qong, B.; Yang, Y.; Wang, Q.L.; Meng, H.; Wei, J.H. Herbal Textual Research on Dalbergiae Odoriferae Lignum in Traditional Chinese Medicine. Mod. Chin. Med. 2023, 25, 178–191. [Google Scholar] [CrossRef]
- Lv, J.Y. Study on Aesthetic Value of Scented Rosewood. Master’s Thesis, Guangxi University, Nanning, China, 2013. [Google Scholar]
- CITES. Amendments to Appendix I and II of CITES. In Proceedings of the CITES Secretary-General’s Remarks at the 4th Regional Dialogue on Combating Trafficking in Wild Fauna and Flora, Bangkok, Thailand, 11–15 September 2017; pp. 11–15. [Google Scholar]
- Liu, F.M.; Zhang, N.N.; Liu, X.J.; Yang, Z.J.; Jia, H.Y.; Xu, D.P. Genetic diversity and population structure analysis of Dalbergia Odorifera germplasm and development of a core collection using microsatellite markers. Genes 2019, 10, 281. [Google Scholar] [CrossRef]
- Meng, H.; Yang, Y.; Feng, J.D. Current status and development of the introductory cultivation of Pterocarpus indicus. Guangdong Agric. Sci. 2010, 37, 79–80. [Google Scholar]
- Ma, Y.H.; Jia, R.F.; Yang, Z.J.; Xu, D.P.; Zhang, N.N.; Liu, X.J. Early growth evaluation on six-year-old Dalbergia odorifera T. Chen families. J. Cent. South For. Univ. Sci. Technol. 2017, 37, 42–47. [Google Scholar]
- Sun, S.; Zeng, X.; Zhang, D.; Guo, S. Diverse fungi associated with partial irregular heartwood of Dalbergia odorifera. Sci. Rep. 2015, 5, 8464. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.J.; Li, J.W.; Meng, H.; He, W.J.; Yang, Y.; Wei, J.H. Effects of ethephon on heartwood formation and related physiological indices of Dalbergia odorifera T. Chen. Front. Plant Sci. 2024, 14, 1281877. [Google Scholar] [CrossRef]
- He, W.J.; Zhang, S.; Zhu, Y.J.; Li, J.W.; Yang, Y.; Meng, H. Analysis of hormone-stimulated heartwood-like substance formation in Dalbergia odorifera. Agric. Sci. Technol. Newsl. 2023, 6, 129–132. [Google Scholar]
- Ingemarsson, B.S.M.; Lundqvist, E.; Eliasson, L. Seasonal variation in ethylene concentration in the wood of Pinus sylvestris L. Tree Physiol. 1991, 8, 273–279. [Google Scholar] [CrossRef]
- Shain, L.; Hillis, W.E. Ethylene production in xylem of Pinus radiata in relation to heartwood formation. Can. J. Bot. 1973, 51, 1331–1335. [Google Scholar] [CrossRef]
- Shigo, A.L.; Hillis, W.E. Heartwood, discolored wood, and microorganisms in living trees. Annu. Rev. Phytopathol. 1973, 11, 197–222. [Google Scholar] [CrossRef]
- Nilsson, M.; Wikman, S.; Eklund, L. Induction of discolored wood in Scots pine (Pinus sylvestris). Tree Physiol. 2002, 22, 331–338. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, C.; Meng, H.; Yu, Z.; Yang, M.; Wei, J. Dalbergia odorifera: A review of its traditional uses, phytochemistry, pharmacology, and quality control. J. Ethnopharmacol. 2020, 248, 112328. [Google Scholar] [CrossRef]
- Ma, R.; Liu, H.; Fu, Y.; Li, Y.; Wei, P.; Liu, Z. Variation of chemical components in sapwood, transition zone, and heartwood of Dalbergia odorifera and its relationship with heartwood formation. Forests 2021, 12, 577. [Google Scholar] [CrossRef]
- Ma, R.; Luo, J.; Wang, W.; Fu, Y. Changes in the physiological activity of parenchyma cells in Dalbergia odorifera xylem and its relationship with heartwood formation. BMC Plant Biol. 2023, 23, 559. [Google Scholar] [CrossRef]
- Nelson, N.D. Xylem ethylene, phenol-oxidizing enzymes, and nitrogen and heartwood formation in walnut and cherry. Can. J. Bot. 1978, 56, 626–634. [Google Scholar] [CrossRef]
- Pallardy, S.G.; Kozlowski, T.T. Physiology of Woody Plants; Elsevier: Amsterdam, The Netherlands; Boston, MA, USA, 2008. [Google Scholar]
- Zhao, X.S.; Zhang, S.H.; Liu, D.; Wei, J.H. Analysis of Dalbergia odorifera saturated solution by GC-MS. Chin. Pharm. J. 2021, 56, 794–800. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Zhu, Y.; Ma, G.; Li, H.; Yang, Y.; Meng, H.; Wei, J. Patterns of Heartwood Formation and Its Key Response Signaling Molecules in Dalbergia odorifera T. Chen. Int. J. Mol. Sci. 2025, 26, 4629. https://doi.org/10.3390/ijms26104629
Li J, Zhu Y, Ma G, Li H, Yang Y, Meng H, Wei J. Patterns of Heartwood Formation and Its Key Response Signaling Molecules in Dalbergia odorifera T. Chen. International Journal of Molecular Sciences. 2025; 26(10):4629. https://doi.org/10.3390/ijms26104629
Chicago/Turabian StyleLi, Jiawen, Yuanjing Zhu, Guangyao Ma, Haoling Li, Yun Yang, Hui Meng, and Jianhe Wei. 2025. "Patterns of Heartwood Formation and Its Key Response Signaling Molecules in Dalbergia odorifera T. Chen" International Journal of Molecular Sciences 26, no. 10: 4629. https://doi.org/10.3390/ijms26104629
APA StyleLi, J., Zhu, Y., Ma, G., Li, H., Yang, Y., Meng, H., & Wei, J. (2025). Patterns of Heartwood Formation and Its Key Response Signaling Molecules in Dalbergia odorifera T. Chen. International Journal of Molecular Sciences, 26(10), 4629. https://doi.org/10.3390/ijms26104629





