Integrated Application of Transcriptomics and Metabolomics Provides Insights into the Different Body-Size Growth in Chinese Mitten Crab (Eriocheir sinensis)
Abstract
1. Introduction
2. Results
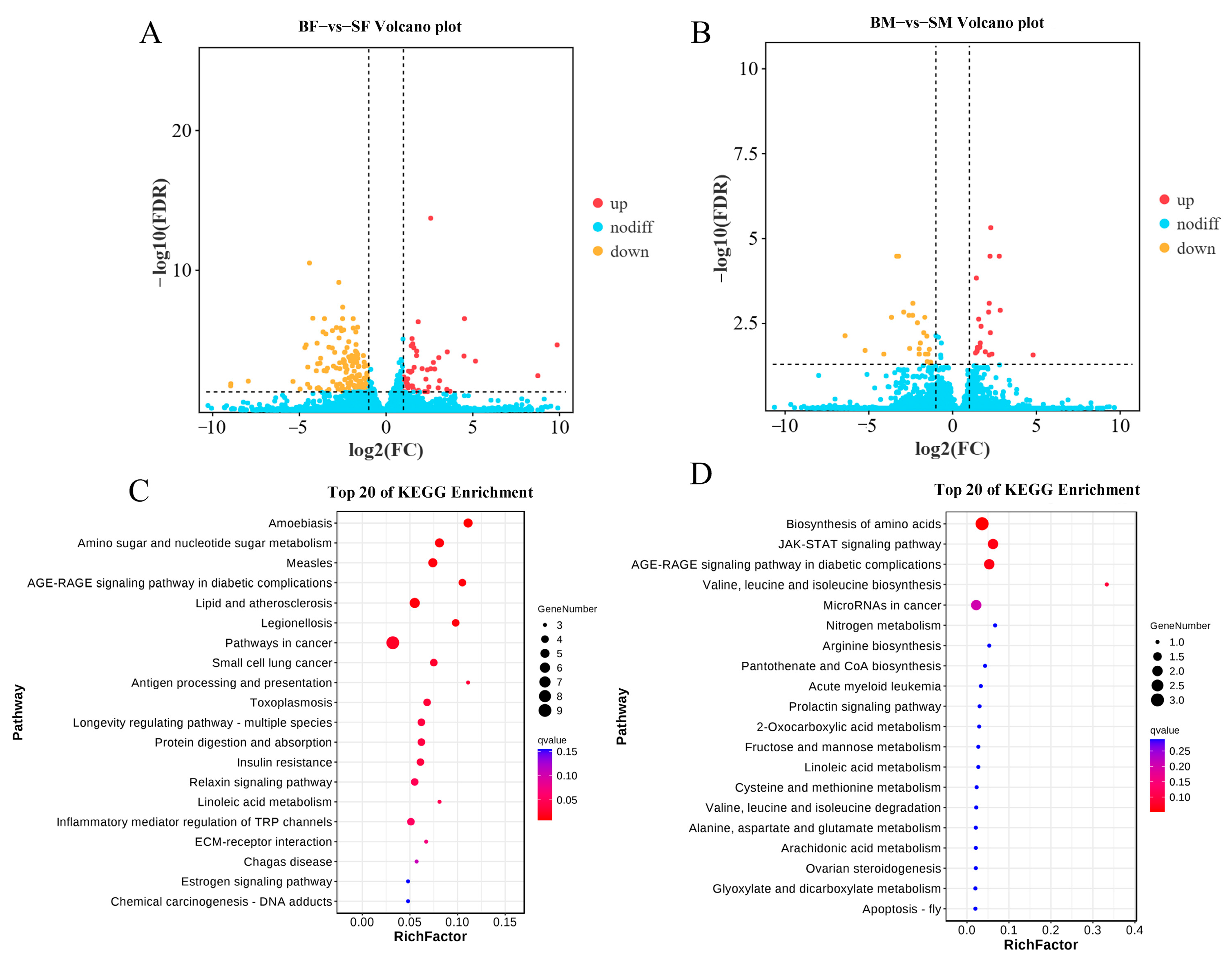
2.1. Changes in Gene Expression of Different Body Sizes of E. sinensis
2.2. Differential Metabolites Analysis
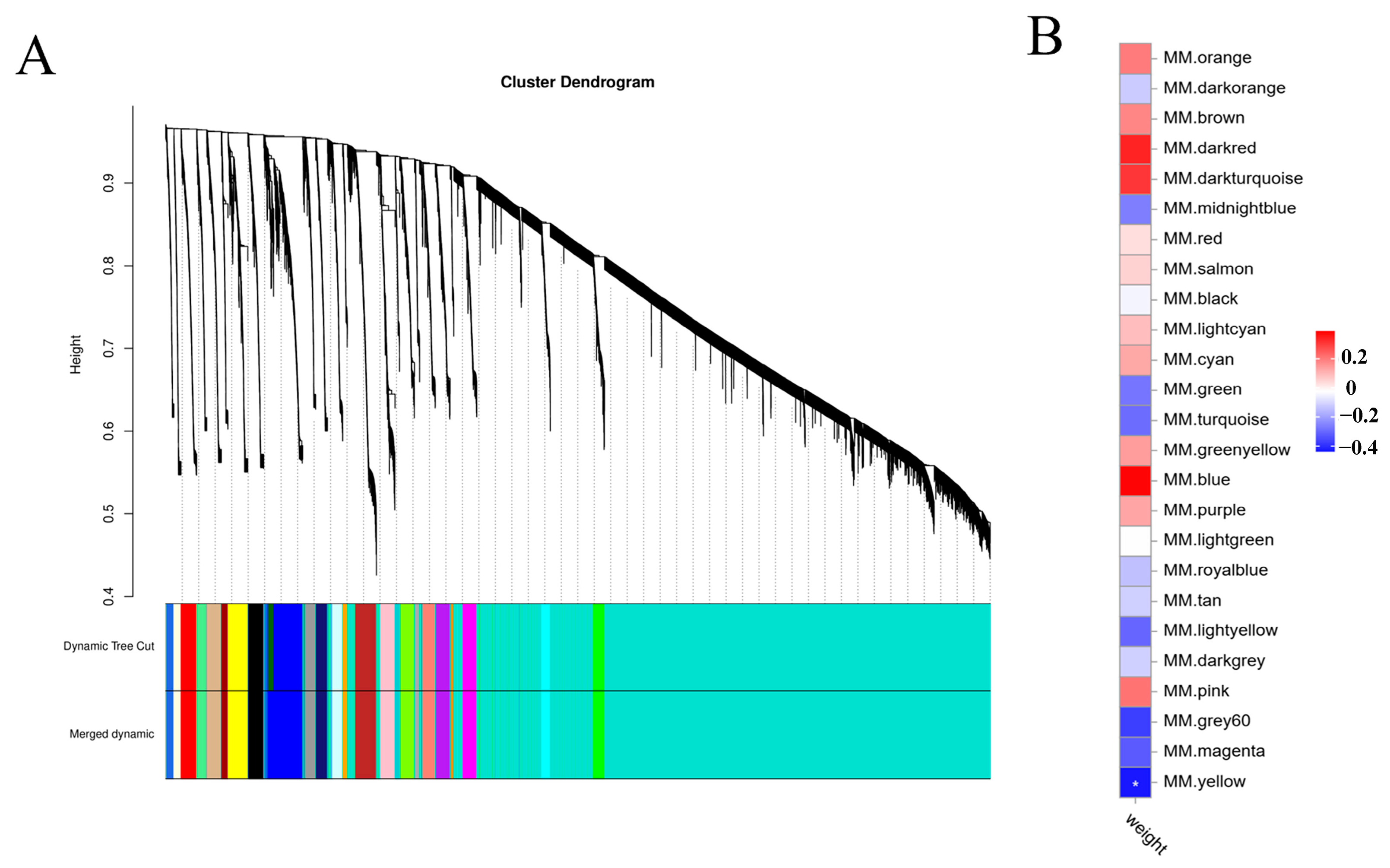
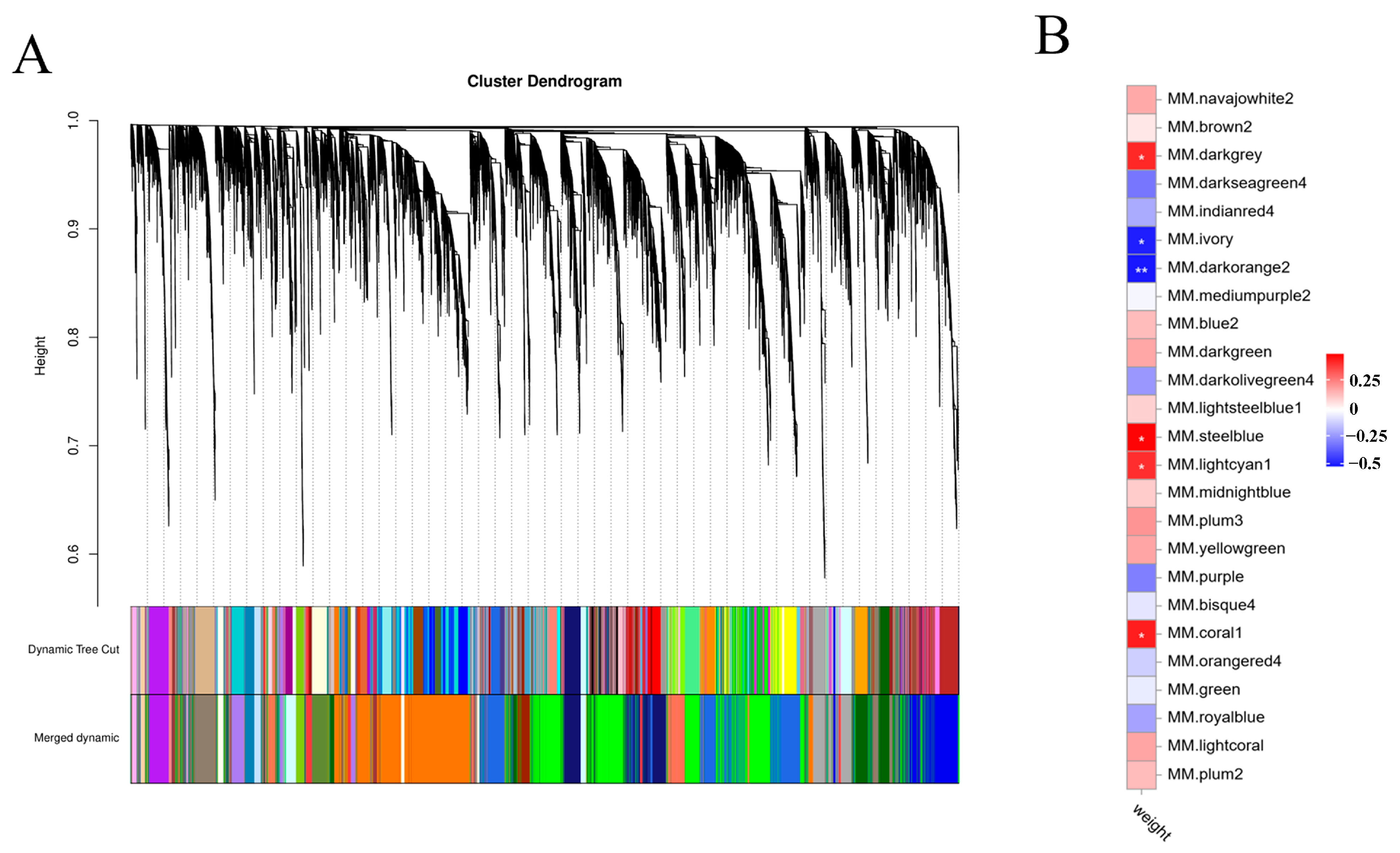
2.3. Weighted Gene Co-Expression Network Analysis (WGCNA)
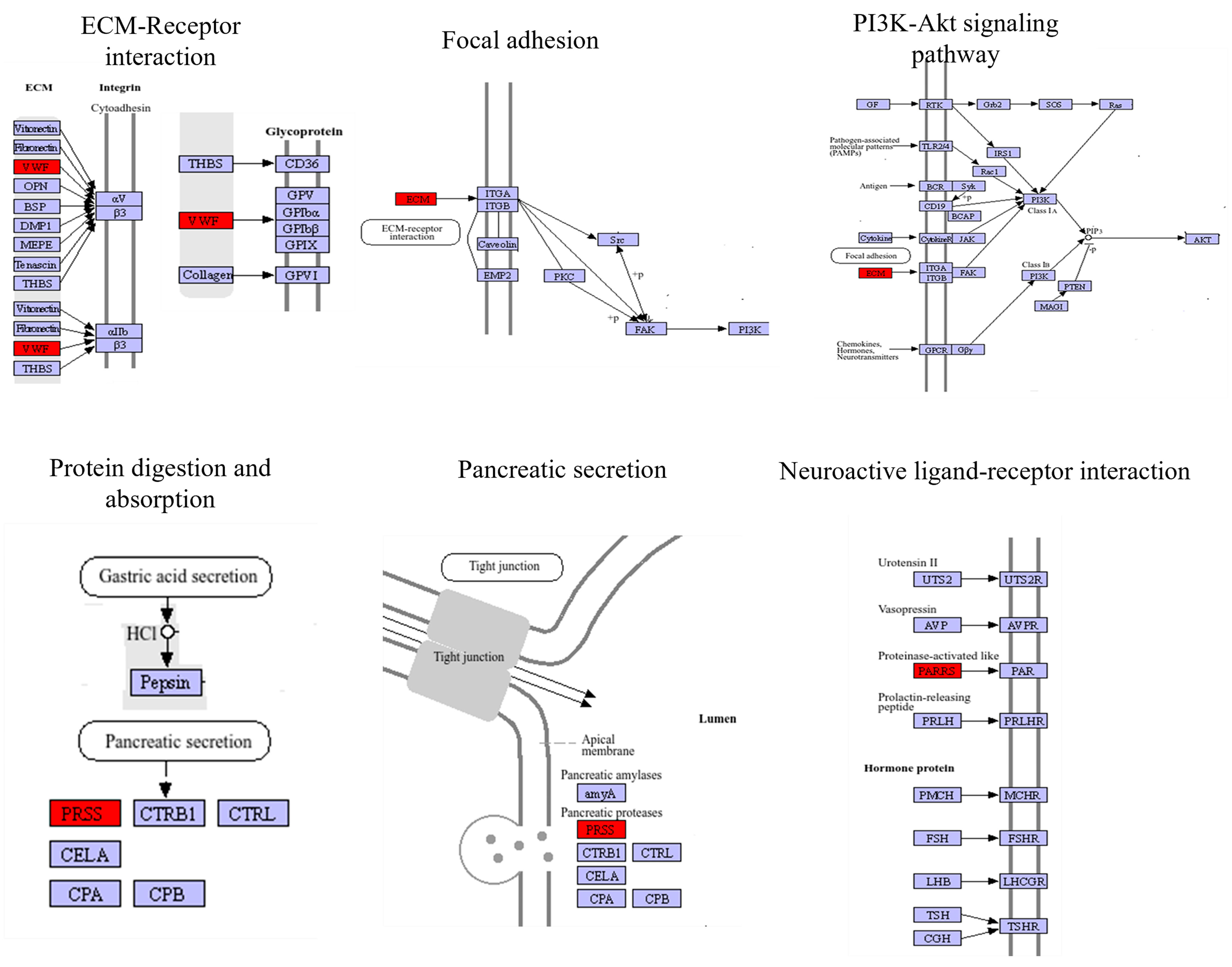
2.4. Transcriptomics-Metabolomics Co-Regulated Network Analysis
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experimental Design and Sample Collection
4.3. Transcriptome Sequencing and Differential Expressed Genes Analysis
4.4. qRT-PCR Validation
4.5. Metabolite Extraction and LC-MS/MS Analysis
4.6. Transcriptomics-Metabolomics Co-Regulated Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sui, L.Y.; Zhang, F.M.; Wang, X.M.; Bossier, P.; Sorgeloos, P.; Hänfling, B. Genetic diversity and population structure of the Chinese mitten crab Eriocheir sinensis in its native range. Mar. Biol. 2009, 156, 1573–1583. [Google Scholar] [CrossRef]
- Cheng, Y.X.; Wu, X.G.; Li, J.Y. Chinese mitten Crab Culture: Current Status and Recent Progress Towards Sustainable Development. Aquaculture 2018, 197–217. [Google Scholar] [CrossRef]
- Xu, F.; Xing, X.M.; Zhang, K.; Zhang, C. Status and prospects of product processing and sustainable utilization of Chinese mitten crab (Eriocheir sinensis). Heliyon 2024, 10, 32922. [Google Scholar] [CrossRef]
- Fishery administration Bureau of the ministry of agriculture and villages; National aquatic products technology extension station; China Society of Fisheries. China Fishery Statistical Yearbook; Chinese Agricultural Press: Beijing, China, 2024; p. 180. [Google Scholar]
- Wu, C.; Shan, J.F.; Feng, J.C.; Jiang, A.; Nie, G.X.; Wang, X.H. Changes in the growth, nutritional quality and gut microbiota of the adult Chinese mitten crab, Eriocheir sinensis, following overwinter rearing. Aquac. Res. 2022, 53, 1348–1362. [Google Scholar] [CrossRef]
- Fruchtman, S.; Jackson, L.; Borski, R. Insulin-like growth factor I disparately regulates prolactin and growth hormone synthesis and secretion: Studies using the teleost pituitary model. Endocrinology 2000, 141, 2886–2894. [Google Scholar] [CrossRef] [PubMed]
- Picha, M.E.; Strom, C.N.; Riley, L.G.; Walker, A.A.; Won, E.T.; Johnstone, W.M.; Borski, R.J. Plasma ghrelin and growth hormone regulation in response to metabolic state in hybrid striped bass: Effects of feeding, ghrelin and insulin-like growth factor-I on in vivo and in vitro GH secretion. Gen. Comp. Endocr. 2009, 161, 365–372. [Google Scholar] [CrossRef]
- Peterson, B.C.; Waldbieser, G.C.; Bilodeau, L. IGF-I and IGF-II mRNA expression in slow and fast-growing families of USDA103 channel catfish (Ictalurus punctatus). Comp. Biochem. Phys. A 2004, 139, 317–323. [Google Scholar] [CrossRef]
- Huang, C.W.; Li, Y.H.; Hu, S.Y.; Chi, J.R.; Lin, G.H.; Lin, C.C.; Gong, H.Y.; Chen, J.Y.; Chen, R.H.; Chang, S.J.; et al. Differential expression patterns of growth-related microRNAs in the skeletal muscle of Nile tilapia (Oreochromis niloticus). J. Anim. Sci. 2012, 90, 4266–4279. [Google Scholar] [CrossRef]
- Salem, M.; Vallejo, R.L.; Leeds, T.D.; Palti, Y.; Liu, S.; Sabbagh, A.; Rexroad, C.E., 3rd; Yao, J. RNA-Seq identifies SNP markers for growth traits in rainbow trout. PLoS ONE 2012, 7, e36264. [Google Scholar] [CrossRef]
- Liu, D.R.; Zhang, J.; Zou, Z.J.; Long, C.; Lin, J.Q.; Zeng, J.Y.; Hou, J.P.; Han, L.Q.; Jiang, Y.L.; Li, S.S.; et al. Identification of SNPs and candidate genes associate with growth performance in all-female mandarin fish (Siniperca chuatsi) by a genome-wide association study. Aquaculture 2024, 586, 740778. [Google Scholar] [CrossRef]
- Ulloa, P.E.; Rincón, G.; Islas-Trejo, A.; Araneda, C.; Iturra, P.; Neira, R.; Medrano, J.F. RNA Sequencing to Study Gene Expression and SNP Variations Associated with Growth in Zebrafish Fed a Plant Protein-Based Diet. Mar. Biotechnol. 2015, 17, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Robledo, D.; Rubiolo, J.A.; Cabaleiro, S.; Martínez, P.; Bouza, C. Differential gene expression and SNP association between fast- and slow-growing turbot (Scophthalmus maximus). Sci. Rep.-UK 2017, 7, 12105. [Google Scholar] [CrossRef]
- Kim, E.Y.; Choi, Y.H. Regulation of adductor muscle growth by the IGF-1/AKT pathway in the triploid Pacific oyster, Crassostrea gigas. Fish. Aquat. Sci. 2019, 22, 19. [Google Scholar] [CrossRef]
- Li, Y.; Fu, H.; Zhang, F.; Ren, L.; Tian, J.; Li, Q.; Liu, S. Insulin-Like Peptide Receptor-Mediated Signaling Pathways Orchestrate Regulation of Growth in the Pacific Oyster (Crassostrea gigas), as Revealed by Gene Expression Profiles. Int. J. Mol. Sci. 2021, 22, 5259. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.C.; Lin, X.R.; Hu, J.H.; Yang, J.J.; Li, X.G.; Wang, G.; Yang, Y.Z.; Chen, Y.C.; Pan, J.L.; Guan, W.B.; et al. Analysis of molecular advantages in offspring of large-sized Chinese mitten crabs based on reproductive lineage. Aquaculture 2025, 596, 741870. [Google Scholar] [CrossRef]
- Zhang, C.; Song, X.Z.; Zhang, Q.; Pang, Y.Y..; Lv, J.H.; Tang, B.P.; Cheng, Y.X.; Yang, X.Z. Changes in bud morphology, growth-related genes and nutritional status during cheliped regeneration in the Chinese mitten crab, Eriocheir sinensis. PLoS ONE 2018, 13, e0209617. [Google Scholar] [CrossRef] [PubMed]
- Riddiford, L.M.; Hiruma, K.; Zhou, X.F.; Nelson, C.A. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Molec. 2003, 33, 1327–1338. [Google Scholar] [CrossRef]
- Perazzolo, L.M.; Barracco, M.A. The prophenoloxidase activating system of the shrimp Penaeus paulensis and associated factors. Dev. Comp. Immunol. 1997, 21, 385–395. [Google Scholar] [CrossRef]
- Alvarez, J.V.; Chung, J.S. The Involvement of Hemocyte Prophenoloxidase in the Shell-Hardening Process of the Blue Crab, Callinectes sapidus. PLoS ONE 2015, 10, e0136916. [Google Scholar] [CrossRef]
- Maszczyk, P.; Brzeziński, T. Body Size, Maturation Size, and Growth Rate of Crustaceans. Life Hist. 2018, 5, 35–65. [Google Scholar]
- Tamayo, D.; Ibarrola, I.; Urrutxurtu, I.; Navarro, E. Physiological basis of extreme growth rate differences in the spat of oyster (Crassostrea gigas). Mar. Biol. 2014, 161, 1627–1637. [Google Scholar] [CrossRef]
- Tamayo, D.; Azpeitia, K.; Markaide, P.; Navarro, E.; Ibarrola, I. Food regime modulates physiological processes underlying size differentiation in juvenile intertidal mussels Mytilus galloprovincialis. Mar. Biol. 2016, 163, 131. [Google Scholar] [CrossRef]
- Guo, X.M.; Luo, T.; Han, D.M.; Zhu, D.F.; Li, Z.Y.; Wu, Z.Y.; Wu, Z.X. Multi-omics analysis revealed room temperature storage affected the quality of litchi by altering carbohydrate metabolism. Sci. Hortic. 2022, 293, 110663. [Google Scholar] [CrossRef]
- Niu, J.; Lin, H.Z.; Jiang, S.G.; Chen, X.; Wu, K.C.; Liu, Y.J.; Wang, S.; Tian, L.X. Comparison of effect of chitin, chitosan, chitosan oligosaccharide and N-acetyl-d-glucosamine on growth performance, antioxidant defenses and oxidative stress status of Penaeus monodon. Aquaculture 2013, 372–375, 1–8. [Google Scholar] [CrossRef]
- Huang, X.S.; Ye, H.H.; Feng, B.Y.; Huang, H.Y. Insights into insulin-like peptide system in invertebrates from studies on IGF binding domain-containing proteins in the female mud crab, Scylla paramamosain. Mol. Cell. Endocrinol. 2015, 416, 36–45. [Google Scholar] [CrossRef]
- Liu, J.D.; Li, E.; Li, X.Y.; Wang, X.D.; Huang, Q.C.; Wang, H.; Miao, Y.X.; Shi, Q.C.; Qin, J.G.; Chen, L.Q. Effects of dietary methionine on the growth and protein synthesis of juvenile Chinese mitten crabs (Eriocheir sinensis) fed fish meal-free diets. Anim. Nutr. 2024, 19, 226–239. [Google Scholar] [CrossRef]
- Deng, H.W.; Xian, D.R.; Lian, T.X.; He, M.Y.; Li, J.J.; Xu, X.P.; Guo, Z.X.; He, J.G.; Weng, S.P. A Dicer2 from Scylla paramamosain activates JAK/STAT signaling pathway to restrain mud crab reovirus. Dev. Comp. Immuno. 2022, 127, 104267. [Google Scholar] [CrossRef] [PubMed]
- Quijada-Rodriguez, A.R.; Allen, G.J.P.; Nash, M.T.; Weihrauch, D. Postprandial nitrogen and acid-base regulation in the seawater acclimated green crab, Carcinus maenas. Comp. Biochem. Phys. A 2022, 267, 111171. [Google Scholar] [CrossRef]
- James, M.O.; Boyle, S.M. Cytochromes P450 in crustacea1This article was invited by Guest Editors Dr John J. Stegeman and Dr David R. Livingstone to be part of a special issue of CBP on cytochrome P450. Comp. Biochem. Phys. C 1998, 121, 157–172. [Google Scholar]
- Cheng, C.H.; Zhang, S.F.; Ma, H.L.; Liu, G.X.; Fan, S.G.; Deng, Y.Q.; Jiang, J.J.; Feng, J.; Guo, Z.X. Essential role of the Cytochrome P450 2 (CYP2) in the mud crab Scylla paramamosain antioxidant defense and immune responses. Fish Shellfish Immun. 2023, 135, 108674. [Google Scholar] [CrossRef]
- Ticho, A.L.; Malhotra, P.; Dudeja, P.K.; Gill, R.K.; Alrefai, W.A. Intestinal Absorption of Bile Acids in Health and Disease. Compr. Physiol. 2019, 10, 21–56. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.T.; Xu, H.Y.; Feng, W.P.; Yang, Y.X.; Han, T.; Wang, J.T. Optimal dietary lysine improves the growth performance, molting frequency, lipid metabolism, immunity and antioxidant of postlarval mud crab Scylla paramamosain. Aquaculture 2024, 579, 740145. [Google Scholar] [CrossRef]
- Sailer, S.; Keller, M.A.; Werner, E.R.; Watschinger, K. The Emerging Physiological Role of AGMO 10 Years after Its Gene Identification. Life 2021, 11, 88. [Google Scholar] [CrossRef]
- Qin, G.K.; Hu, X.; Cebe, P.; Kaplan, D.L. Mechanism of resilin elasticity. Nat. Commun. 2012, 3, 1003. [Google Scholar] [CrossRef]
- Lenkkeri, U.; Männikkö, M.; McCready, P.; Lamerdin, J.; Gribouval, O.; Niaudet, P.M.; Antignac, C.K.; Kashtan, C.E.; Homberg, C.; Olsen, A.; et al. Structure of the gene for congenital nephrotic syndrome of the finnish type (NPHS1) and characterization of mutations. Am. J. Hum. Genet. 1999, 64, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Meneguello, L. Investigating Tolerance Mechanisms to Oncogene-Induced Genome Instability. Ph.D Thesis, University College London, London, UK, 2024. [Google Scholar]
- Palm, W.; Sampaio, J.L.; Brankatschk, M.; Carvalho, M.; Mahmoud, A.; Shevchenko, A.; Eaton, S. Lipoproteins in Drosophila melanogaster-Assembly, Function, and Influence on Tissue Lipid Composition. PloS Genet. 2012, 8, e1002828. [Google Scholar] [CrossRef]
- Kolasa, A.M.; Bhogal, J.K.; DiAngelo, J.R. The heterogeneous nuclear ribonucleoprotein (hnRNP) glorund functions in the Drosophila fat body to regulate lipid storage and transport. Biochem. Biophys. Rep. 2021, 25, 100919. [Google Scholar] [CrossRef]
- Lee, S.; Bao, H.; Ishikawa, Z.; Wang, W.; Lim, H.Y. Cardiomyocyte Regulation of Systemic Lipid Metabolism by the Apolipoprotein B-Containing Lipoproteins in Drosophila. PLoS Genet. 2017, 13, 1006555. [Google Scholar] [CrossRef]
- Velleman, S.G. Role of the extracellular matrix in muscle growth and development. J. Anim. Sci. 2002, 80, E8–E13. [Google Scholar] [CrossRef]
- Hao, R.J.; Du, X.D.; Yang, C.Y.; Deng, Y.W.; Zheng, Z.; Wang, Q.H. Integrated application of transcriptomics and metabolomics provides insights into unsynchronized growth in pearl oyster Pinctada fucata martensii. Sci. Total Environ. 2019, 666, 46–56. [Google Scholar] [CrossRef]
- Stricker, J.; Aratyn-Schaus, Y.; Oakes, P.W.; Gardel, M.L. Spatiotemporal Constraints on the Force-Dependent Growth of Focal Adhesions. Biophys. J. 2011, 100, 2883–2893. [Google Scholar] [CrossRef] [PubMed]
- Flück, M.; Carson, J.A.; Gordon, S.E.; Ziemiecki, A.; Booth, F.W. Focal adhesion proteins FAK and paxillin increase in hypertrophied skeletal muscle. Am. J. Physiol. 1999, 277, C152–C162. [Google Scholar] [CrossRef] [PubMed]
- Guo, S. Molecular Basis of Insulin Resistance: The Role of IRS and Foxo1 in the Control of Diabetes Mellitus and Its Complications. Drug Discov. Today Dis. Mech. 2013, 10, e27–e33. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Yuan, Y.W.; Shi, L.J.; Li, Y.H.; Liu, L.; Sun, D.X. Identification of single nucleotide polymorphisms of PIK3R1 and DUSP1 genes and their genetic associations with milk production traits in dairy cows. J. Anim. Sci. Biotechno. 2019, 10, 81. [Google Scholar] [CrossRef]
- Sofia, V.M.; Da Sacco, L.; Surace, C.; Tomaiuolo, A.C.; Genovese, S.; Grotta, S.; Gnazzo, M.; Ciocca, L.; Petrocchi, S.; Alghisi, F.; et al. Extensive molecular analysis suggested the strong genetic heterogeneity of idiopathic chronic pancreatitis. Mol. Med. 2016, 22, 300–309. [Google Scholar] [CrossRef]
- Berni Canani, R.; Terrin, G.; Cardillo, G.; Tomaiuolo, R.; Castaldo, G. Congenital diarrheal disorders: Improved understanding of gene defects is leading to advances in intestinal physiology and clinical management. J. Pediatr. Gastroenterol Nutr. 2010, 50, 360–366. [Google Scholar] [CrossRef]
- Yarar, D.; To, W.; Abo, A.; Welch, M.D. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr. Biol. 1999, 9, 555–558. [Google Scholar] [CrossRef]
- Uruno, T.; Zhang, P.; Liu, J.; Hao, J.J.; Zhan, X. Haematopoietic lineage cell-specific protein 1 (HS1) promotes actin-related protein (Arp) 2/3 complex-mediated actin polymerization. Biochem. J. 2003, 371 Pt 2, 485–493. [Google Scholar] [CrossRef]
- Uruno, T.; Liu, J.; Zhang, P.; Fan, Y.; Egile, C.; Li, R.; Mueller, S.C.; Zhan, X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 2001, 3, 259–266. [Google Scholar] [CrossRef]
- Gouin, E.; Egile, C.; Dehoux, P.; Villiers, V.; Adams, J.; Gertler, F.; Li, R.; Cossart, P. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 2004, 427, 457–461. [Google Scholar] [CrossRef]
- Mullins, R.D.; Heuser, J.A.; Pollard, T.D. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 1998, 95, 6181–6186. [Google Scholar] [CrossRef]
- Duncan, A.R.; González, D.P.; Del Viso, F.; Robson, A.; Khokha, M.K.; Griffin, J.N. Alkylglycerol monooxygenase, a heterotaxy candidate gene, regulates left-right patterning via Wnt signaling. Dev. Biol. 2019, 456, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Alrayes, N.; Mohamoud, H.S.; Ahmed, S.; Almramhi, M.M.; Shuaib, T.M.; Wang, J.; Al-Aama, J.Y.; Everett, K.; Nasir, J.; Jelani, M. The alkylglycerol monooxygenase (AGMO) gene previously involved in autism also causes a novel syndromic form of primary microcephaly in a consanguineous Saudi family. J. Neurol. Sci. 2016, 363, 240–244. [Google Scholar] [CrossRef]
- Okur, V.; Watschinger, K.; Niyazov, D.; McCarrier, J.; Basel, D.; Hermann, M.; Werner, E.R.; Chung, W.K. Biallelic variants in AGMO with diminished enzyme activity are associated with a neurodevelopmental disorder. Hum. Genet. 2019, 138, 1259–1266. [Google Scholar] [CrossRef]
- Patrakka, J.; Kestilä, M.; Wartiovaara, J.; Ruotsalainen, V.; Tissari, P.; Lenkkeri, U.; Männikkö, M.; Visapää, I.; Holmberg, C.; Rapola, J.; et al. Congenital nephrotic syndrome (NPHS1): Features resulting from different mutations in Finnish patients. Kidney Int. 2000, 58, 972–980. [Google Scholar] [CrossRef]
- Uchida, Y.; Kurimoto, R.; Chiba, T.; Matsushima, T.; Oda, G.; Onishi, I.; Takeuchi, Y.; Gotoh, N.; Asahara, H. RNA binding protein ZCCHC24 promotes tumorigenicity in triple-negative breast cancer. Empo. Rep. 2024, 25, 5352–5382. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.C.; Tai, X.F.; Shao, N.L.; Sun, Y.; Nie, Z.J.; Wang, Y.Y.; Li, Q.J.; Xu, P.; Xu, G.C. Effects of effective microorganisms on the growth performance, nutritional composition and flavour quality of the pond-cultured (Eriocheir sinensis). Aquac. Res. 2021, 52, 871–880. [Google Scholar] [CrossRef]
- Wang, L.M.; Gao, J.C.; Cao, X.; Du, J.L.; Cao, L.P.; Nie, Z.J.; Xu, G.C.; Dong, Z.J. Integrated Analysis of Transcriptomics and Metabolomics Unveil the Novel Insight of One-Year-Old Precocious Mechanism in the Chinese Mitten Crab, Eriocheir sinensis. Int. J. Mol. Sci. 2023, 24, 11171. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.C.; Chen, X.B.; Hou, Y.R.; Jiang, T.; Liu, H.B.; Yang, J. Combined transcriptome and metabolome analysis reveals the mechanism of high nitrite tolerance in freshwater mussel Anodonta woodiana. Comp. Biochem. Phys. D 2024, 52, 101359. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemometr. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics. 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wu, X.Y.; Lai, J.S.; Liu, Y.; Song, M.J.; Li, F.Y.; Gong, Q. Integrated biochemical, transcriptomic and metabolomic analyses provide insight into heat stress response in Yangtze sturgeon (Acipenser dabryanus). Ecotox. Environ. Safe. 2023, 249, 114366. [Google Scholar] [CrossRef]


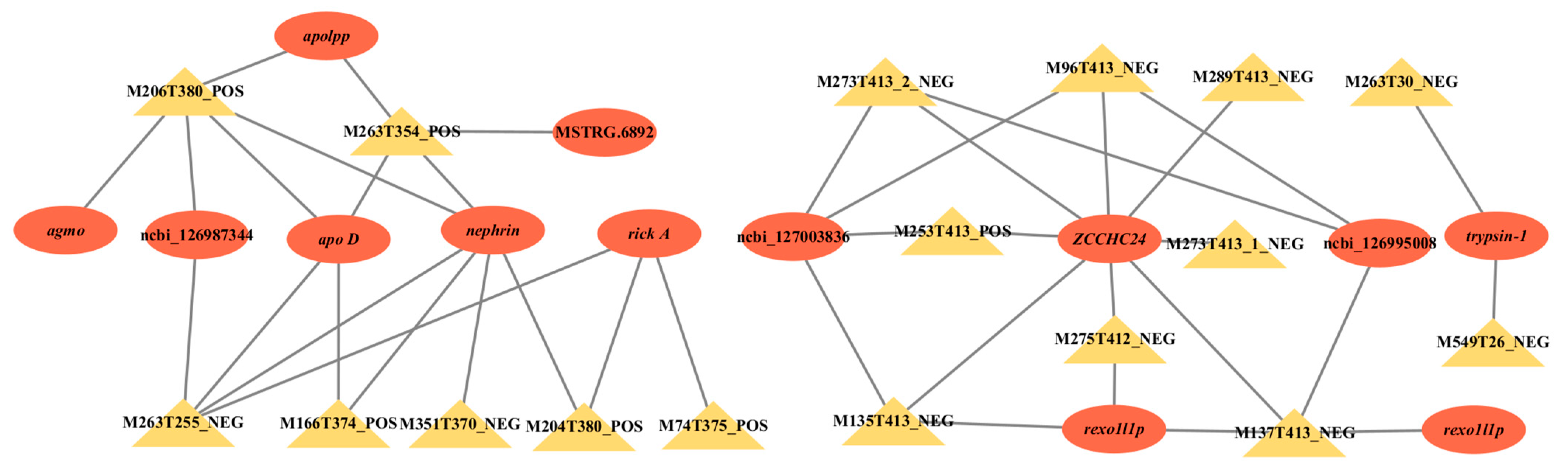
| Modules | |||||
|---|---|---|---|---|---|
| Darkgrey | Ivory | Darkorange2 | Steelblue | Lightcyan1 | Coral1 |
| M934T411_2_POS | M273T413_1_NEG | M1143T298_POS | M74T375_POS | M263T30_NEG | M368T388_NEG |
| M1173T410_POS | M96T413_NEG | M499T298_NEG | M166T374_POS | M527T31_NEG | M513T382_NEG |
| M1162T410_4_POS | M724T155_POS | M376T298_POS | M206T380_POS | M237T31_NEG | M552T387_NEG |
| M1172T410_4_POS | M275T412_NEG | M822T298_POS | M75T374_POS | M551T30_NEG | M697T382_NEG |
| M1172T410_1_POS | M289T413_NEG | M374T298_NEG | M351T370_NEG | M309T28_POS | M144T306_NEG |
| M1162T410_1_POS | M135T413_NEG | M251T298_POS | M506T394_NEG | M573T26_1_NEG | M475T377_NEG |
| M1162T410_3_POS | M253T413_POS | M626T298_POS | M204T380_POS | M249T30_NEG | M551T383_NEG |
| M1181T410_2_POS | M439T153_1_POS | M624T298_NEG | M263T255_NEG | M262T55_NEG | M130T295_POS |
| M1172T410_2_POS | M273T413_2_NEG | M874T298_NEG | M242T404_1_POS | M549T26_NEG | M446T389_POS |
| M1182T410_1_POS | M137T413_NEG | M564T298_POS | M263T354_POS | M187T86_POS | M611T383_NEG |
| Primers | Position | Primer Sequence | Accession No. |
|---|---|---|---|
| ccna2 | Forward | CACCCATATGGTCAAGGAGCTAA | XM_050835160.1 |
| Reverse | CTCTGATATGGAGAACTCCAGGC | ||
| cht2 | Forward | AACACCACCTACACCATGAAGTC | XM_050884678.1 |
| Reverse | TTCTTGTCCACGAACCTCTCAAG | ||
| ago2 | Forward | GATGGAGTAGGAAAGTCAGGCAA | XM_050835160.1 P |
| Reverse | TGATTGTGTTGTCCACTGTTGTG | ||
| fcn2 | Forward | TGAATCACTACGACAACCGACC | XM_050849112.1 |
| Reverse | GTACACTTGGCGAATGCCCTTG | ||
| pim3 | Forward | CGACTTAATGCTACAGGTGGTGA | XM_050869002.1 |
| Reverse | CAGGTTCTCGTCCTTGATGTCG | ||
| tpi1 | Forward | ACAGAACTGCTACAAGGAACCAC | XM_050875155.1 |
| Reverse | CCAAGGATCACCCACTCACATC | ||
| β-actin | Forward | TCATCACCATCGGCAATGA | XM_050843215.1 |
| Reverse | TTGTAAGTGGTCTCGTGGATG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Che, S.; Gao, J.; Zhu, H.; Du, J.; Cao, L.; Zheng, Y.; Xu, G.; Liu, B. Integrated Application of Transcriptomics and Metabolomics Provides Insights into the Different Body-Size Growth in Chinese Mitten Crab (Eriocheir sinensis). Int. J. Mol. Sci. 2025, 26, 4617. https://doi.org/10.3390/ijms26104617
Che S, Gao J, Zhu H, Du J, Cao L, Zheng Y, Xu G, Liu B. Integrated Application of Transcriptomics and Metabolomics Provides Insights into the Different Body-Size Growth in Chinese Mitten Crab (Eriocheir sinensis). International Journal of Molecular Sciences. 2025; 26(10):4617. https://doi.org/10.3390/ijms26104617
Chicago/Turabian StyleChe, Silu, Jiancao Gao, Haojun Zhu, Jinliang Du, Liping Cao, Yao Zheng, Gangchun Xu, and Bo Liu. 2025. "Integrated Application of Transcriptomics and Metabolomics Provides Insights into the Different Body-Size Growth in Chinese Mitten Crab (Eriocheir sinensis)" International Journal of Molecular Sciences 26, no. 10: 4617. https://doi.org/10.3390/ijms26104617
APA StyleChe, S., Gao, J., Zhu, H., Du, J., Cao, L., Zheng, Y., Xu, G., & Liu, B. (2025). Integrated Application of Transcriptomics and Metabolomics Provides Insights into the Different Body-Size Growth in Chinese Mitten Crab (Eriocheir sinensis). International Journal of Molecular Sciences, 26(10), 4617. https://doi.org/10.3390/ijms26104617








