Active Constituent of HQS in T2DM Intervention: Efficacy and Mechanistic Insights
Abstract
1. Introduction
2. Results
2.1. Network Pharmacology
2.1.1. Gene Collection
2.1.2. PPI Network Visualization and Key Target Discovery
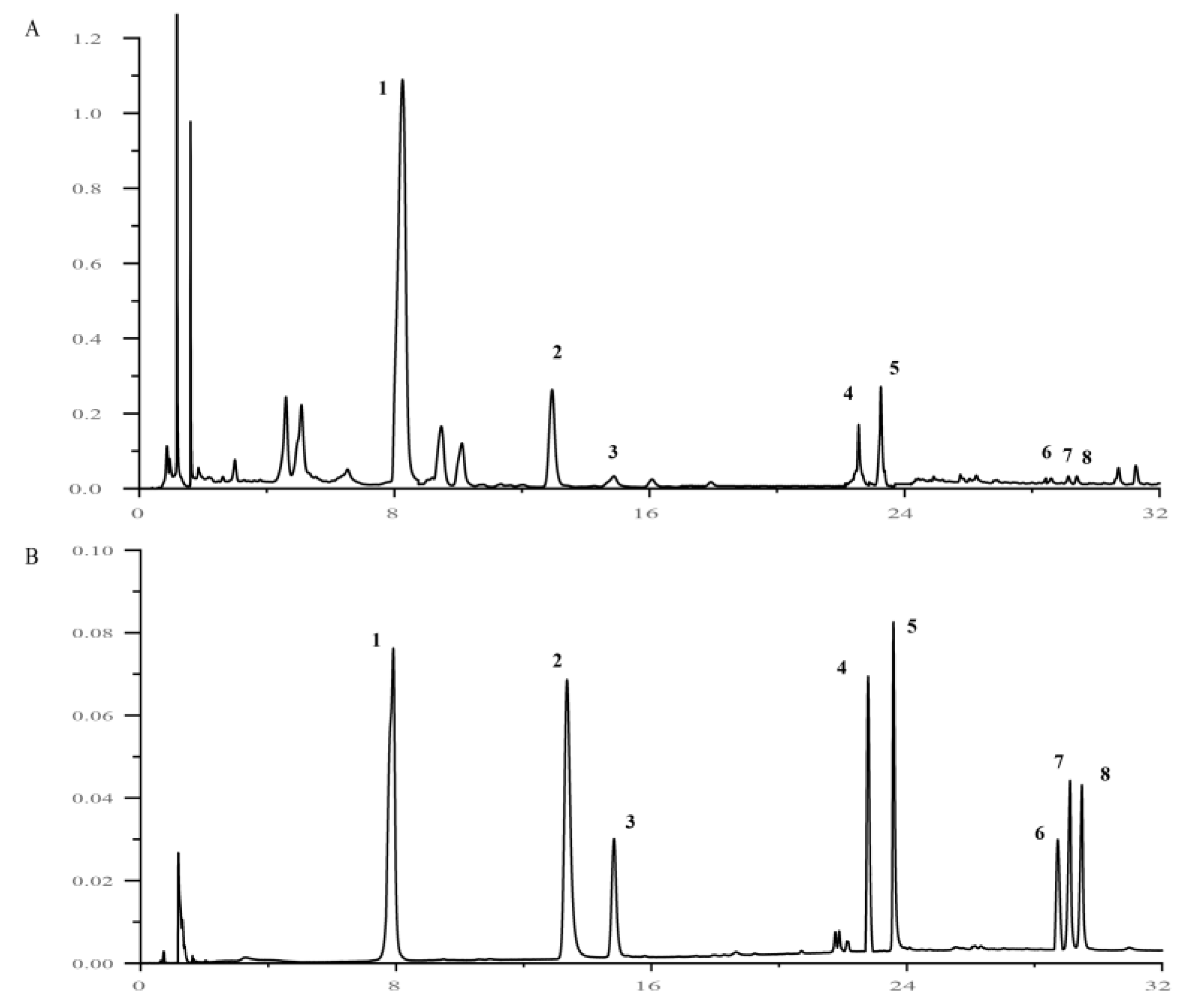
2.2. Spectrum–Effect Correlation Analysis
2.3. Quantitative Analysis of 8 Active Components of HQS
2.4. In Vitro Pharmacological Study of HQS in Alleviating T2DM
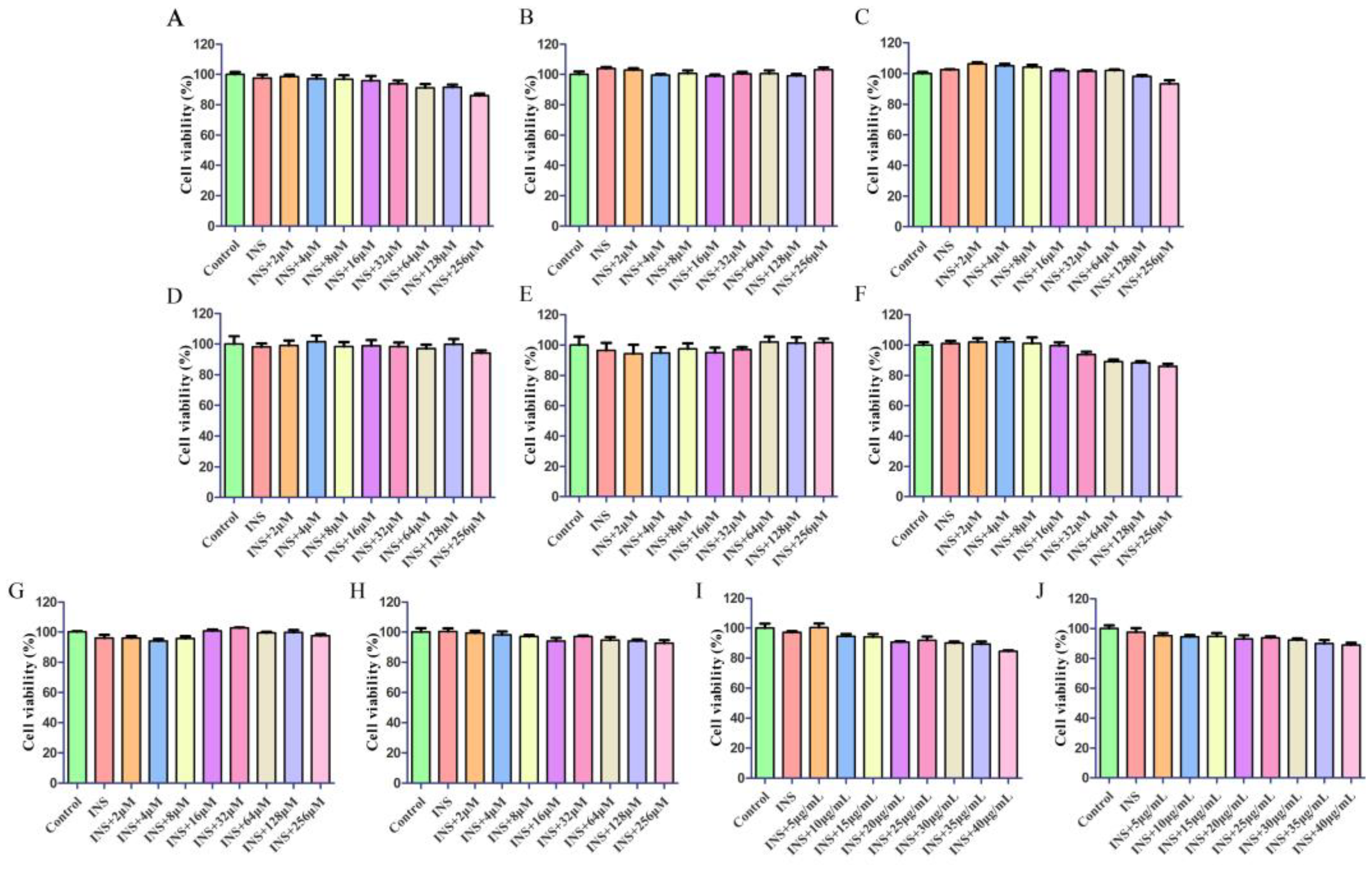
2.4.1. Cytotoxicity Study
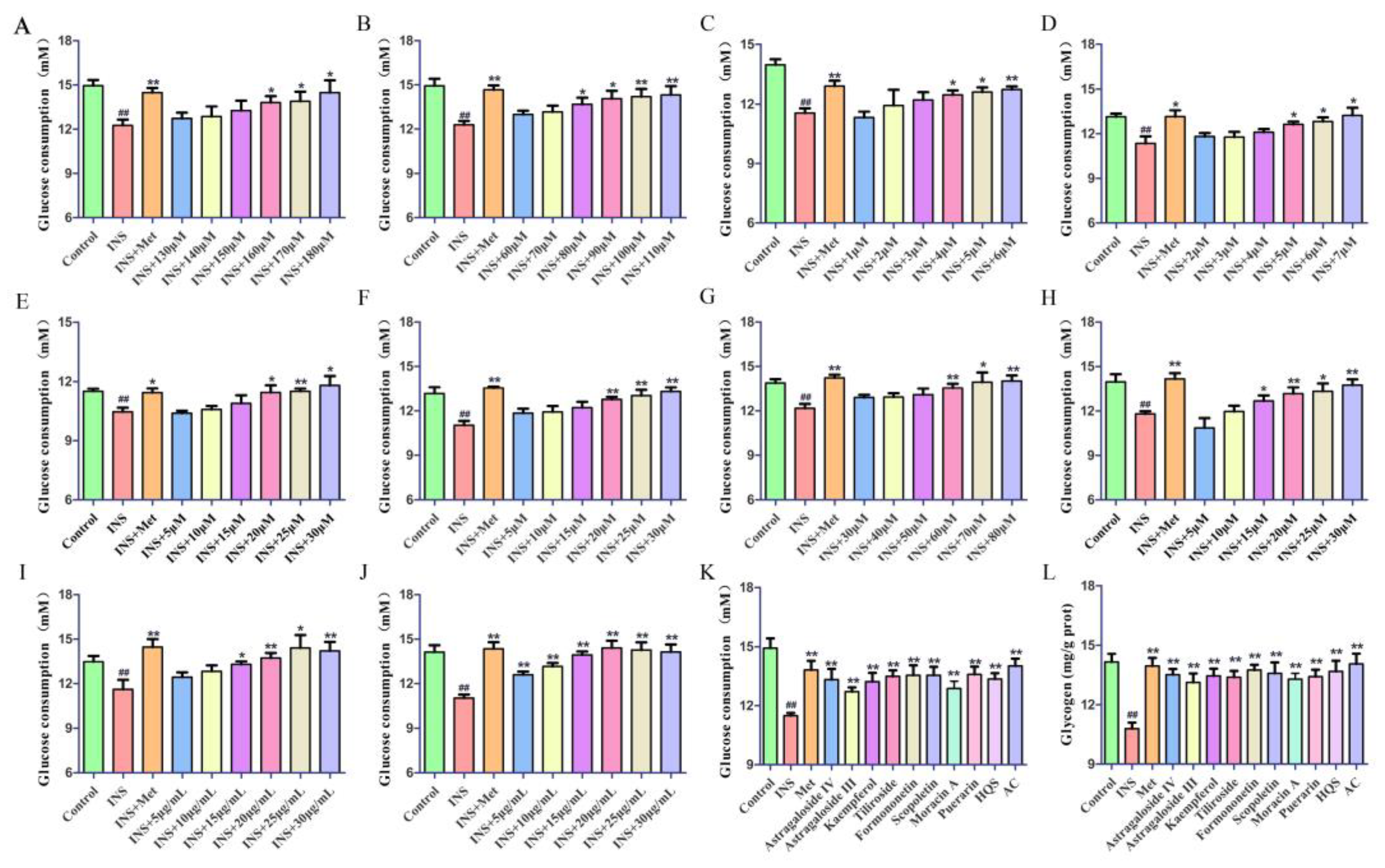
2.4.2. The Hypoglycemic Effect on IR-HepG2 Cells
2.5. In Vivo Pharmacological Study of HQS in Alleviating T2DM
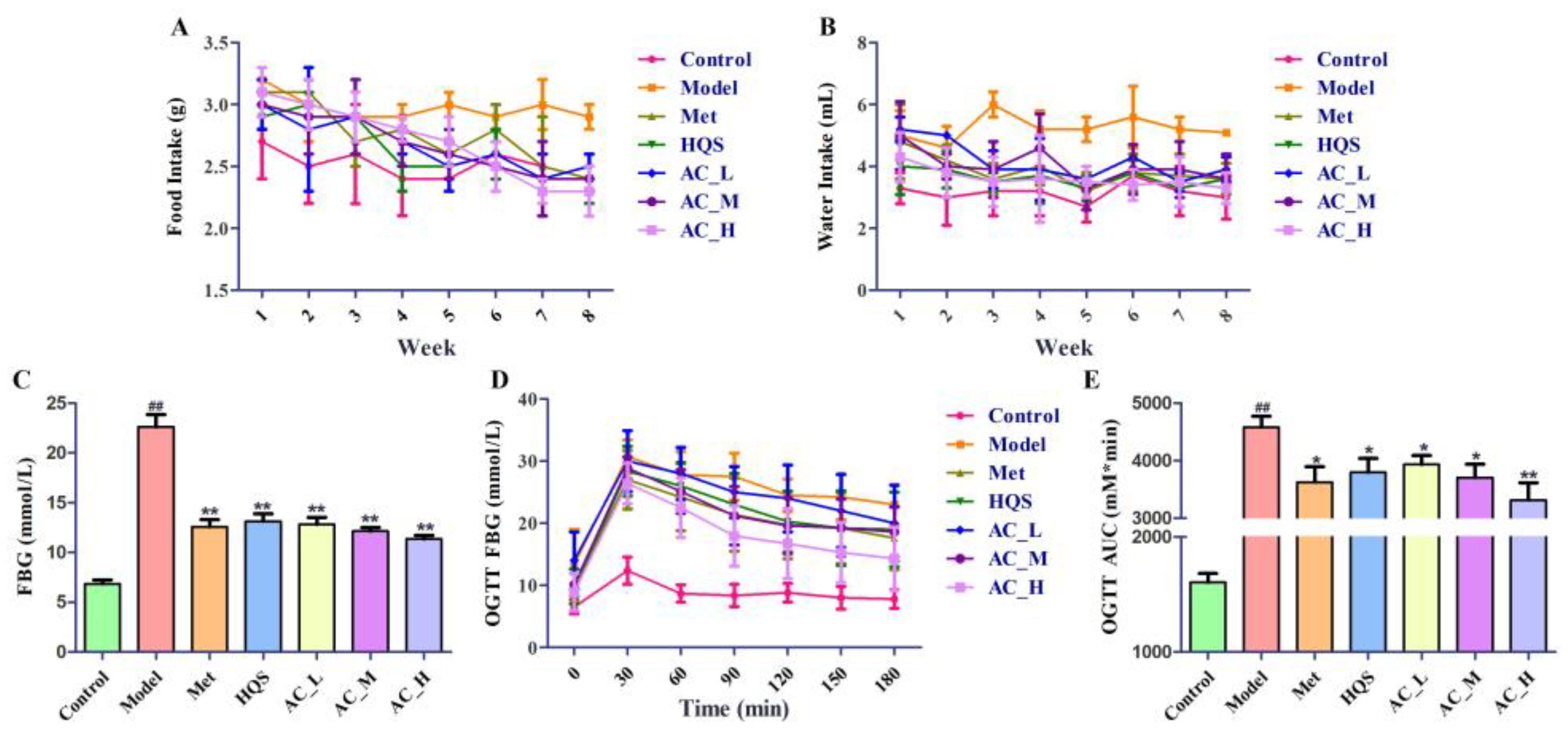
2.5.1. Hypoglycemic Effect of HQS on T2DM Mice
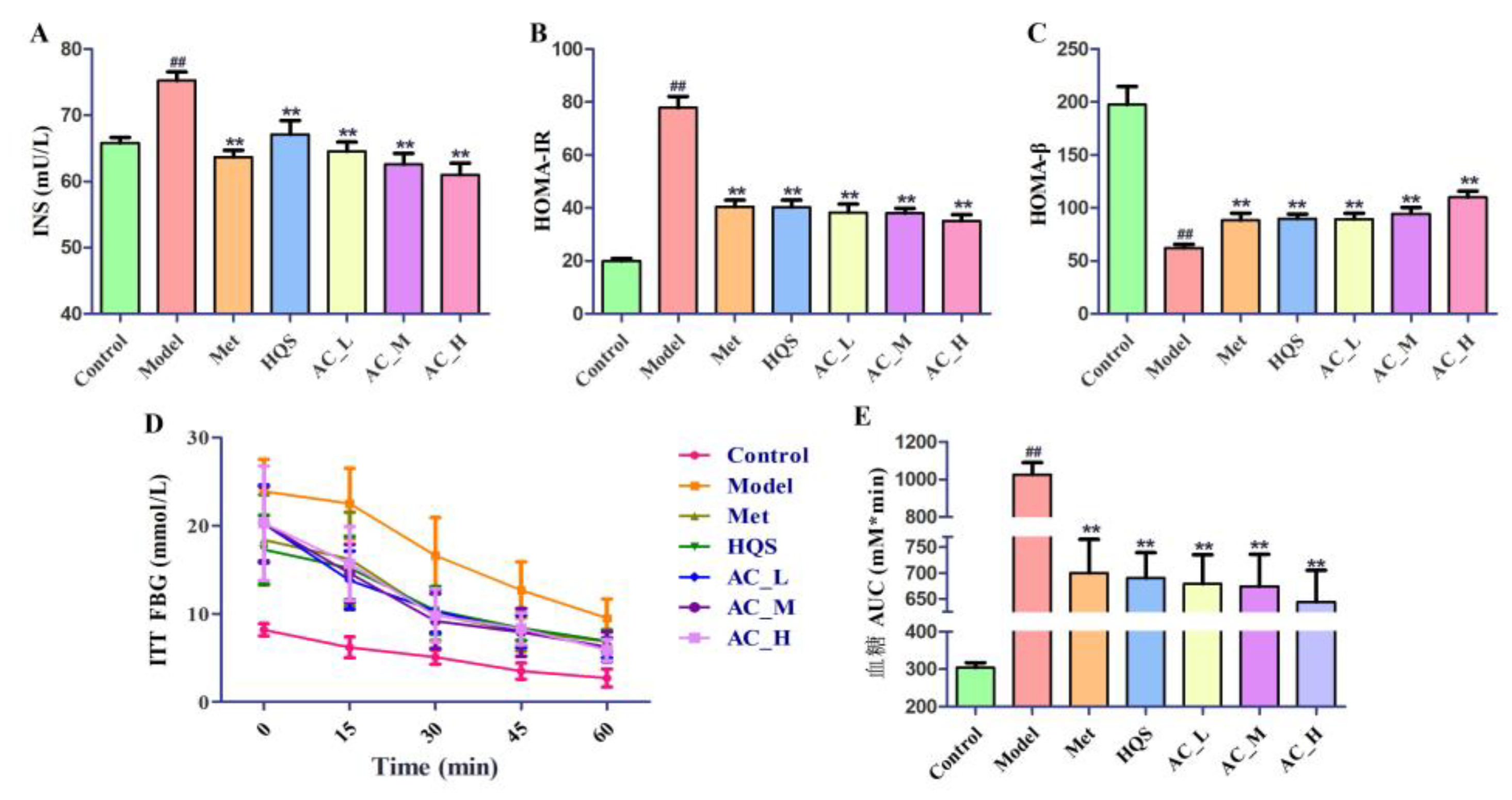
2.5.2. Improvement of Insulin Sensitivity in T2DM Mice
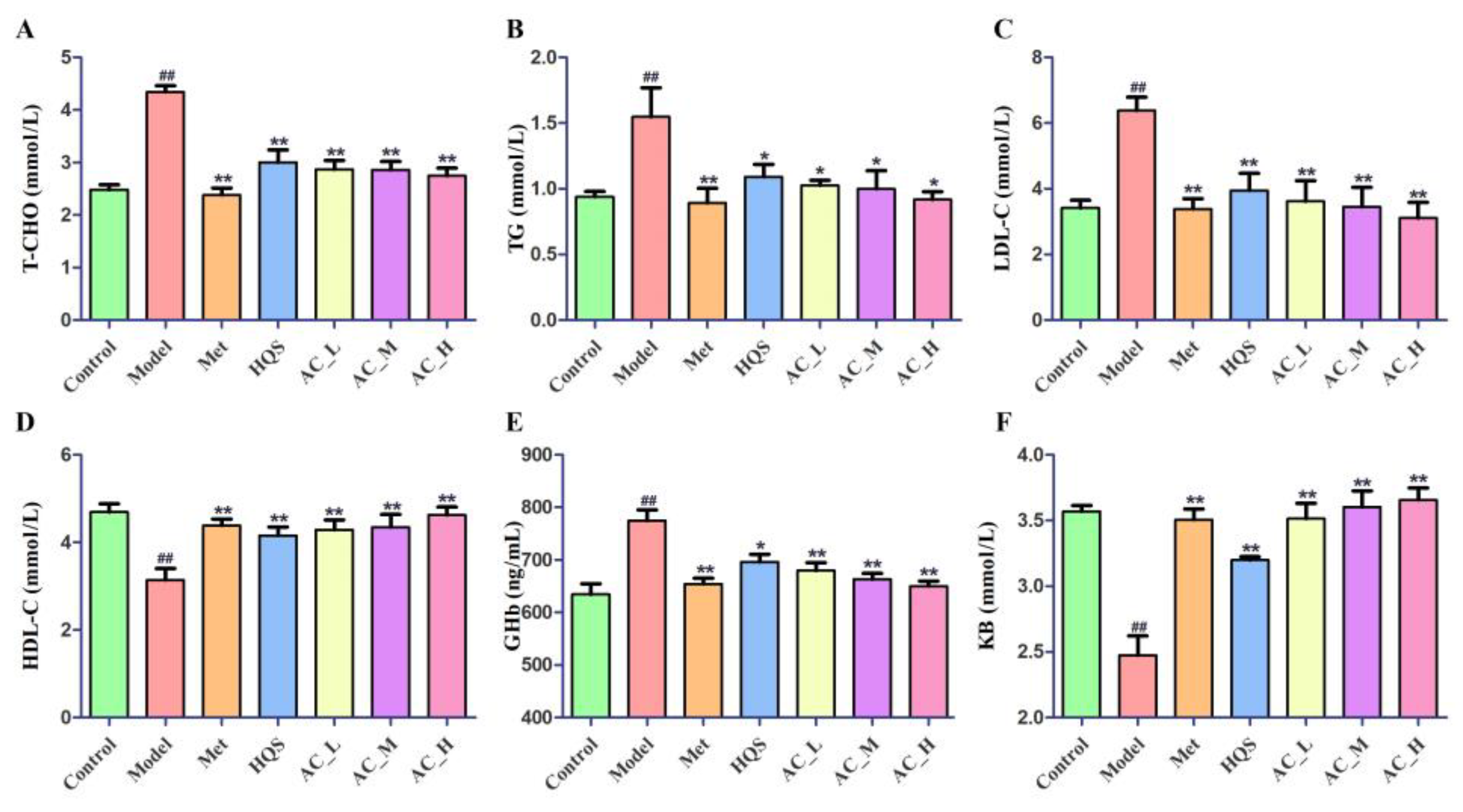
2.5.3. Effects of HQS on Serum Biochemical Indices of T2DM
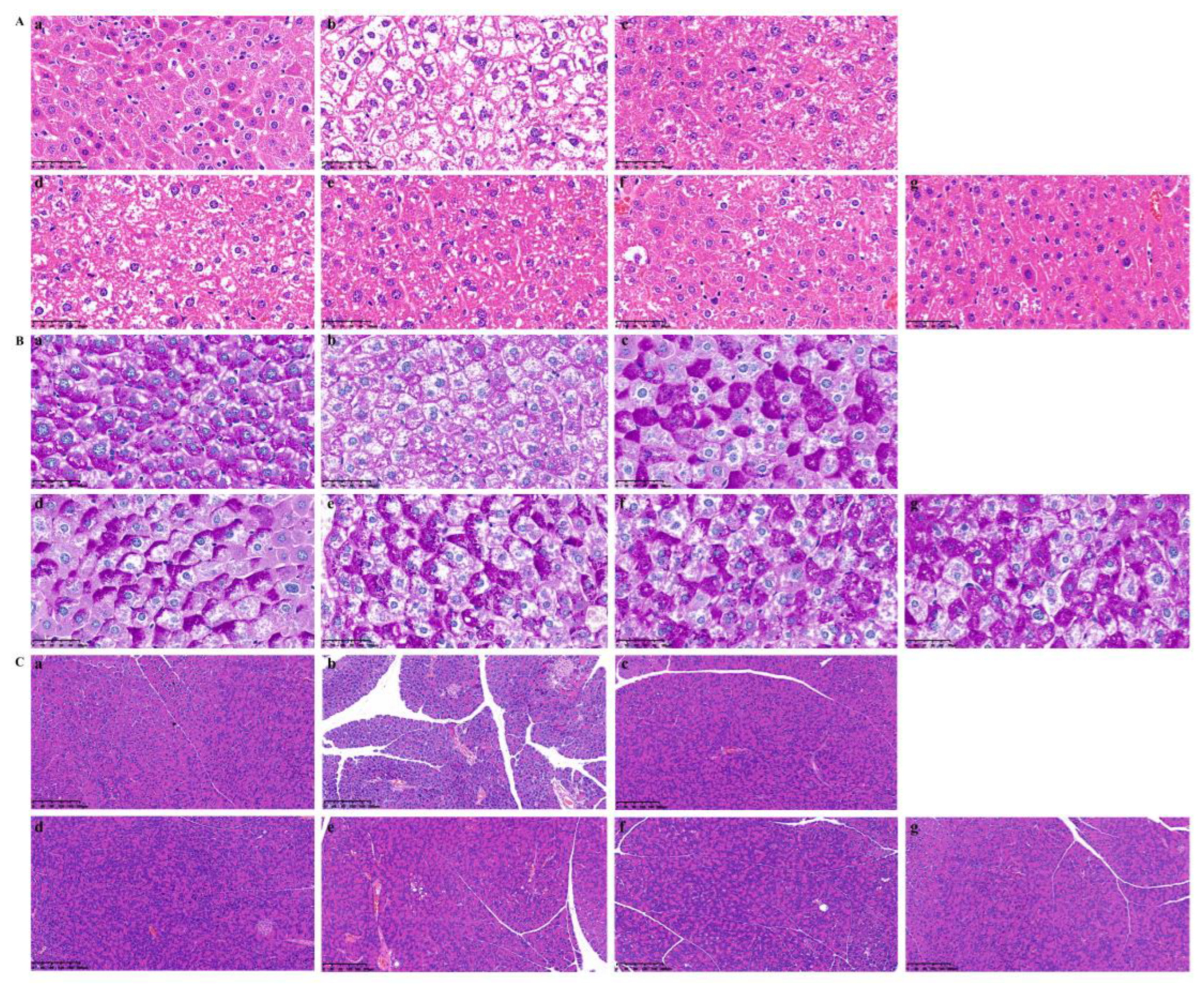
2.5.4. Pathological Changes in Liver and Pancreas Tissue in T2DM Mice
2.6. Metabolomics Analysis
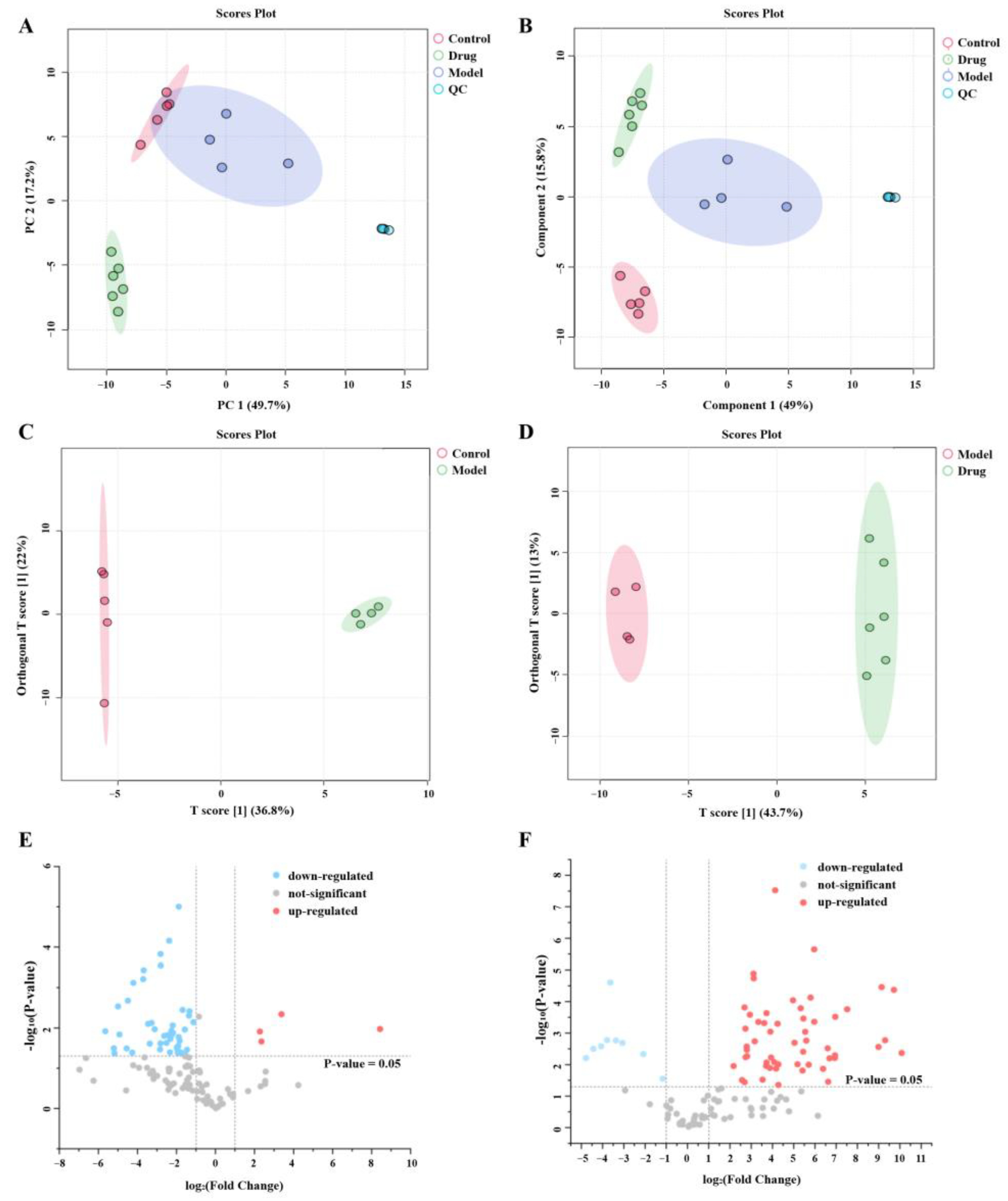
2.6.1. Multivariate Statistical Analysis
2.6.2. Identification of Potential Biomarkers
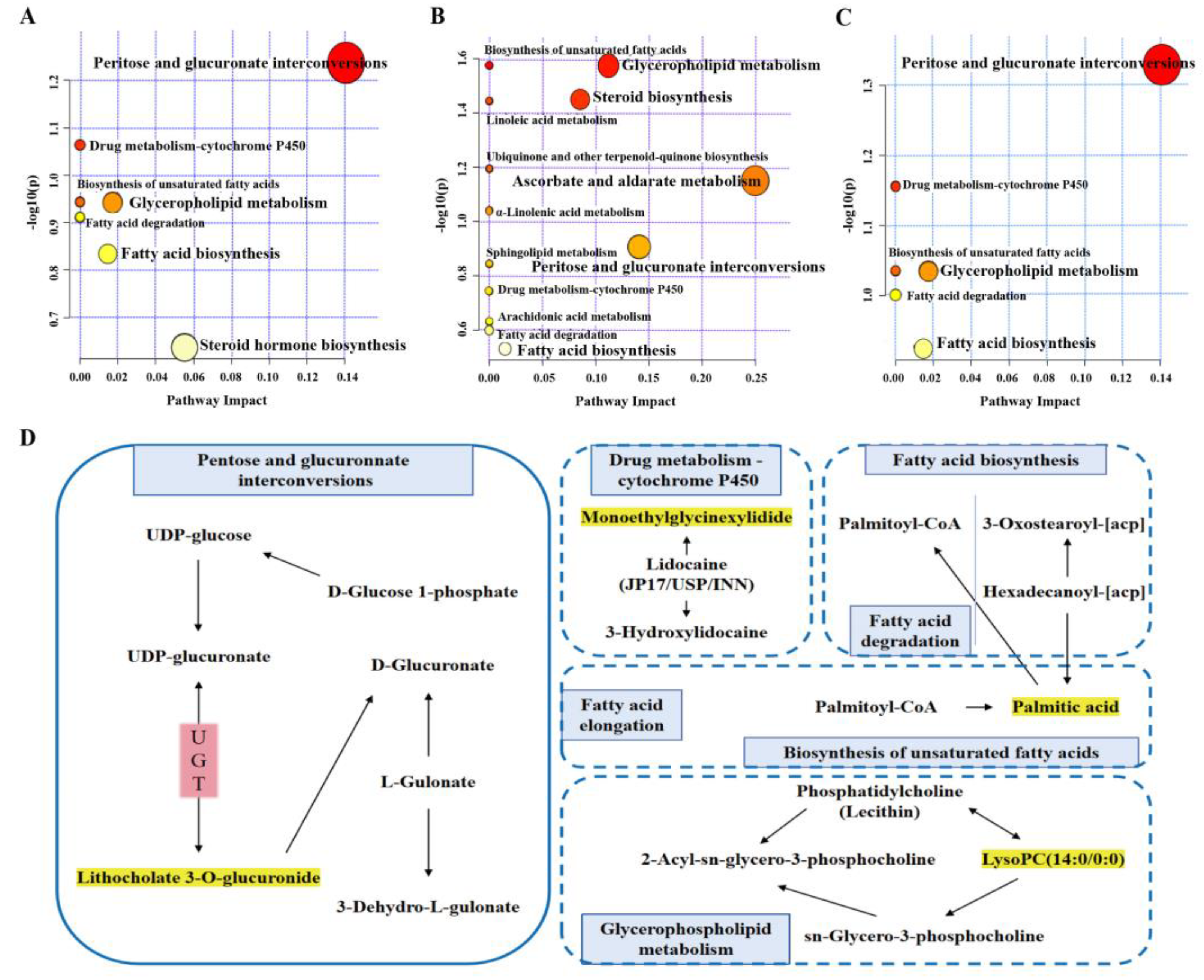
2.6.3. Metabolic Pathway Analysis
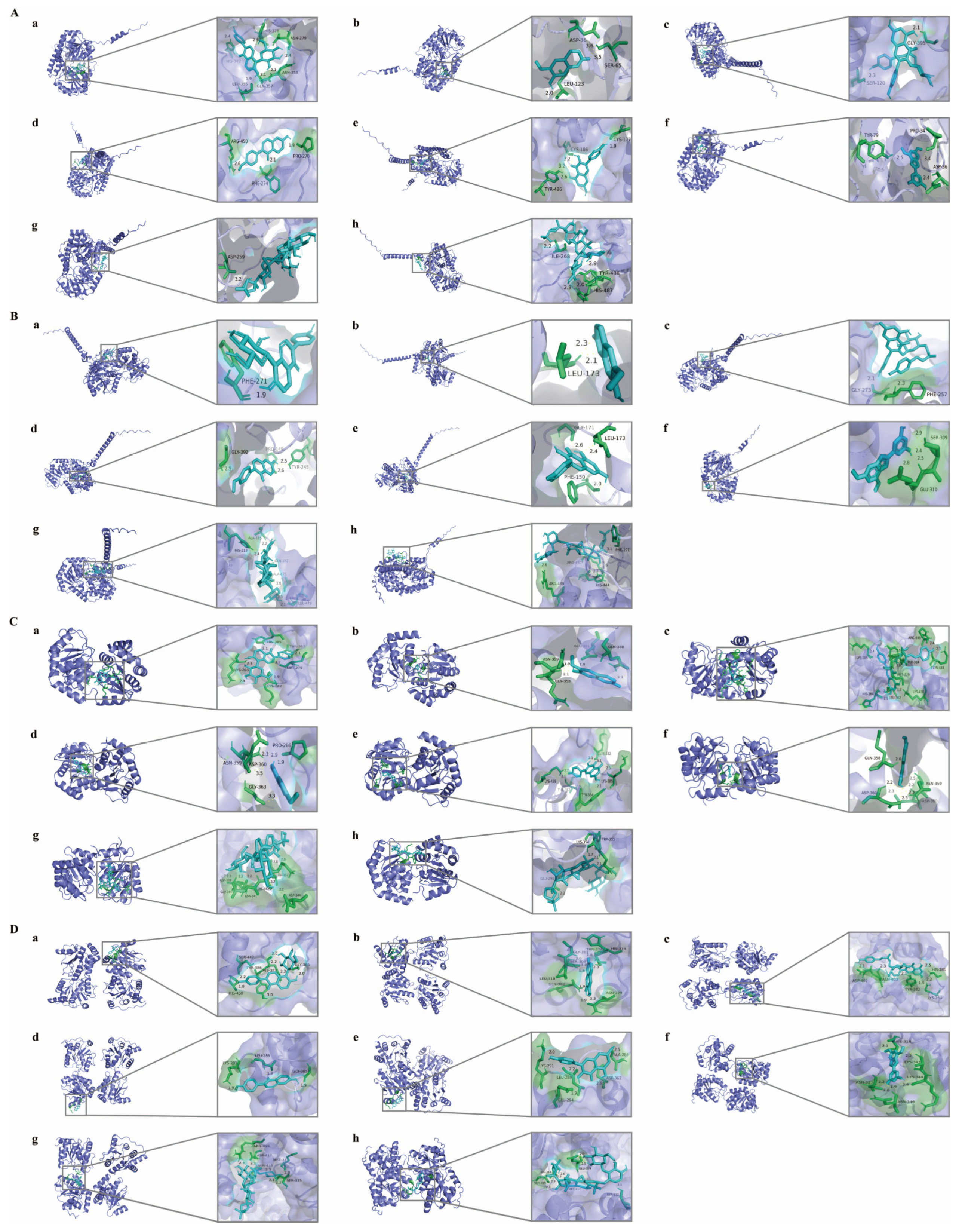
2.7. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Network Pharmacology
4.2.1. Gene Collection of T2DM
4.2.2. Prediction and Identification of HQS Components and Potential Targets
4.2.3. Protein–Protein Interaction (PPI) Network Analysis
4.3. Preparation of the HQS Extract
4.4. Spectrum–Effect Correlation Analysis
4.4.1. Animal Models
4.4.2. Measurement of Biochemical Indicators
4.4.3. UPLC-Q/TOF-MS/MS Analysis
4.4.4. Spectrum–Effect Correlation Analysis
4.5. Determination of 8 Active Components in HQS Extract
4.5.1. Preparation of Standard and Sample Solutions
4.5.2. Quantitative Analysis
4.6. Pharmacological Study of Active Constituent of HQS in the Treatment of T2DM
4.6.1. Cell Culture
4.6.2. Cell Viability
4.6.3. Glucose Consumption Assay
4.6.4. Glycogen Assay
4.6.5. Animal Models
4.6.6. Oral Glucose Tolerance Test (OGTT) and Insulin Tolerance Test (ITT)
4.6.7. Analysis of Serum Biochemical Indicators
4.6.8. Determination of Insulin Sensitivity
4.6.9. Histological Examination
4.7. Metabolomics Analysis
4.8. Molecular Docking
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bacha, F.; Hannon, T.S.; Tosur, M.; Pike, J.M.; Butler, A.; Tommerdahl, K.L.; Zeitler, P.S. Pathophysiology and Treatment of Prediabetes and Type 2 Diabetes in Youth. Diabetes Care 2024, 47, 2038–2049. [Google Scholar] [CrossRef]
- Ruze, R.; Liu, T.; Zou, X.; Song, J.; Chen, Y.; Xu, R.; Yin, X.; Xu, Q. Obesity and Type 2 Diabetes Mellitus: Connections in Epidemiology, Pathogenesis, and Treatments. Front. Endocrinol. 2023, 14, 1161521. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association Professional Practice Committee 2. Diagnosis and Classification of Diabetes: Standards of Care in Diabetes-2024. Diabetes Care 2024, 47, S20–S42. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, J.; Zhao, Y.; Zhou, Q.; Yang, X.; Gao, Y.; Li, Q.; Bai, M.; Liu, J.; Liang, Y.; et al. Study on the Mechanism of Modified Gegen Qinlian Decoction in Regulating the Intestinal Flora-Bile Acid-TGR5 Axis for the Treatment of Type 2 Diabetes Mellitus Based on Macro Genome Sequencing and Targeted Metabonomics Integration. Phytomedicine 2024, 132, 155329. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liu, X.; Tan, J.; Sheng, Q.; Zhang, D.; Li, B.; Zhang, J.; Zhang, F.; Chen, H.; Cui, T.; et al. From Xiaoke to Diabetes Mellitus: A Review of the Research Progress in Traditional Chinese Medicine for Diabetes Mellitus Treatment. Chin. Med. 2023, 18, 75. [Google Scholar] [CrossRef]
- Wu, X.; Zhu, J.-C.; Zhang, Y.; Li, W.-M.; Rong, X.-L.; Feng, Y.-F. Lipidomics Study of Plasma Phospholipid Metabolism in Early Type 2 Diabetes Rats with Ancient Prescription Huang-Qi-San Intervention by UPLC/Q-TOF-MS and Correlation Coefficient. Chem. Biol. Interact. 2016, 256, 71–84. [Google Scholar] [CrossRef]
- Li, Y.; Wang, C.; Jin, Y.; Chen, H.; Cao, M.; Li, W.; Luo, H.; Wu, Z. Huang-Qi San Improves Glucose and Lipid Metabolism and Exerts Protective Effects against Hepatic Steatosis in High Fat Diet-Fed Rats. Biomed. Pharmacother. 2020, 126, 109734. [Google Scholar] [CrossRef]
- Xu, Z.; Huang, J.; Wen, M.; Zhang, X.; Lyu, D.; Li, S.; Xiao, H.; Li, M.; Shen, C.; Huang, H. Gentiopicroside Ameliorates Glucose and Lipid Metabolism in T2DM via Targeting FGFR1. Phytomedicine 2024, 132, 155780. [Google Scholar] [CrossRef]
- Chen, Y.; Wen, Q.; Lin, M.; Yang, B.; Feng, L.; Jia, X. Scientific Validation of Using Active Constituent as Research Focus in Traditional Chinese Medicine: Case Study of Pueraria Lobata Intervention in Type 2 Diabetes. Pharmaceuticals 2024, 17, 1675. [Google Scholar] [CrossRef]
- Zhong, R.-F.; Liu, C.-J.; Hao, K.-X.; Fan, X.-D.; Jiang, J.-G. Polysaccharides from Flos Sophorae Immaturus Ameliorates Insulin Resistance in IR-HepG2 Cells by Co-Regulating Signaling Pathways of AMPK and IRS-1/PI3K/AKT. Int. J. Biol. Macromol. 2024, 280, 136088. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, S.; Zhang, Y.; Wang, H.; Chen, Y.; Lu, H. Activation of NRF2 by Epiberberine Improves Oxidative Stress and Insulin Resistance in T2DM Mice and IR-HepG2 Cells in an AMPK Dependent Manner. J. Ethnopharmacol. 2024, 327, 117931. [Google Scholar] [CrossRef]
- Hu, M.; Yan, H.; Li, H.; Feng, Y.; Sun, W.; Ren, Y.; Ma, L.; Zeng, W.; Huang, F.; Jiang, Z.; et al. Use of Network Pharmacology and Molecular Docking to Explore the Mechanism of Action of Curcuma in the Treatment of Osteosarcoma. Sci. Rep. 2023, 13, 9569. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Miao, J.; Li, X.; Guo, Q.; Deng, Z.; Wu, L. Metabolomics Combined with Systematic Pharmacology Reveals the Therapeutic Effects of Salvia Miltiorrhiza and Radix Pueraria Lobata Herb Pair on Type 2 Diabetes Rats. J. Funct. Foods 2022, 89, 104950. [Google Scholar] [CrossRef]
- Chen, X.; Shi, C.; Wang, Y.; Yu, H.; Zhang, Y.; Zhang, J.; Li, P.; Gao, J. The Mechanisms of Glycolipid Metabolism Disorder on Vascular Injury in Type 2 Diabetes. Front. Physiol. 2022, 13, 952445. [Google Scholar] [CrossRef]
- Akindele, A.J.; Otuguor, E.; Singh, D.; Ota, D.; Benebo, A.S. Hypoglycemic, Antilipidemic and Antioxidant Effects of Valproic Acid in Alloxan-Induced Diabetic Rats. Eur. J. Pharmacol. 2015, 762, 174–183. [Google Scholar] [CrossRef]
- Deng, X.; Lei, H.-Y.; Ren, Y.-S.; Ai, J.; Li, Y.-Q.; Liang, S.; Chen, L.-L.; Liao, M.-C. A Novel Strategy for Active Compound Efficacy Status Identification in Multi-Tropism Chinese Herbal Medicine (Scutellaria Baicalensis Georgi) Based on Multi-Indexes Spectrum-Effect Gray Correlation Analysis. J. Ethnopharmacol. 2023, 300, 115677. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Y.; Gou, S.-H.; Shi, Z.-Q.; Xue, Z.-Y.; Feng, S.-L. Spectrum-Effect Relationship between HPLC Fingerprints and Bioactive Components of Radix Hedysari on Increasing the Peak Bone Mass of Rat. J. Pharm. Anal. 2019, 9, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Wang, X.; Zeng, Z.; Yang, Z.; Lin, Y.; Sun, L.; Lu, Q.; Fan, G. The Improvement of Modified Si-Miao Granule on Hepatic Insulin Resistance and Glycogen Synthesis in Type 2 Diabetes Mellitus Involves the Inhibition of TNF-α/JNK1/IRS-2 Pathway: Network Pharmacology, Molecular Docking, and Experimental Validation. Chin. Med. 2024, 19, 128. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, Y.; Zhang, Q.; Gao, R.; Chang, F.; Li, B.; Huang, K.; Cheng, N.; He, X. Single-Atom Ce-N-C Nanozyme Ameliorates Type 2 Diabetes Mellitus by Improving Glucose Metabolism Disorders and Reducing Oxidative Stress. Biomolecules 2024, 14, 1193. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, H.; Zhu, C.; Deng, J.; Fan, D. Ginsenoside Rg5 Relieves Type 2 Diabetes by Improving Hepatic Insulin Resistance in Db/Db Mice. J. Funct. Foods 2020, 71, 104014. [Google Scholar] [CrossRef]
- Parkman, J.K.; Sklioutovskaya-Lopez, K.; Menikdiwela, K.R.; Freeman, L.; Moustaid-Moussa, N.; Kim, J.H. Effects of High Fat Diets and Supplemental Tart Cherry and Fish Oil on Obesity and Type 2 Diabetes in Male and Female C57BL/6J and TALLYHO/Jng Mice. J. Nutr. Biochem. 2021, 94, 108644. [Google Scholar] [CrossRef] [PubMed]
- Yaikwawong, M.; Jansarikit, L.; Jirawatnotai, S.; Chuengsamarn, S. Curcumin Extract Improves Beta Cell Functions in Obese Patients with Type 2 Diabetes: A Randomized Controlled Trial. Nutr. J. 2024, 23, 119. [Google Scholar] [CrossRef]
- Huang, W.; Huang, G.-P.; Zhang, L.-X.; da Yu, E.; Yang, W.-K.; Ye, M.; Zou, S.-Q.; Ni, L.; He, H.-Q. Lignan-Rich Extract from Cinnamomum Camphora Leaf Attenuates Metabolic Syndrome by Modulating Glycolipid Metabolism and Gut Microbiota in T2DM Mice. Phytomedicine 2024, 135, 156118. [Google Scholar] [CrossRef]
- Liu, Z.; Shang, Q.; Zuo, H.; Li, H.; Fang, D.; Zhang, J.; Huang, H.-D.; Granato, D.; Chen, J.; Chen, J. Cynomorium Songaricum: UHPLC/ESI-LTQ-Orbitrap-MS Analysis and Mechanistic Study on Insulin Sensitivity of a Flavonoid-Enriched Fraction. Phytomedicine 2024, 132, 155862. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Bouscarel, B. Bile Acids and Signal Transduction: Role in Glucose Homeostasis. Cell Signal 2008, 20, 2180–2197. [Google Scholar] [CrossRef]
- Xie, Z.-C.; Li, T.-T.; Gan, B.-L.; Gao, X.; Gao, L.; Chen, G.; Hu, X.-H. Investigation of miR-136-5p Key Target Genes and Pathways in Lung Squamous Cell Cancer Based on TCGA Database and Bioinformatics Analysis. Pathol. Res. Pract. 2018, 214, 644–654. [Google Scholar] [CrossRef]
- Li, X.; Miao, F.; Xin, R.; Tai, Z.; Pan, H.; Huang, H.; Yu, J.; Chen, Z.; Zhu, Q. Combining Network Pharmacology, Molecular Docking, Molecular Dynamics Simulation, and Experimental Verification to Examine the Efficacy and Immunoregulation Mechanism of FHB Granules on Vitiligo. Front. Immunol. 2023, 14, 1194823. [Google Scholar] [CrossRef]
- Ye, L.; Liang, R.; Liu, X.; Li, J.; Yue, J.; Zhang, X. Frailty and Sarcopenia: A Bibliometric Analysis of Their Association and Potential Targets for Intervention. Ageing Res. Rev. 2023, 92, 102111. [Google Scholar] [CrossRef]
- Huddart, R.; Whirl-Carrillo, M.; Altman, R.B.; Klein, T.E. PharmGKB Tutorial for Pharmacogenomics of Drugs Potentially Used in the Context of COVID-19. Clin. Pharmacol. Ther. 2021, 109, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.L.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- Gao, S.; Chen, H.; Zhou, X. Study on the Spectrum-Effect Relationship of the Xanthine Oxidase Inhibitory Activity of Ligustrum Lucidum. J. Sep. Sci. 2019, 42, 3281–3292. [Google Scholar] [CrossRef]
- Zhou, M.; Liu, X.; Wu, Y.; Xiang, Q.; Yu, R. Liver Lipidomics Analysis Revealed the Protective Mechanism of Zuogui Jiangtang Qinggan Formula in Type 2 Diabetes Mellitus with Non-Alcoholic Fatty Liver Disease. J. Ethnopharmacol. 2024, 329, 118160. [Google Scholar] [CrossRef]



| No. | ID | Components | Degree | Betweenness | Closeness |
|---|---|---|---|---|---|
| 1 | B3 | Quercetin | 161 | 0.63 | 0.56 |
| 2 | B2 | Kaempferol | 113 | 0.14 | 0.42 |
| 3 | A1 | Formononetin | 69 | 0.10 | 0.39 |
| 4 | C1 | Beta-sitosterol | 68 | 0.13 | 0.39 |
| 5 | HQ7 | 7-O-Methylisomucronulatol | 40 | 0.08 | 0.39 |
| 6 | HQ5 | Isorhamnetin | 33 | 0.06 | 0.38 |
| 7 | BP14 | Iristectorigenin | 30 | 0.01 | 0.37 |
| 8 | BP18 | Tiliroside | 24 | 0.01 | 0.36 |
| 9 | HQ3 | α-Hederin | 22 | 0.05 | 0.37 |
| 10 | HQ6 | 3′-Hydroxy-6″-O-xylosylpuerarin | 21 | 0.02 | 0.37 |
| 11 | HQ12 | Pulobatones B | 19 | 0.01 | 0.37 |
| 12 | HQ9 | Astragaloside III | 19 | 0.01 | 0.36 |
| 13 | GG3 | Puerarin | 18 | 0.01 | 0.36 |
| 14 | BP8 | Sanggenone F | 15 | 0.01 | 0.36 |
| 15 | BP20 | Kudzusaponin A1 | 13 | 0.02 | 0.36 |
| 16 | BP17 | Moracin A | 12 | 0.00 | 0.36 |
| 17 | HQ2 | Astragaloside IV | 12 | 0.00 | 0.36 |
| 18 | BP9 | Sanggenone H | 10 | 0.01 | 0.35 |
| 19 | BP1 | 6″-O-Acetyldaidzin | 8 | 0.01 | 0.35 |
| 20 | BP21 | Quercetin 3-O-(6″-galloyl)-β-D-glucopyranoside | 8 | 0.00 | 0.35 |
| 21 | BP22 | Scopoletin | 8 | 0.00 | 0.35 |
| 22 | BP3 | M-3′-O-glucopyranoside | 6 | 0.00 | 0.35 |
| 23 | BP10 | Mulberroside C | 5 | 0.00 | 0.35 |
| 24 | BP2 | Mairin | 5 | 0.00 | 0.25 |
| 25 | BP16 | Moracin C | 4 | 0.00 | 0.32 |
| 26 | HQ11 | 3′-Methoxydaidzein | 3 | 0.00 | 0.34 |
| 27 | HQ16 | Corchoionoside C | 3 | 0.00 | 0.35 |
| 28 | B1 | Mairin | 2 | 0.00 | 0.25 |
| No. | Identified Cmponents | M (m/z) | Formular | Error (ppm) | Adduct |
|---|---|---|---|---|---|
| 1 | Astragaloside III-C11H18O9(cleavage) + O-H2 + C10H15N3O6S | 808.401 | C40H63N3O12S | −5.5 | -H |
| 2 | Astragaloside IV-C35H56O8(cleavage) + 2×(+H2) + SO3 | 263.044 | C6H16O9S | −0.9 | -H |
| 3 | Kaempferol + H2 | 287.06 | C15H12O6 | 11.7 | -H |
| 4 | Moracin A | 285.08 | C16H14O5 | 11.1 | -H |
| 5 | Puerarin + H2 | 417.12 | C21H22O9 | 1.7 | -H |
| 6 | Tiliroside-C15H8O5(cleavage) + 2×(+H2) | 375.13 | C15H22O8 | 1.2 | +HCOO |
| 7 | Moracin M-3′-O-glucopyranoside-C6H10O5(cleavage) + O | 257.049 | C14H10O5 | 13.6 | -H |
| 8 | Formononetin-CH2O(cleavage) + 2×(+H2) + SO3 | 321.044 | C15H14O6S | −1 | -H |
| 9 | Kaempferol + 2×(+H2) + C2H2O | 377.086 | C17H16O7 | −3.9 | +HCOO |
| 10 | Scopoletin-CH2O(cleavage) + 2×(-H2) + C2H2O | 199.007 | C11H4O4 | 17.2 | -H |
| 11 | α-Hederin-C35H54O7(cleavage) + 2×(-H2) + SO3 | 222.992 | C6H8O7S | −0.7 | -H |
| 12 | 3′-Methoxydaidzein-H4 | 279.032 | C16H8O5 | 6.8 | -H |
| 13 | Scopoletin + C2H2O | 279.055 | C12H10O5 | 14.9 | +HCOO |
| 14 | Astragaloside I-C36H58O9(cleavage)-H2 | 277.06 | C9H12O7 | 13.1 | +HCOO |
| 15 | 3′-Methoxydaidzein + 2×(+H2) + C2H2O | 329.107 | C18H18O6 | 11.5 | -H |
| 16 | Astragaloside I-C39H60O10(cleavage) + 2×(+O) | 257.048 | C6H12O8 | −11.9 | +HCOO |
| 17 | Polydatin-C14H10O2(cleavage) + H2 | 181.071 | C6H14O6 | −6.1 | -H |
| 18 | Pulobatones B-C26H20O8(cleavage)-H2 | 161.046 | C6H10O5 | 0.9 | -H |
| 19 | Isoastragaloside II-C36H58O10(cleavage)-H2 | 173.045 | C7H10O5 | −4.2 | -H |
| 20 | Astragaloside III-C11H18O9(cleavage) + C10H15N3O6S | 794.426 | C40H65N3O11S | −1.2 | -H |
| 21 | Formononetin-CH2O(cleavage) + 2×(+H2) | 241.082 | C15H14O3 | −20 | -H |
| 22 | 1-Deoxynojirimycin + O+C2H2O | 266.088 | C8H15NO6 | −0.4 | +HCOO |
| 23 | Pulobatones B-C26H20O7(cleavage) + 2×(-H2) + SO3 | 254.983 | C6H8O9S | 4.1 | -H |
| 24 | Kudzusaponin A1-C47H74O18(cleavage) + 2×(+O) | 243.033 | C5H10O8 | −12.4 | +HCOO |
| 25 | 1-Deoxynojirimycin-H2 | 206.067 | C6H11NO4 | 0.5 | +HCOO |
| 26 | Polydatin-O(cleavage)-H4 | 415.104 | C20H18O7 | 0.3 | +HCOO |
| 27 | Astragaloside II-C34H54O10(cleavage)-H2 | 173.045 | C7H10O5 | −4.1 | -H |
| 28 | Quercetin-3-O-sophoroside-C15H8O6(cleavage) + O + H2 | 359.119 | C12H24O12 | −0.8 | -H |
| 29 | 3′-Hydroxy-6″-O-xylosylpuerarin-C21H18O10(cleavage)-H2 + C2H2O | 173.045 | C7H10O5 | −5.5 | -H |
| 30 | Moracin A + H2 | 333.094 | C16H16O5 | −12.1 | +HCOO |
| 31 | Mulberroside C + O + H2 | 475.161 | C24H28O10 | −0.9 | -H |
| 32 | Moracin B-CH2(cleavage) + H2 + C2H2O | 315.09 | C17H16O6 | 8.1 | -H |
| 33 | Scopoletin-H4 | 187.006 | C10H4O4 | 14.9 | -H |
| 34 | Pulobatones B-C26H20O8(cleavage) + 2×(+H2) + SO3 | 293.055 | C6H16O8S | −0.7 | +HCOO |
| 35 | Puerarin | 415.104 | C21H20O9 | 0.3 | -H |
| 36 | Moracin A-CH2(cleavage) | 271.064 | C15H12O5 | 11.4 | -H |
| 37 | Astragaloside I-C39H60O10(cleavage) | 179.055 | C6H12O6 | −6.1 | -H, +HCOO |
| 38 | Astragaloside I-C36H58O9(cleavage) + 2×(+H2) | 283.104 | C9H18O7 | 0.7 | +HCOO |
| 39 | Scopoletin-CH2(cleavage) + O+SO3 | 272.971 | C9H6O8S | 0.2 | -H |
| 40 | Corchoionoside C-C6H10O5(cleavage)-H2 + SO3 | 301.076 | C13H18O6S | 3.3 | -H |
| 41 | Kudzusaponin A1-C47H74O18(cleavage) + O+H2 | 229.053 | C5H12O7 | −15.5 | +HCOO |
| 42 | Polydatin-C6H10O5(cleavage) + O | 243.062 | C14H12O4 | −16.3 | -H |
| 43 | 6″-O-Acetyldaidzin-C15H8O3(cleavage) + H2 | 269.087 | C8H16O7 | −3.9 | +HCOO |
| 44 | Scopoletin-CH2(cleavage) + 2×(-H2) + C2H2O | 215.002 | C11H4O5 | 18.4 | -H |
| 45 | Scopoletin-O(cleavage)-H2 + C2H2O | 215.032 | C12H8O4 | −14.3 | -H |
| 46 | Scopoletin-O(cleavage)-H4 | 171.011 | C10H4O3 | 13.7 | -H |
| 47 | Quercetin 3-O-(6″-galloyl)-β-D-glucopyranoside-C21H18O11(cleavage) + SO3 | 248.973 | C7H6O8S | 8.8 | -H |
| 48 | Sanggenone F + O + C2H2O | 411.108 | C22H20O8 | −1.9 | -H |
| 49 | Mulberroside C-C19H16O4(cleavage) + 2×(+H2) + SO3 | 279.038 | C5H14O8S | −3.1 | +HCOO |
| 50 | Tectorigenin-CH2O(cleavage) | 269.049 | C15H10O5 | 13.2 | -H |
| 51 | Astragaloside IV-C36H58O9(cleavage) | 149.045 | C5H10O5 | −4.6 | -H |
| 52 | Moracin C + H2 + C2H2O | 399.146 | C21H22O5 | 2.1 | +HCOO |
| 53 | 3′-Methoxydaidzein-CH2O(cleavage) + SO3 | 333.008 | C15H10O7S | 1.1 | -H |
| 54 | Tiliroside-C15H8O6(cleavage)-H2 + C2H2O | 395.096 | C17H18O8 | −4.9 | +HCOO |
| 55 | 3′-Methoxydaidzein-CH2O(cleavage) | 253.05 | C15H10O4 | −2.1 | -H |
| 56 | Scopoletin-CH2O(cleavage)-H2 + C2H2O | 201.022 | C11H6O4 | 14.9 | -H |
| 57 | Scopoletin-CH2(cleavage)-H4 | 172.991 | C9H2O4 | 16.7 | -H |
| Compounds | Linear Range (μg) | Regression Equation | r2 | Content (%) |
|---|---|---|---|---|
| Puerarin | 0.33–2.00 | y = 6,259,581x + 7,691,406 | 0.9992 | 42.75 |
| Scopoletin | 0.26–1.58 | y = 50,561,849x – 1,061,227 | 1.0000 | 12.66 |
| Tiliroside | 0.10–0.60 | y = 19,048,338x – 322,554 | 0.9998 | 2.16 |
| Formononetin | 0.11–0.63 | y = 29,669,618x – 394,826 | 0.9997 | 0.25 |
| Kaempferol | 0.14–0.83 | y = 15,037,786x – 719,796 | 0.9995 | 0.25 |
| Moracin A | 0.12–0.73 | y = 18,485,874x – 308,581 | 1.0000 | 0.34 |
| Astragaloside IV | 1.25–7.51 | y = 221,315x – 38,913 | 0.9997 | 0.24 |
| Astragaloside III | 1.28–7.68 | y = 212,790x – 34,537 | 0.9997 | 0.28 |
| No. | tR (min) | Identification | m/z | Formula | MS/MS | |
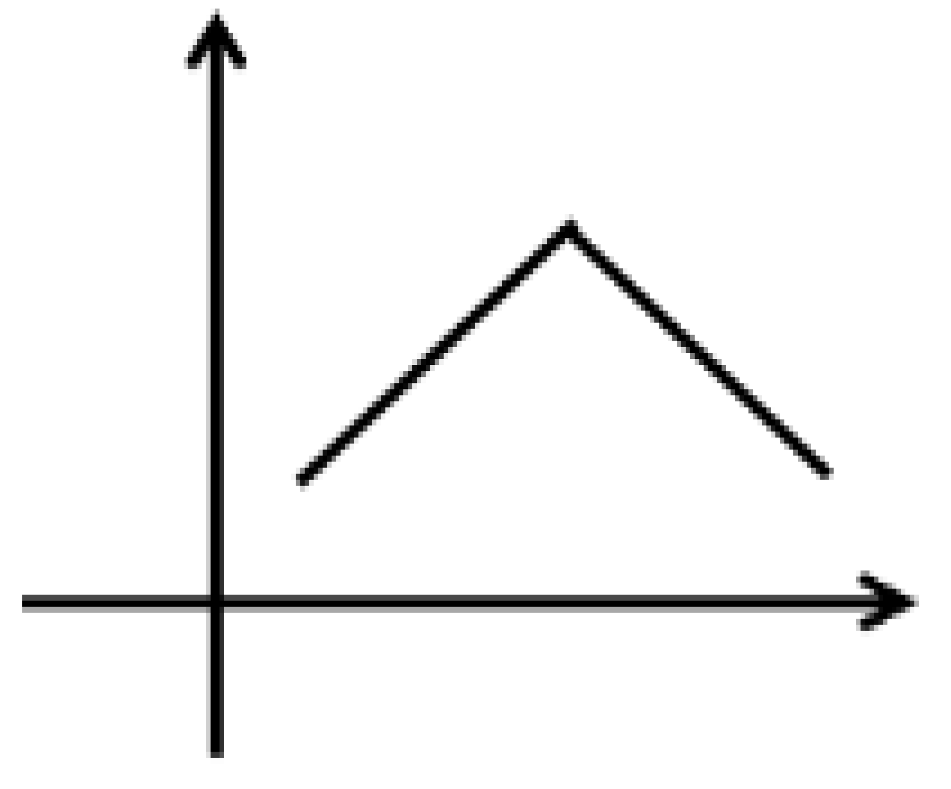 | 1 | 35.18 | Cucurbitacin C | 541.3165 | C32H48O8 | 236.1204, 99.9510, 139.0171, 153.0488 |
| 2 | 35.21 | (3b, 16a, 20R)-3, 16, 20, 22, 25-Pentahydroxy-5-cucurbiten-11-one 3-[rhamnosyl-(1→4)-[glucosyl-(1→6)]-glucoside] | 1035.5399 | C48H80O20 | 293.2168, 813.4589, 975.5179 | |
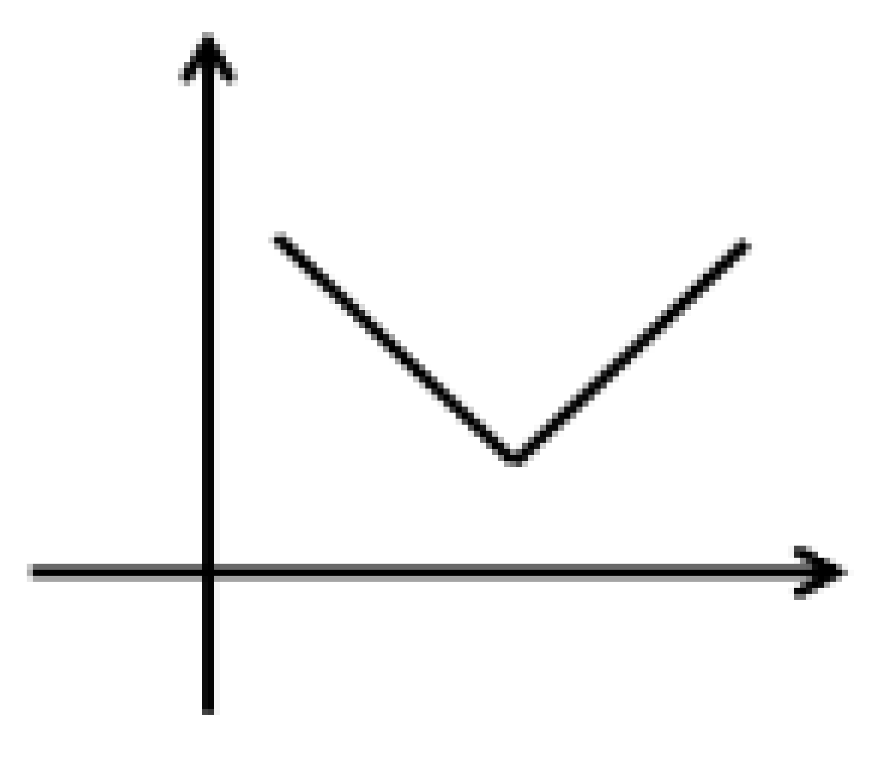 | 3 | 36.36 | Palmitic acid | 301.2389 | C16H32O2 | 171.0523, 161.1419 |
| 4 | 38.91 | 5-Aminopentanal | 302.2422 | C5H11NO | 116.9497, 135.9911 | |
| 5 | 39.94 | Octadecanol | 305.2621 | C18H38O | 205.2485, 177.1300, 179.0922 | |
| 6 | 36.38 | DG (20:5(5Z, 8Z, 11Z, 14Z, 17Z)/18:1(9Z)/0:0) | 319.2491 | C41H68O5 | 258.2428, 291.2491, 251.2767, 257.2859, 265.1507 | |
| 7 | 34.34 | LysoPE (0:0/18:2(9Z, 12Z)) | 476.2755 | C23H44NO7P | 214.0616, 62.0303, 78.9856, 122.0292 | |
| 8 | 32.81 | LysoPC (14:0/0:0) | 502.2679 | C22H46NO7P | 220.9695, 224.1195 | |
| 9 | 32.81 | LysoPE (0:0/16:0) | 512.2963 | C21H44NO7P | 162.9853, 213.0589, 216.8936 | |
| 10 | 34.05 | (17alpha, 23S)-17, 23-Epoxy-29-hydroxy-27-norlanosta-1, 8-diene-3, 15, 24-trione | 527.3059 | C29H40O5 | 243.1236, 221.1888 | |
| 11 | 1.32 | Monoethyl glycinexylidide | 243.0917 | C12H18N2O | 148.0930, 146.9884, 182.0120, 151.0939, 152.0748, 153.0817 | |
| 12 | 33.39 | Doramapimod | 586.3011 | C33H43N5O6 | 217.8901, 218.8950, 219.8961 | |
| 13 | 33.40 | Lithocholate 3-O-glucuronide | 587.3033 | C30H48O9 | 218.9011, 222.8947 | |
| 14 | 40.16 | DG (15:0/22:5(4Z, 7Z, 10Z, 13Z, 16Z)/0:0) | 609.4869 | C40H68O5 | 257.2761, 256.2816, 289.0931, 251.2410, 257.2761, 265.0006 | |
| 15 | 33.41 | Dihydrozeatin-9-N-glucoside-O-glucoside | 658.2158 | C22H35N5O11 | 202.9312, 236.1456 | |
| 16 | 36.39 | PS (18:1(9Z)/20:4(5Z, 8Z, 11Z, 14Z)) | 828.5139 | C44H76NO10P | 291.2666, 321.2870, 280.2745 | |
| 17 | 1.43 | Glutamyl methionine | 277.0879 | C10H18N2O5S | 160.8872, 161.0873, 162.0453 | |
| 18 | 34.42 | AzIV | 1083.5434 | C54H84O22 | 269.2888, 97.0081, 249.0190, 323.2561 |
| No. | Pathway Name | Expected | Raw p | −log(p) | Impact |
|---|---|---|---|---|---|
| 1 | Pentose and glucuronate interconversions | 0.047714 | 0.046913 | 1.3287 | 0.14062 |
| 2 | Drug metabolism—cytochrome P450 | 0.071571 | 0.06974 | 1.1565 | 0 |
| 3 | Biosynthesis of unsaturated fatty acids | 0.095427 | 0.092155 | 1.0355 | 0 |
| 4 | Glycerophospholipid metabolism | 0.095427 | 0.092155 | 1.0355 | 0.01736 |
| 5 | Fatty acid elongation | 0.10338 | 0.099536 | 1.002 | 0 |
| 6 | Fatty acid degradation | 0.10338 | 0.099536 | 1.002 | 0 |
| 7 | Fatty acid biosynthesis | 0.12459 | 0.119 | 0.92446 | 0.01472 |
| Ligand | Receptor | Docking Score (kJ/mol) | Docking Sites |
|---|---|---|---|
| Puerarin | UGT2B15 | −24.23 | Gly-365, His-387, Tyr-386, Ser-447, His-450 |
| Scopoletin | −24.69 | His-375, Thr-374, Gly-311, Ser-309, Asn-379, Gln-360, Leu-310 | |
| Tiliroside | −24.64 | His-281, Lys-284, Tyr-382, Asn-403, Asp-402 | |
| Formononetin | −25.06 | Gly-365, Leu-289, Lys-291 | |
| Kaempferol | −22.34 | Ala-286, Asp-362, Leu-289, Glu-294, Lys-291 | |
| Moracin A | −25.73 | Ile-314, Lys-343, Lys-344, Asn-323, Asn-346 | |
| Astragaloside IV | −19.83 | Ser-315, Met-31, Ser-415, Asp-417, Arg-419 | |
| Astragaloside III | −21.71 | Ser-432, Glu-319, Met-317, Ile-314, Ser-315 | |
| Puerarin | UGT2B10 | −28.62 | His-385, Thr-367, Met-279, Lys-282, Lys-285 |
| Scopoletin | −24.98 | Gln-358, Glu-381, Asn-359 | |
| Tiliroside | −30.33 | Lys-443, Arg-446, Tyr-384, Lys-438, Met-279, Leu-362, Ala-278, His-364, Lys-285 | |
| Formononetin | −26.86 | Pro-286, Asp-360, Gly-363, Asn-359 | |
| Kaempferol | −26.11 | Lys-285, Lys-282, Tyr-384, Thr-367, Lys-438 | |
| Moracin A | −30.12 | Asn-359, Asp-360, Gln-358 | |
| Astragaloside IV | −19.79 | Asp-344, Leu-352, Lys-342, Asn-341, Gly-340, Asp-339 | |
| Astragaloside III | −25.48 | Trp-355, Lys-354, Glu-290 | |
| Puerarin | UGT1A1 | −19.50 | His-372, Leu-355, Gln-357, His-376,Asn-279, Asn-358 |
| Scopoletin | −19.75 | Asp-36, Leu-123, Ser-65 | |
| Tiliroside | −19.66 | Ser-120, Gly-395 | |
| Formononetin | −24.14 | Pro-270, Arg-450, Phe-274 | |
| Kaempferol | −18.95 | Cys-177, Cys-186, Tyr-486 | |
| Moracin A | −19.41 | Asp-36, Pro-34, Tyr-79 | |
| Astragaloside IV | −15.27 | Asp-259 | |
| Astragaloside III | −15.06 | Ile-268, His-487, Tyr-486 | |
| Puerarin | UGT1A9 | −15.48 | Phe-271 |
| Scopoletin | −20.42 | Leu-173 | |
| Tiliroside | −10.04 | Gly-273, Phe-257 | |
| Formononetin | −27.99 | Gly-392, Pro-149, Tyr-245 | |
| Kaempferol | −23.39 | Gly-171, Phe-150, Leu-173 | |
| Moracin A | −20.75 | Ser-309, Glu-310 | |
| Astragaloside IV | −13.01 | His-213, Ala-185, Gln-182, Ala-475,Leu-478 | |
| Astragaloside III | −15.10 | Arg-439, Arg-447, His-444, Phe-271 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wen, Q.; Yang, B.; Feng, L.; Jia, X. Active Constituent of HQS in T2DM Intervention: Efficacy and Mechanistic Insights. Int. J. Mol. Sci. 2025, 26, 4578. https://doi.org/10.3390/ijms26104578
Chen Y, Wen Q, Yang B, Feng L, Jia X. Active Constituent of HQS in T2DM Intervention: Efficacy and Mechanistic Insights. International Journal of Molecular Sciences. 2025; 26(10):4578. https://doi.org/10.3390/ijms26104578
Chicago/Turabian StyleChen, Yaping, Qiuqi Wen, Bing Yang, Liang Feng, and Xiaobin Jia. 2025. "Active Constituent of HQS in T2DM Intervention: Efficacy and Mechanistic Insights" International Journal of Molecular Sciences 26, no. 10: 4578. https://doi.org/10.3390/ijms26104578
APA StyleChen, Y., Wen, Q., Yang, B., Feng, L., & Jia, X. (2025). Active Constituent of HQS in T2DM Intervention: Efficacy and Mechanistic Insights. International Journal of Molecular Sciences, 26(10), 4578. https://doi.org/10.3390/ijms26104578




