Exosomal MicroRNAs as Epigenetic Biomarkers for Endometriosis: A Systematic Review and Bioinformatics Analysis
Abstract
1. Introduction
2. Results
2.1. Study Selection
2.2. Study Characteristics
2.3. Quality Assessment
2.4. miRNA Dysregulation in Endometriosis
2.4.1. Serum
2.4.2. Endometrial Tissues
2.4.3. Tubal Fluid
2.4.4. Uterine Aspirate Fluid
2.4.5. Leukorrhea
2.5. Bioinformatic Analysis
3. Discussion
4. Materials and Methods
4.1. Protocol and Registration
4.2. Eligibility Criteria
- Participants: Women clinically diagnosed with endometriosis;
- Exposure: Exosomal miRNAs;
- Control: Healthy women;
- Outcome: Expression and regulation patterns of exosomal miRNAs in endometriosis patients, along with downstream pathways;
- Study type: Case–control studies.
4.3. Search Strategy
4.4. Selecting Studies
4.5. Data Extraction Process
4.6. Quality Evaluation
4.7. Bioinformatic Assessment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Agarwal, S.K.; Chapron, C.; Giudice, L.C.; Laufer, M.R.; Leyland, N.; Missmer, S.A.; Singh, S.S.; Taylor, H.S. Clinical diagnosis of endometriosis: A call to action. Am. J. Obstet. Gynecol. 2019, 220, 354.e1–354.e12. [Google Scholar] [CrossRef] [PubMed]
- Zondervan, K.T.; Becker, C.M.; Missmer, S.A. Endometriosis. N. Engl. J. Med. 2020, 382, 1244–1256. [Google Scholar] [CrossRef]
- Surrey, E.; Carter, C.M.; Soliman, A.M.; Khan, S.; DiBenedetti, D.B.; Snabes, M.C. Patient-completed or symptom-based screening tools for endometriosis: A scoping review. Arch. Gynecol. Obstet. 2017, 296, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Hudson, Q.J.; Perricos, A.; Wenzl, R.; Yotova, I. Challenges in uncovering non-invasive biomarkers of endometriosis. Exp. Biol. Med. 2020, 245, 437–447. [Google Scholar] [CrossRef]
- Pandey, S. Metabolomics for the identification of biomarkers in endometriosis. Arch. Gynecol. Obstet. 2024; Online ahead of print. [Google Scholar] [CrossRef]
- Agrawal, S.; Tapmeier, T.; Rahmioglu, N.; Kirtley, S.; Zondervan, K.; Becker, C. The miRNA Mirage: How Close Are We to Finding a Non-Invasive Diagnostic Biomarker in Endometriosis? A Systematic Review. Int. J. Mol. Sci. 2018, 19, 599. [Google Scholar] [CrossRef]
- Guerriero, S.; Ajossa, S.; Orozco, R.; Perniciano, M.; Jurado, M.; Melis, G.B.; Alcazar, J.L. Accuracy of transvaginal ultrasound for diagnosis of deep endometriosis in the rectosigmoid: Systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2016, 47, 281–289. [Google Scholar] [CrossRef]
- Kennedy, S.; Bergqvist, A.; Chapron, C.; D’Hooghe, T.; Dunselman, G.; Greb, R.; Hummelshoj, L.; Prentice, A.; Saridogan, E.; ESHRE Special Interest Group for Endometriosis and Endometrium Guideline Development Group. ESHRE guideline for the diagnosis and treatment of endometriosis. Hum. Reprod. 2005, 20, 2698–2704. [Google Scholar] [CrossRef]
- May, K.E.; Conduit-Hulbert, S.A.; Villar, J.; Kirtley, S.; Kennedy, S.H.; Becker, C.M. Peripheral biomarkers of endometriosis: A systematic review. Hum. Reprod. Update 2010, 16, 651–674. [Google Scholar] [CrossRef]
- Mehedintu, C.; Plotogea, M.N.; Ionescu, S.; Antonovici, M. Endometriosis still a challenge. J. Med. Life 2014, 7, 349–357. [Google Scholar]
- Rahmioglu, N.; Nyholt, D.R.; Morris, A.P.; Missmer, S.A.; Montgomery, G.W.; Zondervan, K.T. Genetic variants underlying risk of endometriosis: Insights from meta-analysis of eight genome-wide association and replication datasets. Hum. Reprod. Update 2014, 20, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Nyholt, D.R.; Low, S.K.; Anderson, C.A.; Painter, J.N.; Uno, S.; Morris, A.P.; MacGregor, S.; Gordon, S.D.; Henders, A.K.; Martin, N.G.; et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat. Genet. 2012, 44, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Miao, M.; Miao, Y.; Zhu, Y.; Wang, J.; Zhou, H. Advances in Exosomes as Diagnostic and Therapeutic Biomarkers for Gynaecological Malignancies. Cancers 2022, 14, 4743. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Robbins, P.D. The roles of tumor-derived exosomes in cancer pathogenesis. Clin. Dev. Immunol. 2011, 2011, 842849. [Google Scholar] [CrossRef]
- Javadi, M.; Rad, J.S.; Farashah, M.S.G.; Roshangar, L. An Insight on the Role of Altered Function and Expression of Exosomes and MicroRNAs in Female Reproductive Diseases. Reprod. Sci. 2022, 29, 1395–1407. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef]
- Moreno-Moya, J.M.; Vilella, F.; Simón, C. MicroRNA: Key gene expression regulators. Fertil. Steril. 2014, 101, 1516–1523. [Google Scholar] [CrossRef]
- McGinnis, L.K.; Luense, L.J.; Christenson, L.K. MicroRNA in Ovarian Biology and Disease. Cold Spring Harb. Perspect. Med. 2015, 5, a022962. [Google Scholar] [CrossRef]
- Di Pietro, C. Exosome-mediated communication in the ovarian follicle. J. Assist. Reprod. Genet. 2016, 33, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Vanhie, A.; Caron, E.; Vermeersch, E.; O, D.; Tomassetti, C.; Meuleman, C.; Mestdagh, P.; D’Hooghe, T.M. Circulating microRNAs as Non-Invasive Biomarkers in Endometriosis Diagnosis-A Systematic Review. Biomedicines 2024, 12, 888. [Google Scholar] [CrossRef] [PubMed]
- Harp, D.; Driss, A.; Mehrabi, S.; Chowdhury, I.; Xu, W.; Liu, D.; Garcia-Barrio, M.; Taylor, R.N.; Gold, B.; Jefferson, S.; et al. Exosomes derived from endometriotic stromal cells have enhanced angiogenic effects in vitro. Cell Tissue Res. 2016, 365, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, D.; Zhou, Y.; Peng, C. Identification of a Serum Exosome-Derived lncRNA–miRNA–mRNA ceRNA Network in Patients with Endometriosis. Clin. Exp. Obstet. Gynecol. 2024, 51, 51. [Google Scholar] [CrossRef]
- Jiang, Y.; Chai, X.; Chen, S.; Chen, Z.; Tian, H.; Liu, M.; Wu, X. Exosomes from the Uterine Cavity Mediate Immune Dysregulation via Inhibiting the JNK Signal Pathway in Endometriosis. Biomedicines 2022, 10, 3110. [Google Scholar] [CrossRef]
- Wu, D.; Lu, P.; Mi, X.; Miao, J. Exosomal miR-214 from endometrial stromal cells inhibits endometriosis fibrosis. Mol. Hum. Reprod. 2018, 24, 357–365. [Google Scholar] [CrossRef]
- Wu, Y.; Yuan, W.; Ding, H.; Wu, X. Serum exosomal miRNA from endometriosis patients correlates with disease severity. Arch. Gynecol. Obstet. 2022, 305, 117–127. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.H.; Yuan, M.; Li, D.; Wang, G.Y. Exosomal miR-22-3p derived from peritoneal macrophages enhances proliferation, migration, and invasion of ectopic endometrial stromal cells through regulation of the SIRT1/NF-κB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 571–580. [Google Scholar]
- Zhang, L.; Li, H.; Yuan, M.; Li, D.; Sun, C.; Wang, G. Serum Exosomal MicroRNAs as Potential Circulating Biomarkers for Endometriosis. Dis. Markers 2020, 2020, 2456340. [Google Scholar] [CrossRef]
- Zhang, Y.; Chang, X.; Wu, D.; Deng, M.; Miao, J.; Jin, Z. Down-regulation of Exosomal miR-214-3p Targeting CCN2 Contributes to Endometriosis Fibrosis and the Role of Exosomes in the Horizontal Transfer of miR-214-3p. Reprod. Sci. 2020, 28, 715–727. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, X.; Xia, X.; Fang, X.; Zhang, T.; Huang, F. Endometrial epithelial cells-derived exosomes deliver microRNA-30c to block the BCL9/Wnt/CD44 signaling and inhibit cell invasion and migration in ovarian endometriosis. Cell Death Discov. 2022, 8, 151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; Yan, L.; Liang, G.; Zhu, C.; Wang, Y.; Ji, S.; He, C.; Sun, J.; Zhang, J. Exosomal microRNAs in tubal fluid may be involved in damage to tubal reproductive function associated with tubal endometriosis. Reprod. Biomed. Online 2023, 47, 103249. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Sun, D.F.; Tong, Y. Exosomal miR-202 derived from leukorrhea as a potential biomarker for endometriosis. J. Int. Med. Res. 2023, 51, 3000605221147183. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lian, Y.; Jiang, J.; Wang, L.; Ren, L.; Li, Y.; Yan, X.; Chen, Q. Differential expression of microRNA in exosomes derived from endometrial stromal cells of women with endometriosis-associated infertility. Reprod. Biomed. Online 2020, 41, 170–181. [Google Scholar] [CrossRef]
- Zhang, B.L.; Dong, F.L.; Guo, T.W.; Gu, X.H.; Huang, L.Y.; Gao, D.S. MiRNAs Mediate GDNF-Induced Proliferation and Migration of Glioma Cells. Cell. Physiol. Biochem. 2017, 44, 1923–1938. [Google Scholar] [CrossRef]
- Gentilini, D.; Busacca, M.; Di Francesco, S.; Vignali, M.; Viganò, P.; Di Blasio, A.M. PI3K/Akt and ERK1/2 signalling pathways are involved in endometrial cell migration induced by 17beta-estradiol and growth factors. Mol. Hum. Reprod. 2007, 13, 317–322. [Google Scholar] [CrossRef]
- Cakmak, H.; Guzeloglu-Kayisli, O.; Kayisli, U.A.; Arici, A. Immune-endocrine interactions in endometriosis. Front. Biosci. (Elite Ed.) 2009, 1, 429–443. [Google Scholar]
- Govatati, S.; Kodati, V.L.; Deenadayal, M.; Chakravarty, B.; Shivaji, S.; Bhanoori, M. Mutations in the PTEN tumor gene and risk of endometriosis: A case–control study. Hum. Reprod. 2014, 29, 324–336. [Google Scholar] [CrossRef]
- Stambolic, V.; Suzuki, A.; de la Pompa, J.L.; Brothers, G.M.; Mirtsos, C.; Sasaki, T.; Ruland, J.; Penninger, J.M.; Siderovski, D.P.; Mak, T.W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998, 95, 29–39. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, L.; Li, H.; Ye, J.; Lin, N.; Chen, M.; Pan, D.; Chen, Z. Endometriosis derived exosomal miR-301a-3p mediates macrophage polarization via regulating PTEN-PI3K axis. Biomed. Pharmacother. 2022, 147, 112680. [Google Scholar] [CrossRef]
- Ji, J.; Wang, H.; Yuan, M.; Li, J.; Song, X.; Lin, K. Exosomes from ectopic endometrial stromal cells promote M2 macrophage polarization by delivering miR-146a-5p. Int. Immunopharmacol. 2024, 128, 111573. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; Zhang, J.; Qi, Y.H.; Kong, M.; Liu, S.A.; Hu, J.J. Linc-ROR promotes endometrial cell proliferation by activating the PI3K-Akt pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2218–2225. [Google Scholar] [PubMed]
- Pan, Q.; Luo, X.; Toloubeydokhti, T.; Chegini, N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol. Hum. Reprod. 2007, 13, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.Z.; Yang, Y.; Lang, J.; Sun, P.; Leng, J. Plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis. Hum. Reprod. 2013, 28, 322–330. [Google Scholar] [CrossRef]
- Zhao, W.; Sun, Q.; Yu, Z.; Mao, S.; Jin, Y.; Li, J.; Jiang, Z.; Zhang, Y.; Chen, M.; Chen, P.; et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene 2018, 670, 31–37. [Google Scholar] [CrossRef]
- Hsieh, I.S.; Chang, K.C.; Tsai, Y.T.; Ke, J.Y.; Lu, P.J.; Lee, K.H.; Yeh, S.D.; Hong, T.M.; Chen, Y.L. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis 2013, 34, 530–538. [Google Scholar] [CrossRef]
- Gao, T.; Cao, Y.; Hu, M.; Du, Y. The activation of TGF-β signaling promotes cell migration and invasion of ectopic endometrium by targeting NRP2. Reprod. Biol. 2022, 22, 100697. [Google Scholar] [CrossRef]
- Liang, Y.; Li, S.; Tang, L. MicroRNA 320, an Anti-Oncogene Target miRNA for Cancer Therapy. Biomedicines 2021, 9, 591. [Google Scholar] [CrossRef]
- Angius, A.; Pira, G.; Scanu, A.M.; Uva, P.; Sotgiu, G.; Saderi, L.; Manca, A.; Serra, C.; Uleri, E.; Piu, C.; et al. MicroRNA-425-5p Expression Affects BRAF/RAS/MAPK Pathways In Colorectal Cancers. Int. J. Med. Sci. 2019, 16, 1480–1491. [Google Scholar] [CrossRef]
- Dong, L.; Zhang, L.; Liu, H.; Xie, M.; Gao, J.; Zhou, X.; Zhao, Q.; Zhang, S.; Yang, J. Circ_0007331 knock-down suppresses the progression of endometriosis via miR-200c-3p/HiF-1α axis. J. Cell. Mol. Med. 2020, 24, 12656–12666. [Google Scholar] [CrossRef]
- Zhang, T.T.; Wang, Y.; Zhang, X.W.; Yang, K.Y.; Miao, X.Q.; Zhao, G.H. MiR-200c-3p Regulates DUSP1/MAPK Pathway in the Nonalcoholic Fatty Liver After Laparoscopic Sleeve Gastrectomy. Front. Endocrinol. 2022, 13, 792439. [Google Scholar] [CrossRef] [PubMed]
- Berlanga, P.; Munoz, L.; Piqueras, M.; Sirerol, J.A.; Sánchez-Izquierdo, M.D.; Hervás, D.; Hernández, M.; Llavador, M.; Machado, I.; Llombart-Bosch, A.; et al. miR-200c and phospho-AKT as prognostic factors and mediators of osteosarcoma progression and lung metastasis. Mol. Oncol. 2016, 10, 1043–1053. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Feng, Z.; Zhang, X.; Lan, D.; Wu, Y. Up-regulation of microRNA-200c-3p inhibits invasion and migration of renal cell carcinoma cells via the SOX2-dependent Wnt/β-catenin signaling pathway. Cancer Cell Int. 2019, 19, 231. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Lin, X.; Xu, W.; Lin, X.; Huang, Q.; Shi, L.; Pan, Y.; Zhang, Y.; Zhu, Y.; Li, C.; et al. MiR-210-3p protects endometriotic cells from oxidative stress-induced cell cycle arrest by targeting BARD1. Cell Death Dis. 2019, 10, 144. [Google Scholar] [CrossRef]
- Cumpston, M.; Li, T.; Page, M.J.; Chandler, J.; Welch, V.A.; Higgins, J.P.; Thomas, J. Updated guidance for trusted systematic reviews: A new edition of the Cochrane Handbook for Systematic Reviews of Interventions. Cochrane Database Syst. Rev. 2019, 10, ED000142. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Stang, A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010, 25, 603. [Google Scholar] [CrossRef]
- Hauschild, A.C.; Pastrello, C.; Ekaputeri, G.K.A.; Bethune-Waddell, D.; Abovsky, M.; Ahmed, Z.; Kotlyar, M.; Lu, R.; Jurisica, I. MirDIP 5.2: Tissue context annotation and novel microRNA curation. Nucleic Acids Res. 2023, 51, D217–D225. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]

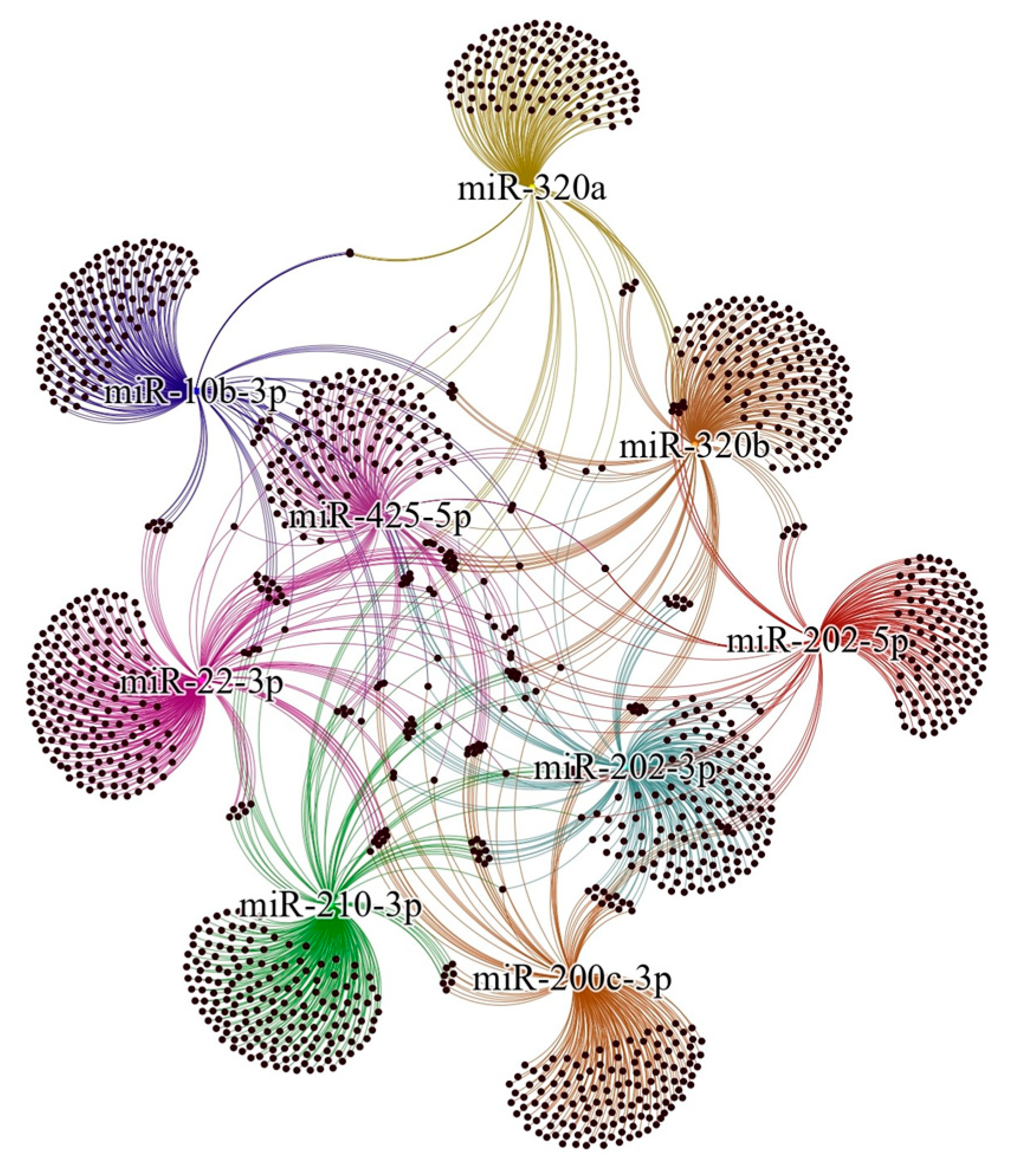
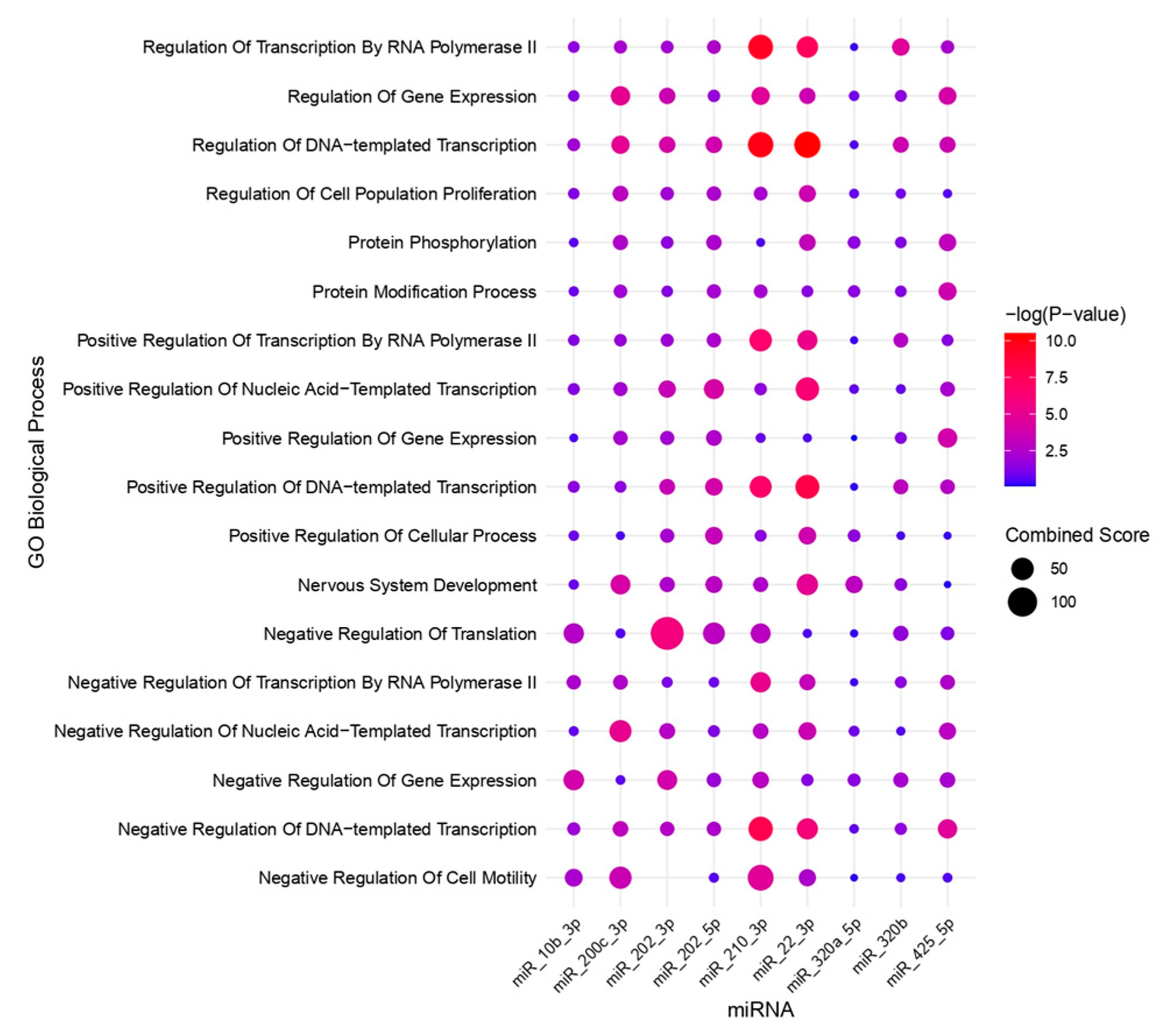
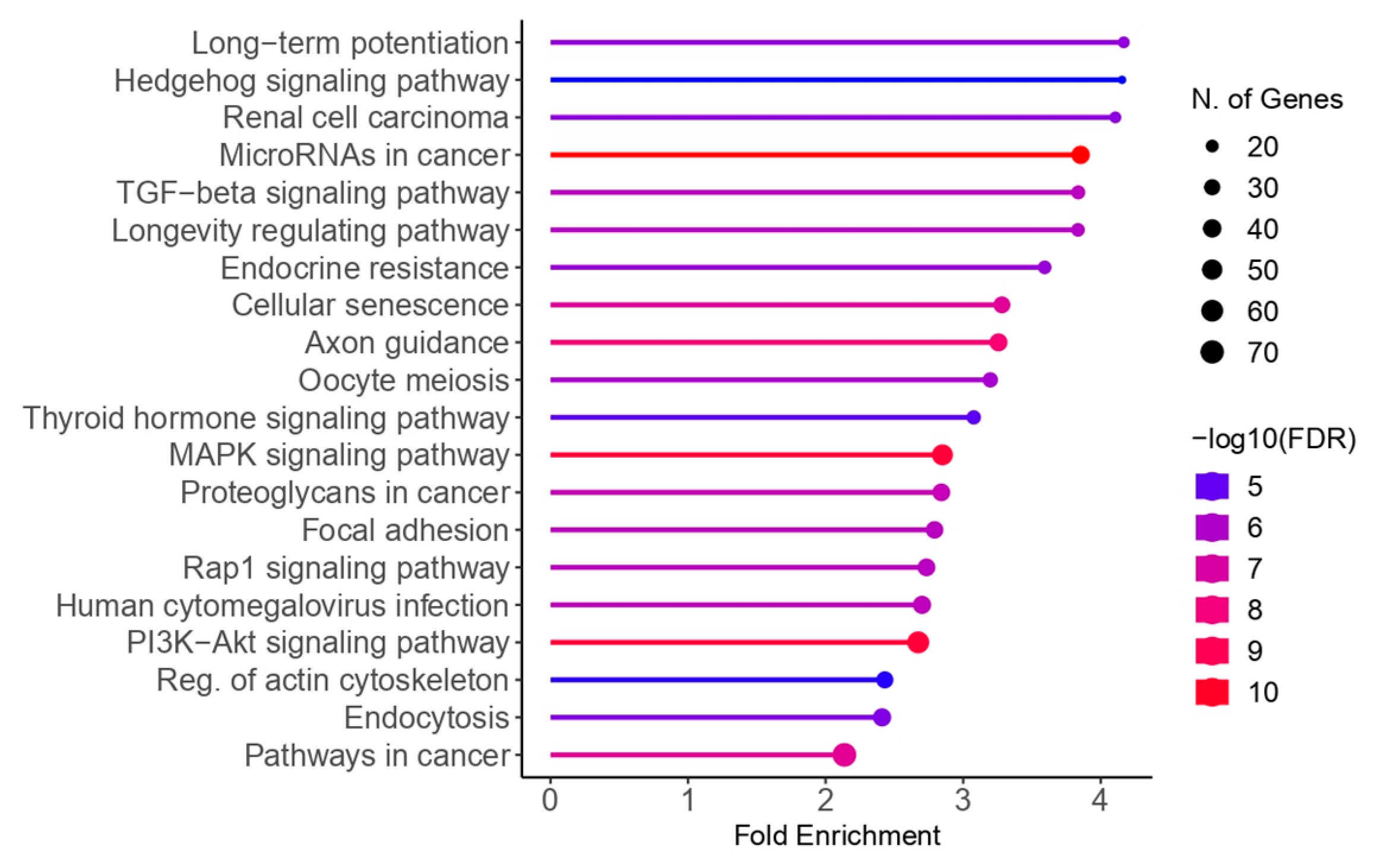
| Author, Year | Country | Sample Size | Sample Type | Staging | Diagnostic Method | Phase of Menstrual Cycle | Method of miRNA Detection | Method of Exosome Detection | Method of Exosome Isolation | |
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||||
| Harp et al., 2016 [23] | USA | 5 | 5 | Eutopic endometrial tissue | - | Laparoscopy | Secretory | RT-qPCR | 1. Nanoparticle tracking 2. Transmission electron microscopy | - |
| Huang et al., 2024 [24] | China | 10 | 10 | Serum | III–IV | Laparoscopy | Proliferative | microarray | 1. Nanoparticle tracking 2. Transmission electron microscopy 3. Western blotting | Ultracentrifugation |
| Jiang et al., 2022 [25] | China | 25 | 25 | 1. Eutopic endometrial tissue 2. Uterine aspirate fluid | III–IV | Laparoscopy | - | RT-qPCR | 1. Nanoparticle tracking 2. Transmission electron microscopy 3. Western blotting | Exosome Isolation Kit (Echobiotech, Beijing, China) |
| Wu et al., 2018 [26] | China | 6 | 6 | Ectopic endometrial tissue | III–IV | Laparoscopy | - | qRT-PCR | 1. Transmission electron microscopy 2. Western blotting | Ultracentrifugation |
| Wu et al., 2022 [27] | China | 42 | 24 | Serum | I–IV | Laparoscopy | Proliferative secretory | 1. Microarray 2. RT-PCR | 1. Transmission electron microscopy 2. Western blotting | Ultracentrifugation |
| Zhang et al., 2020 [28] | China | 20 | 20 | Ectopic endometrial tissue | - | Laparoscopy + Histopathologic examination | - | 1. Microarray 2. qRT-PCR | 1. Nanoparticle tracking 2. Transmission electron microscopy 3. Western blotting | Differential centrifugation |
| Zhang et al., 2020 [29] | China | 20 | 20 | Serum | I–IV | Laparoscopy + Histopathologic examination | - | 1. Microarray 2. qRT-PCR | 1. Nanoparticle tracking 2. Transmission electron microscopy 3. Western blotting | Ultracentrifugation |
| Zhang et al., 2020 [30] | China | 20 | 20 | Ectopic endometrial tissue | III | Laparoscopy | Proliferative | qRT-PCR | 1. Transmission electron microscopy 2. Western blotting | SBI ExoQuick-TC |
| Zhang et al., 2022 [31] | China | 24 | 20 | 1. Ectopic endometrial tissue 2. Eutopic endometrial tissue | - | Laparoscopy | Proliferative | 1. Microarray 2. qRT-PCR | 1. Nanoparticle tracking 2. Transmission electron microscopy 3. Western blotting | Ultracentrifugation |
| Zhang et al., 2023 [32] | China | 5 | 5 | Tubal fluid | - | MRI + Laparoscopy + Histopathologic examination | Secretory | 1. Microarray 2. qRT-PCR | 1. Nanoparticle tracking 2. Transmission electron microscopy 3. Western blotting | Ultracentrifugation |
| Zheng et al., 2023 [33] | China | 11 | 11 | 1. Ectopic endometrial tissue 2. Leukorrhea | - | Laparoscopy + Histopathological examination | - | 1. Microarray 2. RT-qPCR | 1. Transmission electron microscopy 2. Western blotting | Ultracentrifugation |
| Zhou et al., 2020 [34] | China | 3 | 3 | Eutopic endometrial tissue | II–IV | Laparoscopy | Secretory | qRT-PCR | Transmission electron microscopy | ExoQuick-TC Exosome Precipitation Solution (System Biosciences) |
| Reference | Selection | Comparability | Exposure | Overall Quality Score | |||||
|---|---|---|---|---|---|---|---|---|---|
| An Adequate Definition of Cases | Representativeness of Cases | Selection of Controls | Definition of Controls | Comparability of Cases and Controls Based on Design or Analysis | Ascertainment of Exposure | Same Method for Ascertainment of Cases and Controls | Non-Response Rate | ||
| Harp et al., 2016 [23] | * | - | * | * | - | * | * | - | 5/9 |
| Huang et al., 2024 [24] | * | * | * | * | - | * | * | - | 6/9 |
| Jiang et al., 2022 [25] | * | * | * | * | - | * | * | - | 6/9 |
| Wu et al., 2018 [26] | * | * | - | * | ** | * | * | - | 7/9 |
| Wu et al., 2022 [27] | * | * | * | * | ** | * | * | - | 8/9 |
| Zhang et al., 2020 [28] | * | * | * | * | - | * | * | - | 6/9 |
| Zhang et al., 2020 [29] | * | * | - | * | - | * | * | - | 5/9 |
| Zhang et al., 2020 [30] | * | * | * | - | - | - | * | - | 4/9 |
| Zhang et al., 2022 [31] | * | * | * | * | ** | * | * | - | 8/9 |
| Zhang et al., 2023 [32] | * | * | - | - | - | * | * | - | 4/9 |
| Zheng et al., 2023 [33] | * | * | * | * | - | * | * | - | 6/9 |
| Zhou et al., 2020 [34] | * | * | * | - | - | * | * | - | 5/9 |
| Reference | Number of Differential microRNA | Differential miRNA Expression Criteria | Dysregulated Exosomal microRNA |
|---|---|---|---|
| Tubal fluid | |||
| Zhang et al., 2023 [32] | 14 | Fold change >2 p < 0.05 | Upregulated (↑): miR-6087; miR-4443; miR-5194; miR-6834-3p. Downregulated (↓): miR-6747-5p; miR-1273f; miR-5699-5p; miR-10b-3p; miR-3911; miR-4419a; miR-4441; miR-4655-3p; miR-6778-5p; miR-6845-5p. |
| Serum | |||
| Huang et al., 2024 [24] | 50 | Fold change >0.5 p < 0.05 | Upregulated (↑): miR-4689; miR-4651; miR-6086; miR-6836; miR-551b-5p; miR-3124-5p; miR-671-5p. Downregulated (↓): miR-4497; miR-6779-5p; miR-185-5p; let-7i-5p; miR-27b-3p; miR-22-3p; miR-19b-3p; miR-221-3p; miR-3135b; miR-18a-5p; miR-423-3p; miR-27a-3p; miR-4454; miR-151a-3p; miR-1273g-3p; miR-4429; miR-423-5p; miR-320d; miR-191-5p; miR-151a-5p; miR-23b-3p; miR-24-3p; miR-17-5p; miR-107; miR-103a-3p; miR-320c; miR-320a; miR-320b; miR-199a-3p; miR-199b-3p; miR-20a-5p; miR-26a-5p; miR-23a-3p; miR-139-5p; miR-93-5p; miR-361-5p; let-7g-5p; let-7f-5p; miR-584-5p; miR-223-3p; miR-151b; let-7e-5p; miR-25-3p. |
| Wu et al., 2022 [27] | 45 | Fold change >2 p < 0.05 | Upregulated (↑): miR-6795-3p; miR-6889-3p; miR-4731-3p; miR-6731-3p; miR-6760-3p; miR-6870-3p; miR-7114-3p; miR-424-5p; miR-6813-3p; miR-3940-3p; miR-1271-5p; miR-1303; miR-6785-3p; miR-3675-3p; miR-1273g-3p; miR-3180-5p; miR-4475; miR-146b-3p; miR-500a-3p; miR-877-5p; miR-885-3p; miR-6818-3p; miR-6751-5p; miR-539-5p; miR-32-3p; miR-4505. Downregulated (↓): miR-128-1-5p; miR-215-5p; miR-26b-5p; miR-4453; miR-510-3p; miR-3140-3p; miR-3929; miR-3678-3p; miR-4303; miR-6743-3p; miR-514a-3p; miR-4315; miR-3074-5p; miR-628-3p; miR-6836-5p; miR-659-5p; miR-323b-5p; miR-5091; miR-3910. |
| Zhang et al., 2020 [29] | 24 | Fold change >1 p < 0.05 | Upregulation (↑): miR-197-5p; miR-22-3p; miR-320a; miR-320b; miR-3692-5p; miR-4476; miR-4530; miR-4532; miR-4721; miR-4758-5p; miR-494-3p; miR-6126; miR-6734-5p; miR-6776-5p; miR-6780b-5p; miR-6785-5p; miR-6791-5p; miR-939-5p. Downregulated (↓): miR-134-5p; miR-3141; miR-4499; miR-6088; miR-6165; miR-6728-5p. |
| Endometrial tissue | |||
| Harp et al., 2016 [23] | - | p < 0.05 | Upregulation (↑): miR-21 |
| Jiang et al., 2022 [25] | 21 | Fold change ≥1.50 p < 0.05 | Upregulation (↑): miR-210-3p; miR-30d-3p; miR-141-5p; miR-200c-3p; miR-224-5p; miR-4521; miR-29b-3p; miR-30b-5p; miR-16-2-3p; miR-345-5p; miR-375-3p; miR-9-3p; miR-9-5p; miR-190b-5p; miR-34c-5p. Downregulation (↓): miR-708-5p; miR-143-5p; miR-132-3p. |
| Wu et al., 2018 [26] | 1 | - | Downregulation (↓): miR-214. |
| Zhang et al., 2020 [28] | 20 | Fold change >1.5 p < 0.05 | Upregulation (↑): miR-22-3p; miR-28-5p; miR-302a; miR-320b; miR-3118; miR-3168; miR-425-5p; miR-4256; miR-4447; miR-507; miR-596; miR-5582; miR-610; miR-663a; miR-6720; miR-133. Downregulation (↓): miR-296; miR-1912; miR-2113; miR-3188. |
| Zhang et al., 2020 [30] | - | p < 0.05 | Downregulation (↓): miR-214-3p. |
| Zhang et al., 2022 [31] | - | p < 0.05 | Downregulation (↓): miR-30c. |
| Zheng et al., 2023 [33] | 217 | p < 0.05 | Upregulation (↑): miR-202-3p; miR-202-5p. |
| Zhou et al., 2020 [34] | 49 | Fold change >1.5 p < 0.05 | Upregulation (↑): miR-10b-3p; miR-1468-5p; miR-125b-2-3p; miR-2682-5p; miR-494-5p; miR-200c-3p; miR-345-3p; miR-450a-2-3p; miR-3180; miR-615-3p; miR-196a-5p; miR-6873-3p; miR-4483; miR-3661; miR-379-3p; miR-411-3p; miR-142-3p; miR-4532; miR-6131; miR-3195; let-7c-3p; miR-1343-3p; miR-1299; miR-99a-5p; let-7c-5p; miR-10b-5p. Downregulation (↓): miR-425-5p; miR-4671-3p; miR-4664-3p; miR-4473; miR-6500-3p; miR-3653-3p; miR-4421; miR-378h; miR-29c-5p; miR-4262; miR-1269a; miR-124-3p; miR-1273h-3p; miR-1273h-5p; miR-548am-3p; miR-548f-3p; miR-6859-5p; miR-548j-3p; miR-4467; miR-4648; miR-365a-3p; miR-365b-3p; miR-486-3p. |
| Uterine aspirate fluid | |||
| Jiang et al., 2022 [25] | 9 | Fold change ≥1.50 p < 0.05 | Upregulation (↑): miR-210-3p; miR-20b-5p; miR-625-5p; miR-342-5p; miR-155-5p; miR-146a-5p; miR-130b-3p. Downregulation (↓): miR-335-3p; miR-132-5p. |
| Leukorrhea | |||
| Zheng et al., 2023 [33] | 217 | p < 0.05 | Upregulation (↑): miR-202-3p; miR-202-5p. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, C.M.d.A.M.; Souza, A.T.B.d.; Neta, A.P.R.; Freire, L.V.P.; Sarmento, A.C.A.; Medeiros, K.S.d.; Luchessi, A.D.; Cobucci, R.N.; Gonçalves, A.K.; Crispim, J.C.d.O. Exosomal MicroRNAs as Epigenetic Biomarkers for Endometriosis: A Systematic Review and Bioinformatics Analysis. Int. J. Mol. Sci. 2025, 26, 4564. https://doi.org/10.3390/ijms26104564
Santos CMdAM, Souza ATBd, Neta APR, Freire LVP, Sarmento ACA, Medeiros KSd, Luchessi AD, Cobucci RN, Gonçalves AK, Crispim JCdO. Exosomal MicroRNAs as Epigenetic Biomarkers for Endometriosis: A Systematic Review and Bioinformatics Analysis. International Journal of Molecular Sciences. 2025; 26(10):4564. https://doi.org/10.3390/ijms26104564
Chicago/Turabian StyleSantos, Cristina Maria de Araújo Medeiros, Amaxsell Thiago Barros de Souza, Antonia Pereira Rosa Neta, Liziane Virginia Pereira Freire, Ayane Cristine Alves Sarmento, Kleyton Santos de Medeiros, André Ducati Luchessi, Ricardo Ney Cobucci, Ana Katherine Gonçalves, and Janaina Cristiana de Oliveira Crispim. 2025. "Exosomal MicroRNAs as Epigenetic Biomarkers for Endometriosis: A Systematic Review and Bioinformatics Analysis" International Journal of Molecular Sciences 26, no. 10: 4564. https://doi.org/10.3390/ijms26104564
APA StyleSantos, C. M. d. A. M., Souza, A. T. B. d., Neta, A. P. R., Freire, L. V. P., Sarmento, A. C. A., Medeiros, K. S. d., Luchessi, A. D., Cobucci, R. N., Gonçalves, A. K., & Crispim, J. C. d. O. (2025). Exosomal MicroRNAs as Epigenetic Biomarkers for Endometriosis: A Systematic Review and Bioinformatics Analysis. International Journal of Molecular Sciences, 26(10), 4564. https://doi.org/10.3390/ijms26104564






