Profile of Rat Adrenal microRNAs Induced by Gonadectomy and Testosterone or Estradiol Replacement
Abstract
1. Introduction
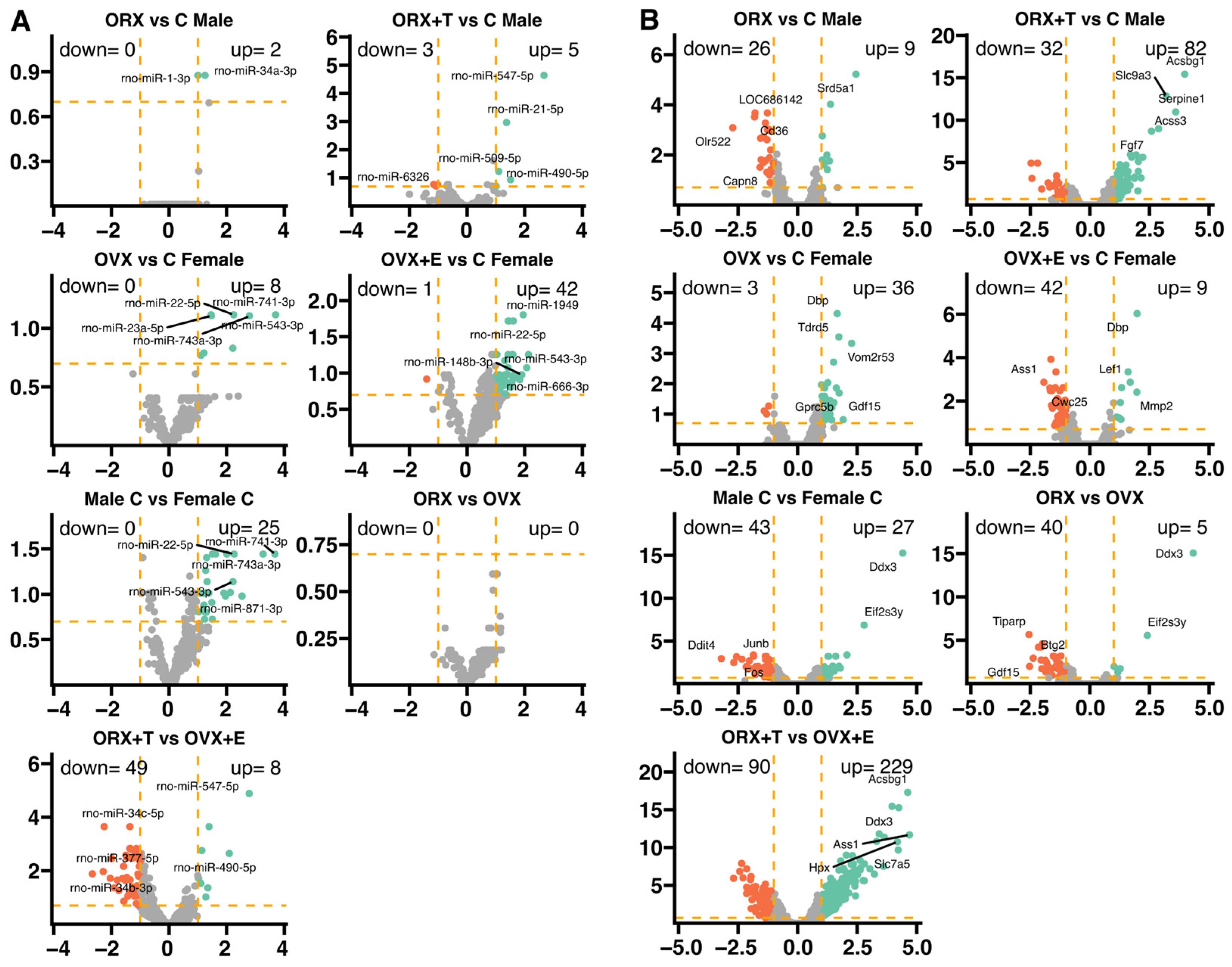
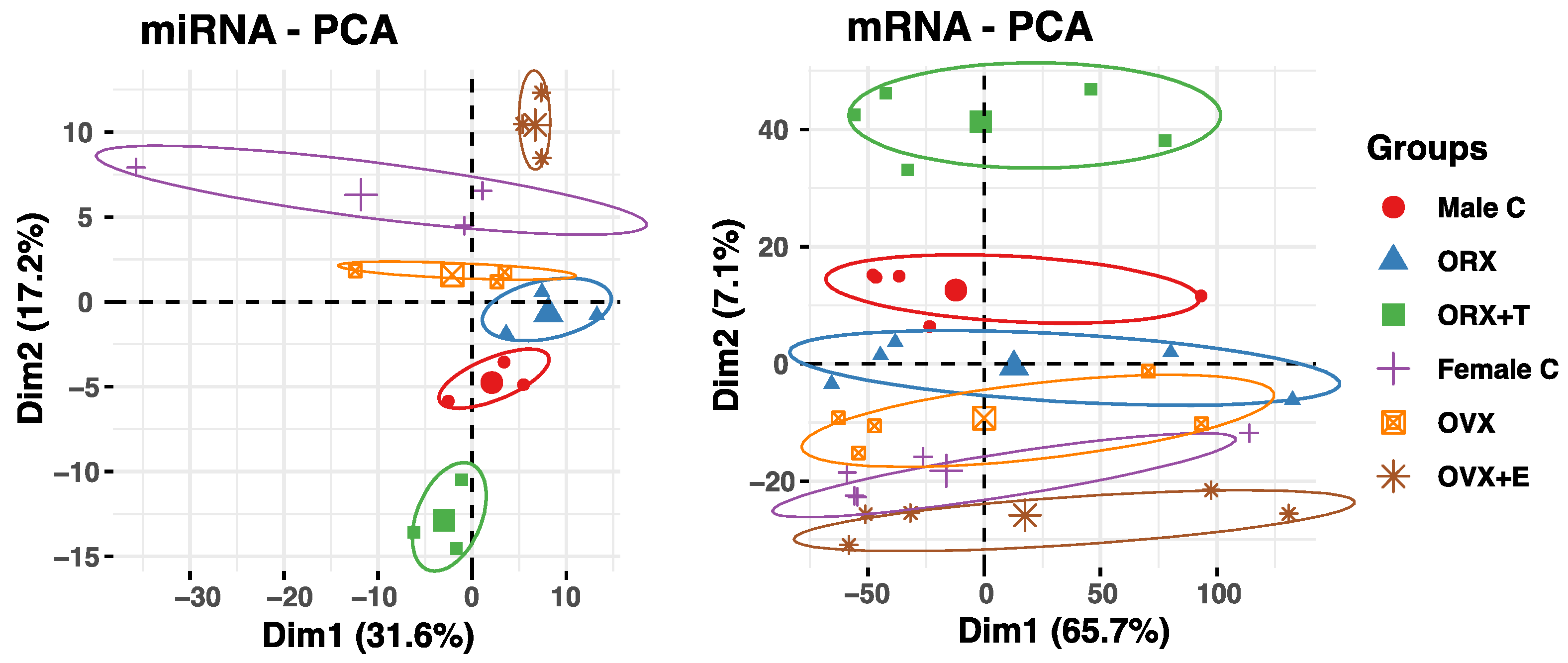
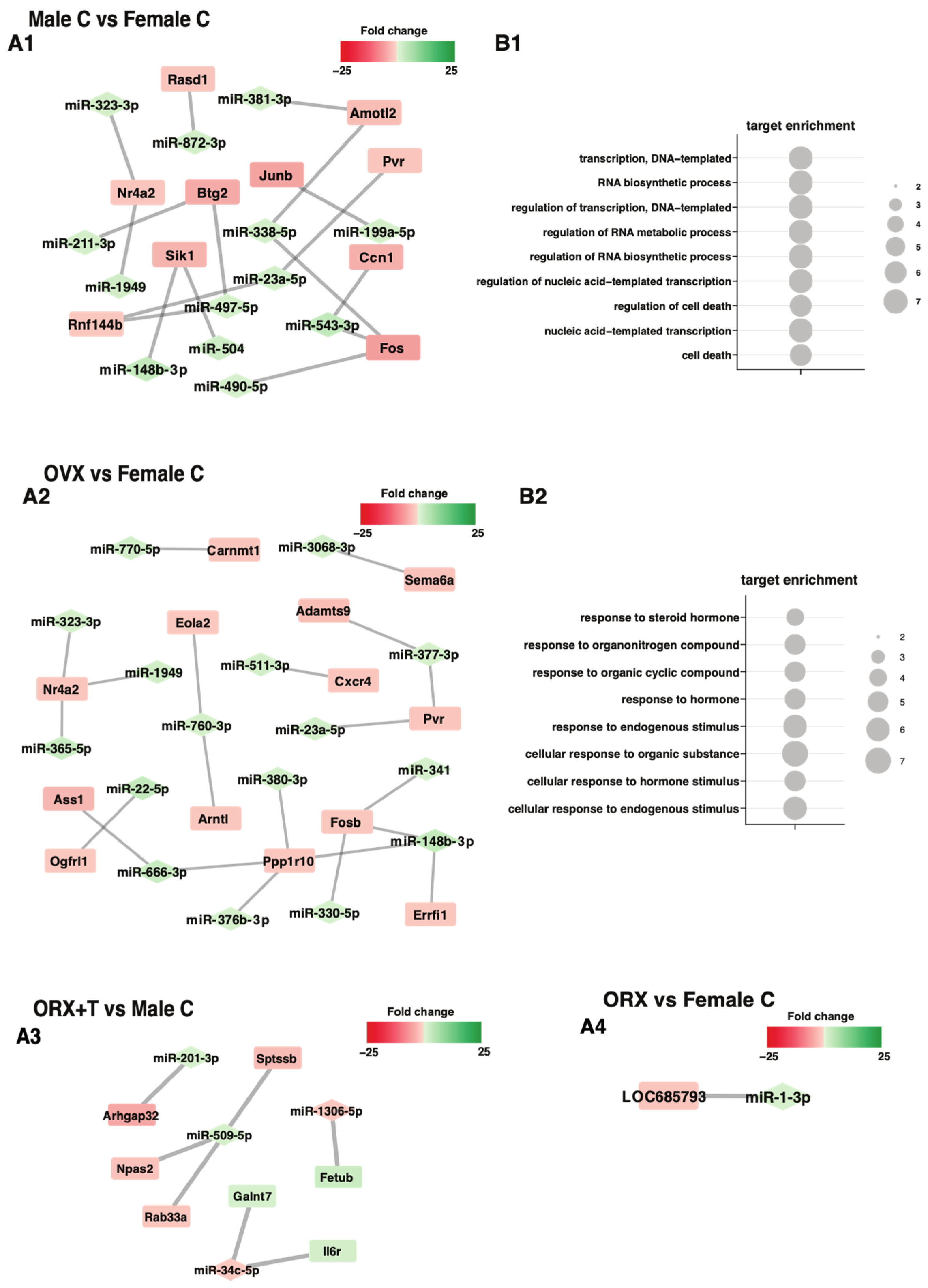
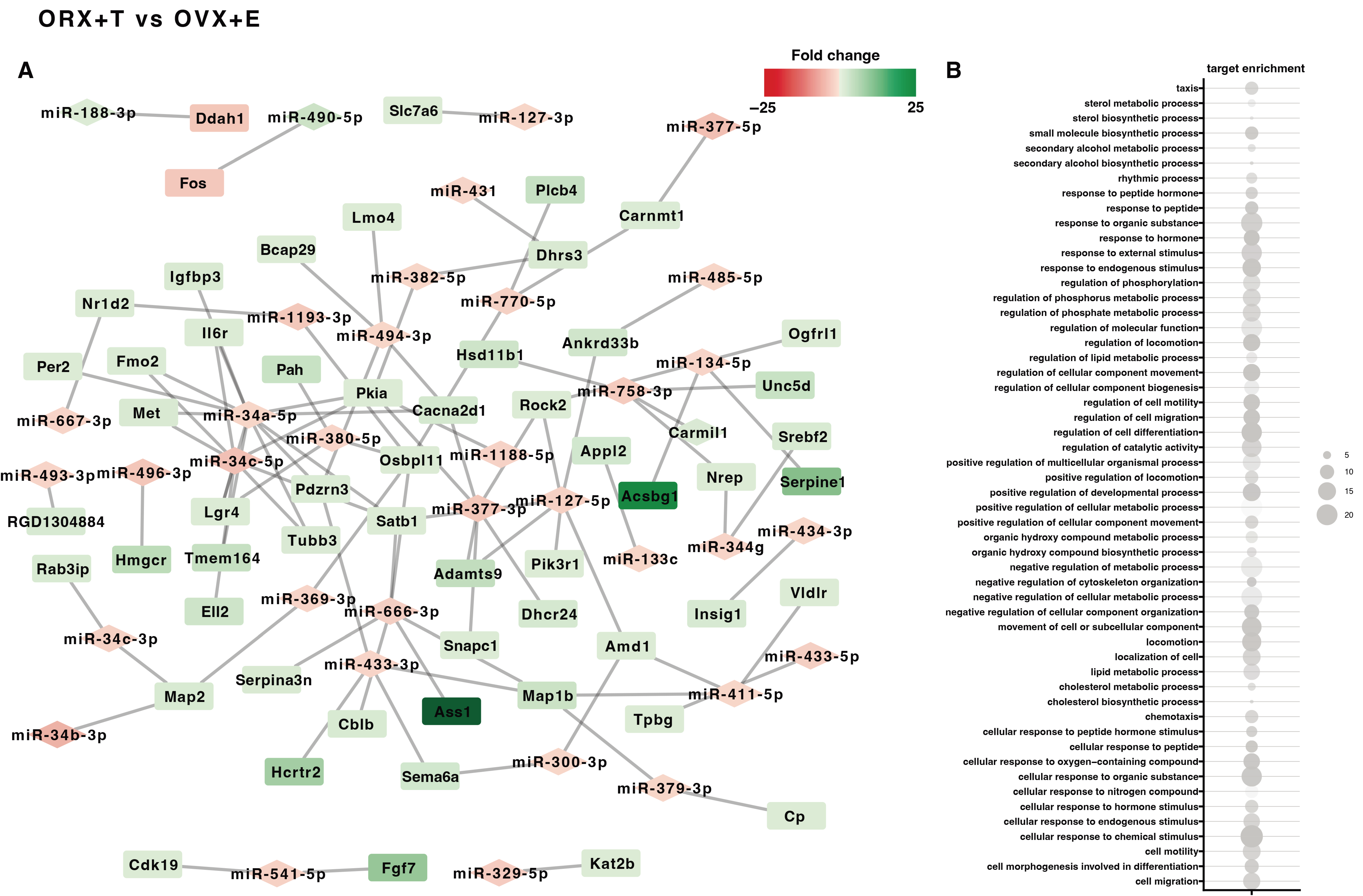
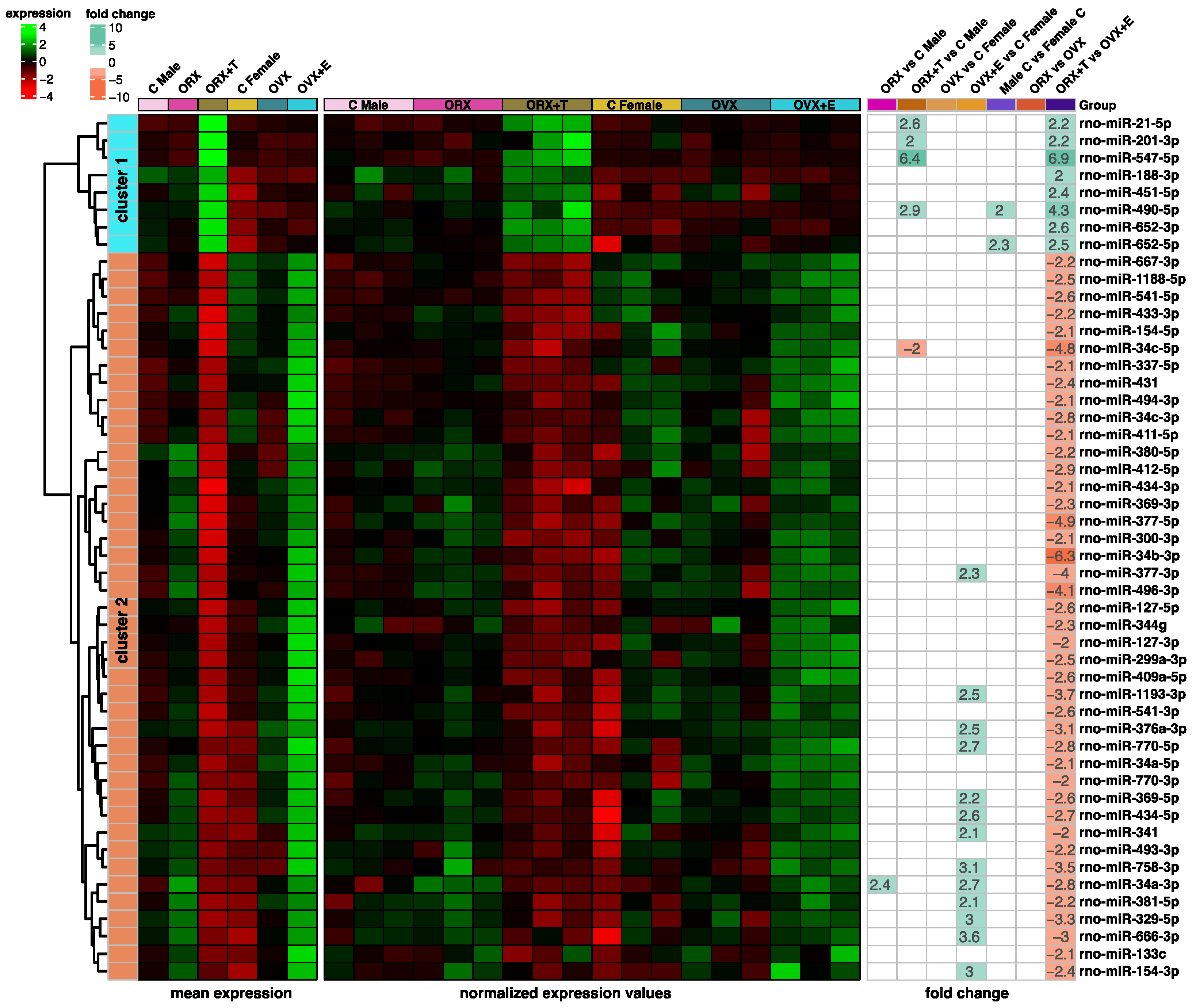
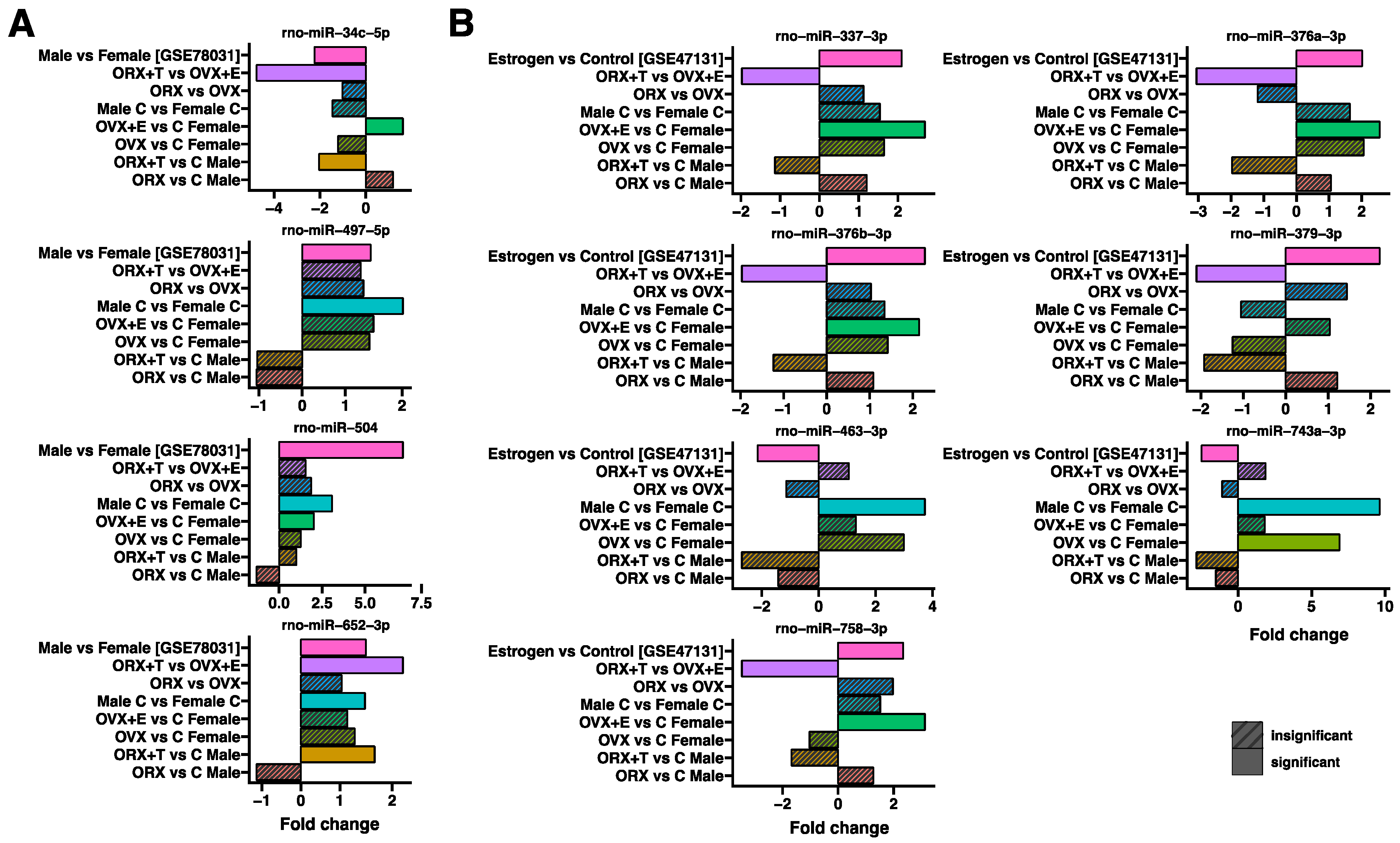
2. Results
3. Discussion
4. Materials and Methods
4.1. Animals and Experiments
4.2. miRNA Isolation
4.3. Microarray Preparation, Hybridization, and Scanning
4.4. Microarray Data Analysis
4.5. miRNA-Target Gene Identification
4.6. miRNA Co-Expression Analysis–Clustering of miRNA Data
4.7. Comparative Analysis to Other Data
4.8. Validation of miRNA Expression by qPCR
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kitay, J.I. Effects of estradiol on pituitary-adrenal function in male and female rats. Endocrinology 1963, 72, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Nussdorfer, G.G. Cytophysiology of the adrenal cortex. Int. Rev. Cytol. 1986, 98, 1–405. [Google Scholar]
- Malendowicz, L.K. Cytophysiology of the Mammalian Adrenal Cortex as Related to Sex, Gonadectomy and Gonadal Hormones; The PTPN Publishing House: Poznań, Poland, 1994; p. 232. [Google Scholar]
- Goel, N.; Workman, J.L.; Lee, T.T.; Innala, L.; Viau, V. Sex differences in the HPA axis. Compr. Physiol. 2014, 4, 1121–1155. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L.; White, P.C. History of Adrenal Research: From Ancient Anatomy to Contemporary Molecular Biology. Endocr. Rev. 2023, 44, 70–116. [Google Scholar] [CrossRef] [PubMed]
- Trejter, M.; Hochol, A.; Tyczewska, M.; Ziolkowska, A.; Jopek, K.; Szyszka, M.; Malendowicz, L.K.; Rucinski, M. Sex-related gene expression profiles in the adrenal cortex in the mature rat: Microarray analysis with emphasis on genes involved in steroidogenesis. Int. J. Mol. Med. 2015, 35, 702–714. [Google Scholar] [CrossRef]
- Jopek, K.; Celichowski, P.; Szyszka, M.; Tyczewska, M.; Milecka, P.; Malendowicz, L.K.; Rucinski, M. Transcriptome Profile of Rat Adrenal Evoked by Gonadectomy and Testosterone or Estradiol Replacement. Front. Endocrinol. 2017, 8, 26. [Google Scholar] [CrossRef]
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008, 135, 1201–1214. [Google Scholar] [CrossRef]
- Hutvagner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef]
- Zeng, Y.; Wagner, E.J.; Cullen, B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 2002, 9, 1327–1333. [Google Scholar] [CrossRef]
- Zeng, Y.; Cullen, B.R. Sequence requirements for micro RNA processing and function in human cells. RNA 2003, 9, 112–123. [Google Scholar] [CrossRef]
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. siRNAs can function as miRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Kamanu, T.K.; Radovanovic, A.; Archer, J.A.; Bajic, V.B. Exploration of miRNA families for hypotheses generation. Sci. Rep. 2013, 3, 2940. [Google Scholar] [CrossRef]
- Zou, Q.; Mao, Y.; Hu, L.; Wu, Y.; Ji, Z. miRClassify: An advanced web server for miRNA family classification and annotation. Comput. Biol. Med. 2014, 45, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Mathelier, A.; Carbone, A. Large scale chromosomal mapping of human microRNA structural clusters. Nucleic Acids Res. 2013, 41, 4392–4408. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kuhn, D.E.; Martin, M.M.; Feldman, D.S.; Terry, A.V., Jr.; Nuovo, G.J.; Elton, T.S. Experimental validation of miRNA targets. Methods 2008, 44, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Slezak-Prochazka, I.; Durmus, S.; Kroesen, B.J.; van den Berg, A. MicroRNAs, macrocontrol: Regulation of miRNA processing. RNA 2010, 16, 1087–1095. [Google Scholar] [CrossRef]
- de Planell-Saguer, M.; Rodicio, M.C. Analytical aspects of microRNA in diagnostics: A review. Anal. Chim. Acta 2011, 699, 134–152. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Kilikevicius, A.; Meister, G.; Corey, D.R. Reexamining assumptions about miRNA-guided gene silencing. Nucleic Acids Res. 2022, 50, 617–634. [Google Scholar] [CrossRef] [PubMed]
- Gangaraju, V.K.; Lin, H. MicroRNAs: Key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009, 10, 116–125. [Google Scholar] [CrossRef]
- Pauley, K.M.; Chan, E.K. MicroRNAs and their emerging roles in immunology. Ann. N. Y. Acad. Sci. 2008, 1143, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Sorrentino, A.; Condorelli, G.; Peschle, C. Role of microRNAs in haemopoiesis, heart hypertrophy and cancer. Biochem. Soc. Trans. 2008, 36, 1206–1210. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. MicroRNAs in neurodegeneration. Curr. Opin. Neurobiol. 2008, 18, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, D.; Jopek, K.; Sterzynska, K.; Ginter-Matuszewska, B.; Nowicki, M.; Rucinski, M.; Januchowski, R. The Significance of MicroRNAs Expression in Regulation of Extracellular Matrix and Other Drug Resistant Genes in Drug Resistant Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2020, 21, 2619. [Google Scholar] [CrossRef]
- Kazmierczak, D.; Jopek, K.; Sterzynska, K.; Nowicki, M.; Rucinski, M.; Januchowski, R. The Profile of MicroRNA Expression and Potential Role in the Regulation of Drug-Resistant Genes in Cisplatin- and Paclitaxel-Resistant Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2022, 23, 526. [Google Scholar] [CrossRef]
- Kulcenty, K.; Wroblewska, J.P.; Rucinski, M.; Kozlowska, E.; Jopek, K.; Suchorska, W.M. MicroRNA Profiling During Neural Differentiation of Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2019, 20, 3651. [Google Scholar] [CrossRef]
- Shenouda, S.K.; Alahari, S.K. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009, 28, 369–378. [Google Scholar] [CrossRef]
- Fasoulakis, Z.; Daskalakis, G.; Diakosavvas, M.; Papapanagiotou, I.; Theodora, M.; Bourazan, A.; Alatzidou, D.; Pagkalos, A.; Kontomanolis, E.N. MicroRNAs Determining Carcinogenesis by Regulating Oncogenes and Tumor Suppressor Genes During Cell Cycle. MicroRNA 2020, 9, 82–92. [Google Scholar] [CrossRef]
- Kontomanolis, E.N.; Koutras, A.; Syllaios, A.; Schizas, D.; Mastoraki, A.; Garmpis, N.; Diakosavvas, M.; Angelou, K.; Tsatsaris, G.; Pagkalos, A.; et al. Role of Oncogenes and Tumor-suppressor Genes in Carcinogenesis: A Review. Anticancer Res. 2020, 40, 6009–6015. [Google Scholar] [CrossRef]
- Cherradi, N. microRNAs as Potential Biomarkers in Adrenocortical Cancer: Progress and Challenges. Front. Endocrinol. 2015, 6, 195. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.; Zhao, J.T.; Sidhu, S.B. The role of microRNAs in the pathophysiology of adrenal tumors. Mol. Cell Endocrinol. 2017, 456, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Yamazaki, Y.; Felizola, S.J.; Ise, K.; Morimoto, R.; Satoh, F.; Arai, Y.; Sasano, H. Adrenocortical carcinoma: Review of the pathologic features, production of adrenal steroids, and molecular pathogenesis. Endocrinol. Metab. Clin. North Am. 2015, 44, 399–410. [Google Scholar] [CrossRef]
- Baquedano, M.S.; Belgorosky, A. Human Adrenal Cortex: Epigenetics and Postnatal Functional Zonation. Horm. Res. Paediatr. 2018, 89, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Tombol, Z.; Turai, P.I.; Decmann, A.; Igaz, P. MicroRNAs and Adrenocortical Tumors: Where do we Stand on Primary Aldosteronism? Horm. Metab. Res. 2020, 52, 394–403. [Google Scholar] [CrossRef]
- Azhar, S.; Dong, D.; Shen, W.J.; Hu, Z.; Kraemer, F.B. The role of miRNAs in regulating adrenal and gonadal steroidogenesis. J. Mol. Endocrinol. 2020, 64, R21–R43. [Google Scholar] [CrossRef]
- Midan, H.M.; Helal, G.K.; Abulsoud, A.I.; Elshaer, S.S.; El-Husseiny, A.A.; Fathi, D.; Abdelmaksoud, N.M.; Abdel Mageed, S.S.; Elballal, M.S.; Zaki, M.B.; et al. The potential role of miRNAs in the pathogenesis of adrenocortical carcinoma—A focus on signaling pathways interplay. Pathol. Res. Pract. 2023, 248, 154690. [Google Scholar] [CrossRef]
- Patel, D.; Boufraqech, M.; Jain, M.; Zhang, L.; He, M.; Gesuwan, K.; Gulati, N.; Nilubol, N.; Fojo, T.; Kebebew, E. MiR-34a and miR-483-5p are candidate serum biomarkers for adrenocortical tumors. Surgery 2013, 154, 1224–1228, discussion 1229. [Google Scholar] [CrossRef]
- Chabre, O.; Libe, R.; Assie, G.; Barreau, O.; Bertherat, J.; Bertagna, X.; Feige, J.J.; Cherradi, N. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocr. Relat. Cancer 2013, 20, 579–594. [Google Scholar] [CrossRef]
- Hu, Z.; Shen, W.J.; Cortez, Y.; Tang, X.; Liu, L.F.; Kraemer, F.B.; Azhar, S. Hormonal regulation of microRNA expression in steroid producing cells of the ovary, testis and adrenal gland. PLoS ONE 2013, 8, e78040. [Google Scholar] [CrossRef]
- Robertson, S.; MacKenzie, S.M.; Alvarez-Madrazo, S.; Diver, L.A.; Lin, J.; Stewart, P.M.; Fraser, R.; Connell, J.M.; Davies, E. MicroRNA-24 is a novel regulator of aldosterone and cortisol production in the human adrenal cortex. Hypertension 2013, 62, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.G.; Plonczynski, M.W.; Carvajal, C.A.; Gomez-Sanchez, E.P.; Gomez-Sanchez, C.E. Microribonucleic acid-21 increases aldosterone secretion and proliferation in H295R human adrenocortical cells. Endocrinology 2008, 149, 2477–2483. [Google Scholar] [CrossRef]
- Hu, Z.; Shen, W.J.; Kraemer, F.B.; Azhar, S. Regulation of adrenal and ovarian steroidogenesis by miR-132. J. Mol. Endocrinol. 2017, 59, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Longhurst, P.A.; Kauer, J.; Leggett, R.E.; Levin, R.M. The influence of ovariectomy and estradiol replacement on urinary bladder function in rats. J. Urol. 1992, 148, 915–919. [Google Scholar] [CrossRef]
- Cagen, L.M.; Baer, P.G. Effects of gonadectomy and steroid treatment on renal prostaglandin 9-ketoreductase activity in the rat. Life Sci. 1987, 40, 95–100. [Google Scholar] [CrossRef]
- Schulte-Beerbuhl, M.; Nieschlag, E. Comparison of testosterone, dihydrotestosterone, luteinizing hormone, and follicle-stimulating hormone in serum after injection of testosterone enanthate of testosterone cypionate. Fertil. Steril. 1980, 33, 201–203. [Google Scholar] [CrossRef]
- Link, J.C.; Hasin-Brumshtein, Y.; Cantor, R.M.; Chen, X.; Arnold, A.P.; Lusis, A.J.; Reue, K. Diet, gonadal sex, and sex chromosome complement influence white adipose tissue miRNA expression. BMC Genom. 2017, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Graham, I.; Hastings, R.; Gunewardena, S.; Brinkmeier, M.L.; Conn, P.M.; Camper, S.A.; Kumar, T.R. Gonadotrope-specific deletion of Dicer results in severely suppressed gonadotropins and fertility defects. J. Biol. Chem. 2015, 290, 2699–2714. [Google Scholar] [CrossRef]
- Noutsios, G.T.; Thorenoor, N.; Zhang, X.; Phelps, D.S.; Umstead, T.M.; Durrani, F.; Floros, J. SP-A2 contributes to miRNA-mediated sex differences in response to oxidative stress: Pro-inflammatory, anti-apoptotic, and anti-oxidant pathways are involved. Biol. Sex Differ. 2017, 8, 37. [Google Scholar] [CrossRef]
- Ye, L.; Wang, J.; Yi, K.; Wang, F.; Wang, J.; Wu, H.; Yang, H.; Yang, Z.; Zhang, Q. Recent findings on miR-370 expression, regulation and functions in cancer (Review). Oncol. Rep. 2023, 49, 79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, X.; Zhang, S.; Wu, S.; Xiao, Q.; Gu, Y.; Guo, X.; Lin, X.; Chen, L.; Zhao, Y.; et al. miR-370-3p Regulates Adipogenesis through Targeting Mknk1. Molecules 2021, 26, 6926. [Google Scholar] [CrossRef]
- Chen, Y.; Xia, X.; Fang, M.; Chen, G.; Cao, J.; Qu, H.; Wang, H. Maternally derived low glucocorticoid mediates adrenal developmental programming alteration in offspring induced by dexamethasone. Sci. Total Environ. 2021, 797, 149084. [Google Scholar] [CrossRef]
- Veronez, L.C.; Fedatto, P.F.; Correa, C.A.P.; Lira, R.C.P.; Baroni, M.; da Silva, K.R.; Santos, P.; Antonio, D.S.M.; Queiroz, R.P.S.; Antonini, S.R.R.; et al. MicroRNA expression profile predicts prognosis of pediatric adrenocortical tumors. Pediatr. Blood Cancer 2022, 69, e29553. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Caramuta, S.; Velazquez-Fernandez, D.; Akcakaya, P.; Xie, H.; Hoog, A.; Zedenius, J.; Backdahl, M.; Larsson, C.; Lui, W.O. The role of microRNA deregulation in the pathogenesis of adrenocortical carcinoma. Endocr. Relat. Cancer 2011, 18, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.E.; Holloway, A.K.; Weng, J.; Fojo, T.; Kebebew, E. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer 2011, 117, 1630–1639. [Google Scholar] [CrossRef]
- Koperski, L.; Kotlarek, M.; Swierniak, M.; Kolanowska, M.; Kubiak, A.; Gornicka, B.; Jazdzewski, K.; Wojcicka, A. Next-generation sequencing reveals microRNA markers of adrenocortical tumors malignancy. Oncotarget 2017, 8, 49191–49200. [Google Scholar] [CrossRef]
- Blatkiewicz, M.; Kaminski, K.; Szyszka, M.; Al-Shakarchi, Z.; Olechnowicz, A.; Stelcer, E.; Komarowska, H.; Tyczewska, M.; Klimont, A.; Karczewski, M.; et al. The Enhanced Expression of ZWILCH Predicts Poor Survival of Adrenocortical Carcinoma Patients. Biomedicines 2023, 11, 1233. [Google Scholar] [CrossRef]
- Soon, P.S.; Tacon, L.J.; Gill, A.J.; Bambach, C.P.; Sywak, M.S.; Campbell, P.R.; Yeh, M.W.; Wong, S.G.; Clifton-Bligh, R.J.; Robinson, B.G.; et al. miR-195 and miR-483-5p Identified as Predictors of Poor Prognosis in Adrenocortical Cancer. Clin. Cancer Res. 2009, 15, 7684–7692. [Google Scholar] [CrossRef]
- Babinska, A.; Peksa, R.; Wisniewski, P.; Swiatkowska-Stodulska, R.; Sworczak, K. Diagnostic and prognostic role of SF1, IGF2, Ki67, p53, adiponectin, and leptin receptors in human adrenal cortical tumors. J. Surg. Oncol. 2017, 116, 427–433. [Google Scholar] [CrossRef]
- Ribeiro, T.C.; Latronico, A.C. Insulin-like growth factor system on adrenocortical tumorigenesis. Mol. Cell Endocrinol. 2012, 351, 96–100. [Google Scholar] [CrossRef]
- Salvianti, F.; Canu, L.; Poli, G.; Armignacco, R.; Scatena, C.; Cantini, G.; Di Franco, A.; Gelmini, S.; Ercolino, T.; Pazzagli, M.; et al. New insights in the clinical and translational relevance of miR483-5p in adrenocortical cancer. Oncotarget 2017, 8, 65525–65533. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, P.; Kazmierczak, D.; Jopek, K.; Nowicki, M.; Rucinski, M.; Januchowski, R. The Profile of MicroRNA Expression and Potential Role in the Regulation of Drug-Resistant Genes in Doxorubicin and Topotecan Resistant Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2022, 23, 5846. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Kauffmann, A.; Gentleman, R.; Huber, W. arrayQualityMetrics—A bioconductor package for quality assessment of microarray data. Bioinformatics 2009, 25, 415–416. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R. The Genefilter: Methods for Filtering Genes from Microarray Experiments. 2025. Available online: https://www.bioconductor.org/packages/devel/bioc/manuals/genefilter/man/genefilter.pdf (accessed on 12 December 2024).
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Dawson, C. ggprism: A ‘ggplot2’ Extension Inspired by ‘GraphPad Prism’. 2024. Available online: https://csdaw.github.io/ggprism/articles/ggprism.html (accessed on 12 December 2024).
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. 2020. Available online: https://rpkgs.datanovia.com/factoextra/index.html (accessed on 12 December 2024).
- Pajak, M.; Simpson, T.I. miRNAtap: microRNA Targets—Aggregated Predictions. R Package Version 1.40.0. 2024. Available online: https://bioconductor.org/packages/release/bioc/html/miRNAtap.html (accessed on 12 December 2024).
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Jopek, K.; Tyczewska, M.; Celichowski, P.; Malendowicz, L.K.; Rucinski, M. Transcriptome Profile in Unilateral Adrenalectomy-Induced Compensatory Adrenal Growth in the Rat. Int. J. Mol. Sci. 2018, 19, 1111. [Google Scholar] [CrossRef]
- Fresno, C.; Fernandez, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef]
- Benjamini, Y.; Cohen, R. Weighted false discovery rate controlling procedures for clinical trials. Biostatistics 2017, 18, 91–104. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jopek, K.; Tyczewska, M.; Blatkiewicz, M.; Olechnowicz, A.; Szyszka, M.; Stelcer, E.; Ciesiółka, S.; Jopek, M.; Malendowicz, L.K.; Ruciński, M. Profile of Rat Adrenal microRNAs Induced by Gonadectomy and Testosterone or Estradiol Replacement. Int. J. Mol. Sci. 2025, 26, 4543. https://doi.org/10.3390/ijms26104543
Jopek K, Tyczewska M, Blatkiewicz M, Olechnowicz A, Szyszka M, Stelcer E, Ciesiółka S, Jopek M, Malendowicz LK, Ruciński M. Profile of Rat Adrenal microRNAs Induced by Gonadectomy and Testosterone or Estradiol Replacement. International Journal of Molecular Sciences. 2025; 26(10):4543. https://doi.org/10.3390/ijms26104543
Chicago/Turabian StyleJopek, Karol, Marianna Tyczewska, Małgorzata Blatkiewicz, Anna Olechnowicz, Marta Szyszka, Ewelina Stelcer, Sylwia Ciesiółka, Maria Jopek, Ludwik K. Malendowicz, and Marcin Ruciński. 2025. "Profile of Rat Adrenal microRNAs Induced by Gonadectomy and Testosterone or Estradiol Replacement" International Journal of Molecular Sciences 26, no. 10: 4543. https://doi.org/10.3390/ijms26104543
APA StyleJopek, K., Tyczewska, M., Blatkiewicz, M., Olechnowicz, A., Szyszka, M., Stelcer, E., Ciesiółka, S., Jopek, M., Malendowicz, L. K., & Ruciński, M. (2025). Profile of Rat Adrenal microRNAs Induced by Gonadectomy and Testosterone or Estradiol Replacement. International Journal of Molecular Sciences, 26(10), 4543. https://doi.org/10.3390/ijms26104543





