Metabolomic Plasma Profile of Chronic Obstructive Pulmonary Disease Patients
Abstract
1. Introduction
2. Results
2.1. Main Characteristics of Participants
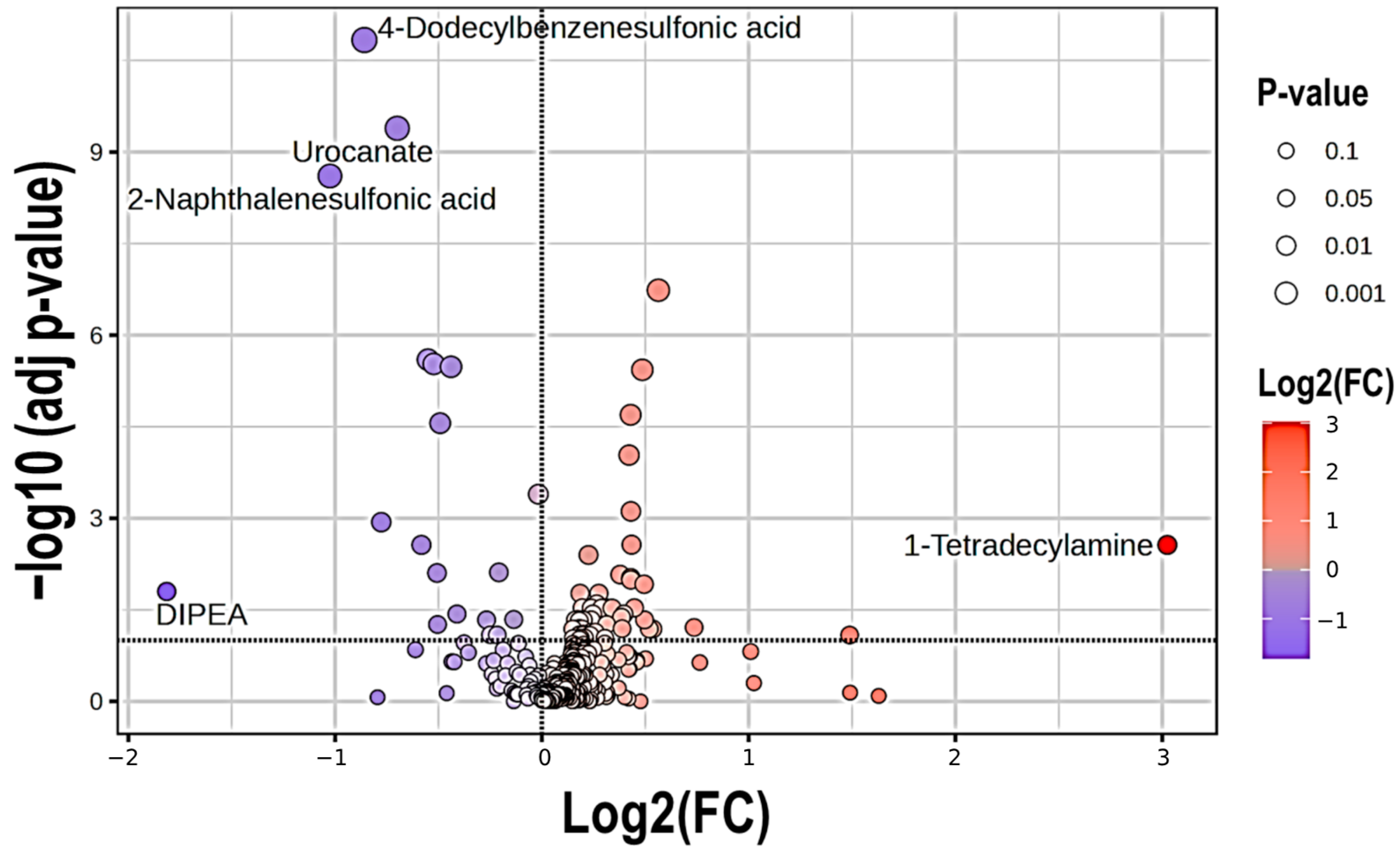
2.2. Metabolite Profile in COPD Patients
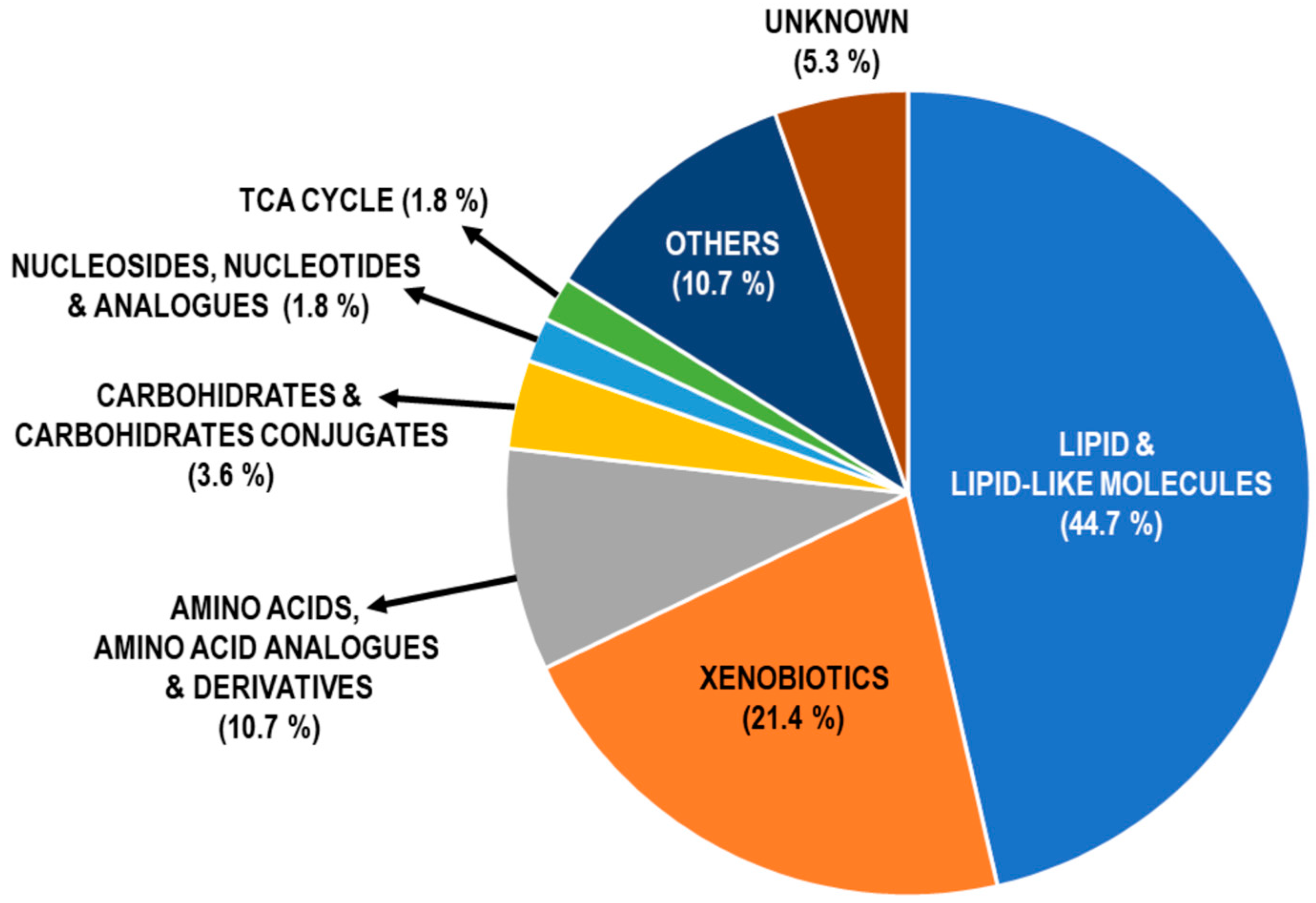
2.3. Pathway Enrichment Analysis of DAMs in COPD
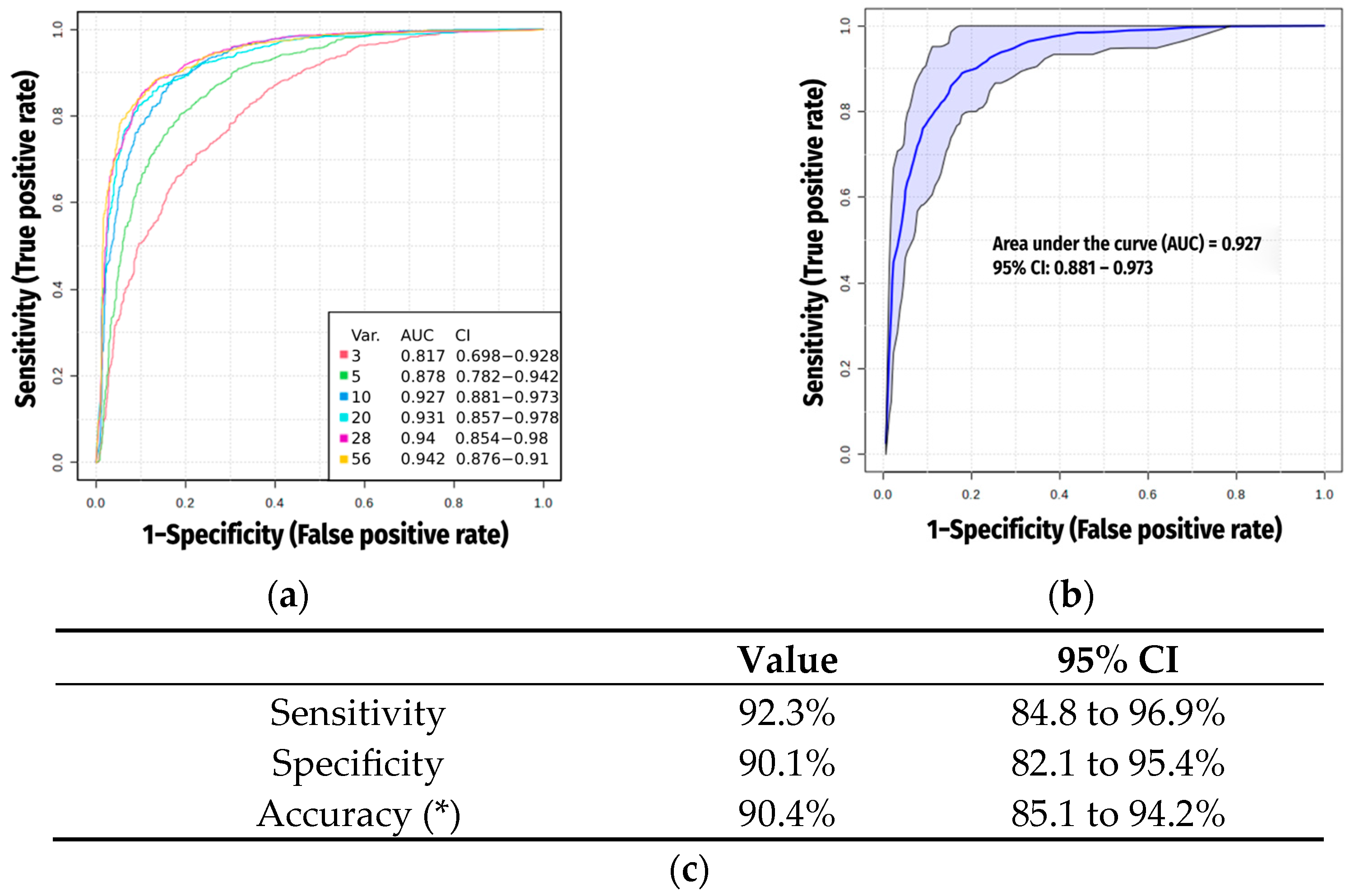
2.4. Multivariate Analysis and AI Modeling
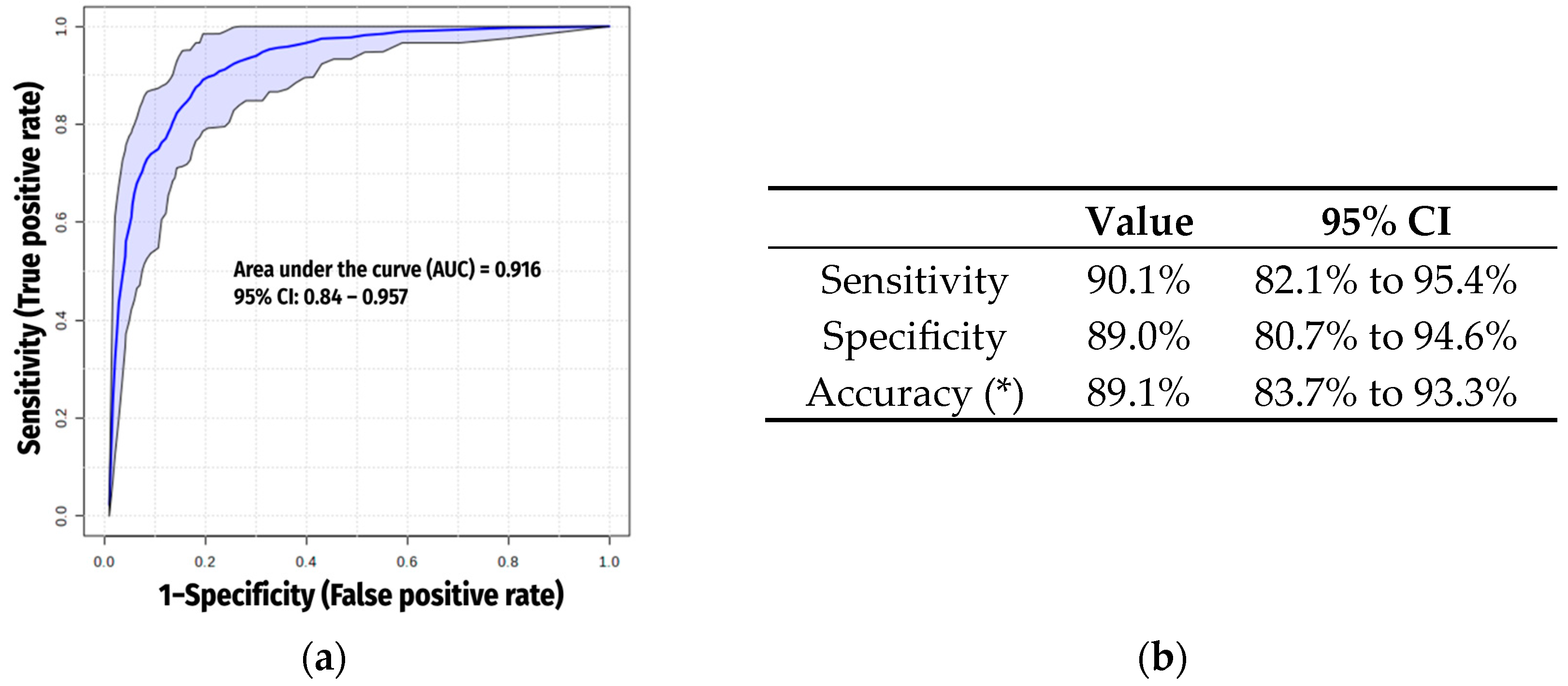
2.5. Exclusion of Xenobiotics
3. Discussion
3.1. Previous Studies
3.2. Interpretation of Novel Findings
3.2.1. Lipids and Lipid-like Molecules
3.2.2. Xenobiotics
3.2.3. Amino Acids, Analogues and Derivates
3.2.4. Carbohydrates
3.3. Potential Confounding Factors
3.4. Final Metabolomic Signature for COPD (Excluding Xenobiotics)
3.5. Strengths and Potential Limitations
4. Materials and Methods
4.1. Participants and Ethics
4.2. Collection of Blood Samples
4.3. Metabolomics Procedure
4.3.1. Metabolite Extraction
4.3.2. LC-MS Run Parameters
4.3.3. Metabolite Identification and Quantification
Semi-Targeted
Untargeted
4.4. Data Analysis
4.4.1. Statistical Analysis of Clinical Data
4.4.2. Metabolomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| ATP | Adenosine triphosphate |
| AUC | Area under the curve |
| a-KG | α-ketoglutarate |
| BMI | Body mass index |
| CI | Confidence interval |
| CMPF | 3-carboxy-4-methyl-5-propyl-2-furanpropionate |
| CO | Carbon monoxide |
| COPD | Chronic Obstructive Pulmonary Disease |
| CRP | C-reactive protein |
| CT | Computed tomography |
| DAMs | Differentially abundant metabolites |
| DEA | Diethanolamine |
| DEHP | Bis(2-ethylhexyl) phthalate |
| DIPEA | Diisopropylethylamine |
| DLco | Diffusion capacity for carbon monoxide |
| EDTA | Ethylenediaminetetraacetic acid |
| FC | Fold change |
| FDR | False discovery rate |
| FEV1 | Forced Expiratory Volume in one second |
| FVC | Forced Vital Capacity |
| GOLD | Global Initiative for Chronic Obstructive Pulmonary Disease |
| HC | Controls (asymptomatic smokers without airflow limitation) |
| HICA | 2-hydroxyisocaproic acid |
| HMDB | Human Metabolome Database |
| HPLC | High-performance liquid chromatography |
| IQR | Interquartile range |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LCFA | Long-chain fatty acid |
| LC-MS | Liquid Chromatography-Mass Spectrometry |
| MCFA | Medium-chain fatty acid |
| MS | Multi-scale |
| NAD | Nicotinamide adenine dinucleotide |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| Post-BD | Post-bronchodilator |
| PPAR-γ | Peroxisome proliferator-activated receptor gamma |
| PPG n5 | Pentapropylen glycol |
| PPG n4 | Tetrapropylene glycol |
| ROC | Receiver operating characteristic |
| SCFA | Short-chain fatty acid |
| SD | Standard deviation |
| SMPDB | Small Molecule Pathway Database |
| SVM | Support Vector machine |
| TCA | Tricarboxylic (or citric) acid cycle |
| UPLC | Ultra-high-performance liquid chromatography |
| VLCFA | Very long-chain fatty acid |
| vWF | Von Willebrand factor |
| 3HBA | 3-hydroxibenzaldehyde |
| 4HBA | 4-hydroxibenzaldehyde |
| 11-dehTxB2 | 11-dehydrothromboxane B2 |
References
- Agustí, A.; Celli, B.R.; Criner, G.J.; Halpin, D.; Anzueto, A.; Barnes, P.; Bourbeau, J.; Han, M.K.; Martinez, F.J.; de Oca, M.M.; et al. Global Initiative for Chronic Obstructive Lung Disease 2023 Report: GOLD Executive Summary. Respirology 2023, 28, 316–338. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Inflammatory Mechanisms in Patients with Chronic Obstructive Pulmonary Disease. J. Allergy Clin. Immunol. 2016, 138, 16–27. [Google Scholar] [CrossRef]
- Mariniello, D.F.; D’Agnano, V.; Cennamo, D.; Conte, S.; Quarcio, G.; Notizia, L.; Pagliaro, R.; Schiattarella, A.; Salvi, R.; Bianco, A.; et al. Comorbidities in COPD: Current and Future Treatment Challenges. J. Clin. Med. 2024, 13, 743. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.H.; Lee, J.H.; Kim, M.; Kim, S.J.; Kim, K.H.; Park, J.-S.; Kim, T.-H.; Kim, Y.I.; Lee, E.W.; Kim, J.O.; et al. Determinants of Respiratory Symptom Development in Patients with Chronic Airflow Obstruction. Respir. Med. 2006, 100, 2170–2176. [Google Scholar] [CrossRef]
- Danielsson, P.; Ólafsdóttir, I.S.; Benediktsdóttir, B.; Gíslason, T.; Janson, C. The Prevalence of Chronic Obstructive Pulmonary Disease in Uppsala, Sweden—The Burden of Obstructive Lung Disease (BOLD) Study: Cross-Sectional Population-Based Study. Clin. Respir. J. 2012, 6, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.; Cusack, R.P.; Chaudhary, N.; Satia, I.; Kurmi, O.P. Under- and over-Diagnosis of COPD: A Global Perspective. Breathe 2019, 15, 24–35. [Google Scholar] [CrossRef]
- Kurmi, O.P.; Li, L.; Smith, M.; Augustyn, M.; Chen, J.; Collins, R.; Guo, Y.; Han, Y.; Qin, J.; Xu, G.; et al. Regional Variations in the Prevalence and Misdiagnosis of Air Flow Obstruction in China: Baseline Results from a Prospective Cohort of the China Kadoorie Biobank (CKB). BMJ Open Respir. Res. 2014, 1, e000025. [Google Scholar] [CrossRef]
- Laniado-Laborin, R.; Rendón, A.; Bauerle, O. Chronic Obstructive Pulmonary Disease Case Finding in Mexico in an At-Risk Population. Int. J. Tuberc. Lung Dis. 2011, 15, 818–823. [Google Scholar] [CrossRef]
- Martinez, C.H.; Mannino, D.M.; Jaimes, F.A.; Curtis, J.L.; Han, M.K.; Hansel, N.N.; Diaz, A.A. Undiagnosed Obstructive Lung Disease in the United States. Associated Factors and Long-Term Mortality. Ann. Am. Thorac. Soc. 2015, 12, 1788–1795. [Google Scholar] [CrossRef]
- Miravitlles, M.; Soriano, J.B.; García-Río, F.; Muñoz, L.; Duran-Tauleria, E.; Sanchez, G.; Sobradillo, V.; Ancochea, J. Prevalence of COPD in Spain: Impact of Undiagnosed COPD on Quality of Life and Daily Life Activities. Thorax 2009, 64, 863–868. [Google Scholar] [CrossRef]
- Queiroz, M.C.d.C.A.M.d.; Moreira, M.A.C.; Rabahi, M.F. Underdiagnosis of COPD at Primary Health Care Clinics in the City of Aparecida de Goiânia, Brazil. J. Bras. Pneumol. 2012, 38, 692–699. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Soriano, J.B.; Alfageme, I.; Miravitlles, M.; De Lucas, P.; Soler-Cataluña, J.J.; García-Río, F.; Casanova, C.; Rodríguez González-Moro, J.M.; Cosío, B.G.; Sánchez, G.; et al. Prevalence and Determinants of COPD in Spain: EPISCAN II. Arch. Bronconeumol. 2021, 57, 61–69. [Google Scholar] [CrossRef]
- Hughes, M.J.; McGettrick, H.M.; Sapey, E. Shared Mechanisms of Multimorbidity in COPD, Atherosclerosis and Type-2 Diabetes: The Neutrophil as a Potential Inflammatory Target. Eur. Respir. Rev. 2020, 29, 190102. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, C.; Quero, S.; Millares, L.; Faner, R.; Cosío, B.G.; Peces-Barba, G.; Castro-Acosta, A.; Montón, C.; Palou, A.; Pascual-Guardia, S.; et al. Relationship between Respiratory Microbiome and Systemic Inflammatory Markers in COPD: A Pilot Study. Int. J. Mol. Sci. 2024, 25, 8467. [Google Scholar] [CrossRef]
- Lira-Lucio, J.A.; Falfán-Valencia, R.; Ramírez-Venegas, A.; Buendía-Roldán, I.; Rojas-Serrano, J.; Mejía, M.; Pérez-Rubio, G. Lung Microbiome Participation in Local Immune Response Regulation in Respiratory Diseases. Microorganisms 2020, 8, 1059. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Yue, Y.; Zhang, Q. Imbalance of Gut Microbiota Is Involved in the Development of Chronic Obstructive Pulmonary Disease: A Review. Biomed. Pharmacother. 2023, 165, 115150. [Google Scholar] [CrossRef]
- Agarwal, A.R.; Kadam, S.; Brahme, A.; Agrawal, M.; Apte, K.; Narke, G.; Kekan, K.; Madas, S.; Salvi, S. Systemic Immuno-Metabolic Alterations in Chronic Obstructive Pulmonary Disease (COPD). Respir. Res. 2019, 20, 171. [Google Scholar] [CrossRef]
- Dasgupta, S.; Ghosh, N.; Bhattacharyya, P.; Roy Chowdhury, S.; Chaudhury, K. Metabolomics of Asthma, COPD, and Asthma-COPD Overlap: An Overview. Crit. Rev. Clin. Lab. Sci. 2023, 60, 153–170. [Google Scholar] [CrossRef]
- Gea, J.; Enríquez-Rodríguez, C.J.; Pascual-Guardia, S. Metabolomics in COPD. Arch. Bronconeumol. 2023, 59, 311–321. [Google Scholar] [CrossRef]
- Godbole, S.; Bowler, R.P. Metabolome Features of COPD: A Scoping Review. Metabolites 2022, 12, 621. [Google Scholar] [CrossRef]
- Ran, N.; Pang, Z.; Gu, Y.; Pan, H.; Zuo, X.; Guan, X.; Yuan, Y.; Wang, Z.; Guo, Y.; Cui, Z.; et al. An Updated Overview of Metabolomic Profile Changes in Chronic Obstructive Pulmonary Disease. Metabolites 2019, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Gea, J.; Casadevall, C.; Nebot, P.; Enríquez Rodríguez, C.J.; Faner, M.R.; Cosio, B.G.; Haro, N.; Pascual-Guardia, S.; Peces-Barba, G.; Monsó, E.; et al. Aging and Metabolic Changes in COPD Patients. In Proceedings of the B70. COPD in the Spotlight: Insights into Disease Pathogenesis, San Diego, CA, USA, 6 May 2024; American Thoracic Society: New York, NY, USA; p. A4314. [Google Scholar]
- Lochner, M.; Berod, L.; Sparwasser, T. Fatty Acid Metabolism in the Regulation of T Cell Function. Trends Immunol. 2015, 36, 81–91. [Google Scholar] [CrossRef]
- Chen, H.; Li, Z.; Dong, L.; Wu, Y.; Shen, H.; Chen, Z. Lipid Metabolism in Chronic Obstructive Pulmonary Disease. Int. J. Chron. Obstruct. Pulmon. Dis. 2019, 14, 1009–1018. [Google Scholar] [CrossRef]
- Yedgar, S.; Krimsky, M.; Cohen, Y.; Flower, R.J. Treatment of Inflammatory Diseases by Selective Eicosanoid Inhibition: A Double-Edged Sword? Trends Pharmacol. Sci. 2007, 28, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, P.; Hanaoka, M.; Droma, Y.; Kubo, K. Enhanced Levels of Prostaglandin E2 and Matrix Metalloproteinase-2 Correlate with the Severity of Airflow Limitation in Stable COPD. Respirology 2008, 13, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Bowler, R.P.; Jacobson, S.; Cruickshank, C.; Hughes, G.J.; Siska, C.; Ory, D.S.; Petrache, I.; Schaffer, J.E.; Reisdorph, N.; Kechris, K. Plasma Sphingolipids Associated with Chronic Obstructive Pulmonary Disease Phenotypes. Am. J. Respir. Crit. Care Med. 2015, 191, 275–284. [Google Scholar] [CrossRef]
- Agarwal, A.R.; Yin, F.; Cadenas, E. Short-Term Cigarette Smoke Exposure Leads to Metabolic Alterations in Lung Alveolar Cells. Am. J. Respir. Cell Mol. Biol. 2014, 51, 284–293. [Google Scholar] [CrossRef]
- Titz, B.; Boué, S.; Phillips, B.; Talikka, M.; Vihervaara, T.; Schneider, T.; Nury, C.; Elamin, A.; Guedj, E.; Peck, M.J.; et al. Effects of Cigarette Smoke, Cessation, and Switching to Two Heat-Not-Burn Tobacco Products on Lung Lipid Metabolism in C57BL/6 and Apoe−/− Mice—An Integrative Systems Toxicology Analysis. Toxicol. Sci. 2016, 149, 441–457. [Google Scholar] [CrossRef]
- Charalampous, N.; Antonopoulou, M.; Chasapis, C.T.; Vlastos, D.; Dormousoglou, M.; Dailianis, S. New Insights into the Oxidative and Cytogenotoxic Effects of Tetraglyme on Human Peripheral Blood Cells. Sci. Total Environ. 2024, 954, 176484. [Google Scholar] [CrossRef]
- Millares, L.; Pascual, S.; Montón, C.; García-Núñez, M.; Lalmolda, C.; Faner, R.; Casadevall, C.; Setó, L.; Capilla, S.; Moreno, A.; et al. Relationship between the Respiratory Microbiome and the Severity of Airflow Limitation, History of Exacerbations and Circulating Eosinophils in COPD Patients. BMC Pulm. Med. 2019, 19, 112. [Google Scholar] [CrossRef]
- Eshwarnath, V. Biosurfactants from Bacteria Isolated from Cold Climatic Regions and Its Evaluation for Biomedical and Non-Medical Applications. Sathyabama Institute of Science and Technology. Master’s Thesis, Sathyabama Institute of Science and Technology, Chennai, Tamil Nadu, India, 2023. [Google Scholar]
- Lee, J.; Choi, J.-W.; Han, H.Y.; Kim, W.S.; Song, H.-Y.; Byun, E.-B.; Byun, E.-H.; Lee, Y.-H.; Yuk, J.-M. 4-Hydroxybenzaldehyde Restricts the Intracellular Growth of Toxoplasma gondii by Inducing SIRT1-Mediated Autophagy in Macrophages. Korean J. Parasitol. 2020, 58, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.S.; Im, S.J.; Lee, Y.J.; Cho, Y.H.; Do, Y.R.; Byun, J.W.; Ku, C.R.; Lee, E.J. Vasculoprotective Effects of 3-Hydroxybenzaldehyde against VSMCs Proliferation and ECs Inflammation. PLoS ONE 2016, 11, e0149394. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Sasaki, K. 3-Hydroxybutyrate Could Serve as a Principal Energy Substrate for Human Microbiota. Med. Hypotheses 2024, 182, 111217. [Google Scholar] [CrossRef]
- Debigaré, R.; Côté, C.H.; Maltais, F. Ubiquitination and Proteolysis in Limb and Respiratory Muscles of Patients with Chronic Obstructive Pulmonary Disease. Proc. Am. Thorac. Soc. 2010, 7, 84–90. [Google Scholar] [CrossRef]
- Gea, J.; Pascual, S.; Casadevall, C.; Orozco-Levi, M.; Barreiro, E. Muscle Dysfunction in Chronic Obstructive Pulmonary Disease: Update on Causes and Biological Findings. J. Thorac. Dis. 2015, 7, E418–E438. [Google Scholar] [CrossRef]
- Pandey, K.C.; De, S.; Mishra, P.K. Role of Proteases in Chronic Obstructive Pulmonary Disease. Front. Pharmacol. 2017, 8, 512. [Google Scholar] [CrossRef] [PubMed]
- Puig-Vilanova, E.; Rodriguez, D.A.; Lloreta, J.; Ausin, P.; Pascual-Guardia, S.; Broquetas, J.; Roca, J.; Gea, J.; Barreiro, E. Oxidative Stress, Redox Signaling Pathways, and Autophagy in Cachectic Muscles of Male Patients with Advanced COPD and Lung Cancer. Free Radic. Biol. Med. 2015, 79, 91–108. [Google Scholar] [CrossRef]
- Gea, J.; Sancho-Muñoz, A.; Chalela, R. Nutritional Status and Muscle Dysfunction in Chronic Respiratory Diseases: Stable Phase versus Acute Exacerbations. J. Thorac. Dis. 2018, 10, S1332–S1354. [Google Scholar] [CrossRef]
- Goldstein, S.; Askanazi, J.; Weissman, C.; Thomashow, B.; Kinney, J.M. Energy Expenditure in Patients with Chronic Obstructive Pulmonary Disease. Chest 1987, 91, 222–224. [Google Scholar] [CrossRef]
- Wilson, D.O.; Donahoe, M.; Rogers, R.M.; Pennock, B.E. Metabolic Rate and Weight Loss in Chronic Obstructive Lung Disease. JPEN J. Parenter. Enter. Nutr. 1990, 14, 7–11. [Google Scholar] [CrossRef]
- Enríquez-Rodríguez, C.J.; Casadevall, C.; Faner, R.; Castro-Costa, A.; Pascual-Guàrdia, S.; Seijó, L.; López-Campos, J.L.; Peces-Barba, G.; Monsó, E.; Barreiro, E.; et al. COPD: Systemic Proteomic Profiles in Frequent and Infrequent Exacerbators. ERJ Open Res. 2024, 10, 4–2024. [Google Scholar] [CrossRef] [PubMed]
- Enríquez-Rodríguez, C.J.; Casadevall, C.; Faner, R.; Pascual-Guardia, S.; Castro-Acosta, A.; López-Campos, J.L.; Peces-Barba, G.; Seijo, L.; Caguana-Vélez, O.A.; Monsó, E. A Pilot Study on Proteomic Predictors of Mortality in Stable COPD. Cells 2024, 13, 1351. [Google Scholar] [CrossRef] [PubMed]
- Enríquez-Rodríguez, C.J.; Pascual-Guardia, S.; Casadevall, C.; Caguana-Vélez, O.A.; Rodríguez-Chiaradia, D.; Barreiro, E.; Gea, J. Proteomic Blood Profiles Obtained by Totally Blind Biological Clustering in Stable and Exacerbated COPD Patients. Cells 2024, 13, 866. [Google Scholar] [CrossRef] [PubMed]
- Prokić, I.; Lahousse, L.; de Vries, M.; Liu, J.; Kalaoja, M.; Vonk, J.M.; van der Plaat, D.A.; van Diemen, C.C.; van der Spek, A.; Zhernakova, A.; et al. A Cross-Omics Integrative Study of Metabolic Signatures of Chronic Obstructive Pulmonary Disease. BMC Pulm. Med. 2020, 20, 193. [Google Scholar] [CrossRef]
- Gea, J.; Enríquez-Rodríguez, C.J.; Agranovich, B.; Pascual-Guardia, S. Update on Metabolomic Findings in COPD Patients. ERJ Open Res. 2023, 9, 180–2023. [Google Scholar] [CrossRef]
- Li, X.; Cheng, J.; Shen, Y.; Chen, J.; Wang, T.; Wen, F.; Chen, L. Metabolomic Analysis of Lung Cancer Patients with Chronic Obstructive Pulmonary Disease Using Gas Chromatography-Mass Spectrometry. J. Pharm. Biomed. Anal. 2020, 190, 113524. [Google Scholar] [CrossRef]
- Pinto-Plata, V.; Casanova, C.; Divo, M.; Tesfaigzi, Y.; Calhoun, V.; Sui, J.; Polverino, F.; Priolo, C.; Petersen, H.; de Torres, J.P.; et al. Plasma Metabolomics and Clinical Predictors of Survival Differences in COPD Patients. Respir. Res. 2019, 20, 219. [Google Scholar] [CrossRef]
- Ghosh, N.; Choudhury, P.; Subramani, E.; Saha, D.; Sengupta, S.; Joshi, M.; Banerjee, R.; Roychowdhury, S.; Bhattacharyya, P.; Chaudhury, K. Metabolomic Signatures of Asthma-COPD Overlap (ACO) Are Different from Asthma and COPD. Metabolomics 2019, 15, 87. [Google Scholar] [CrossRef]
- Ghosh, N.; Choudhury, P.; Kaushik, S.R.; Arya, R.; Nanda, R.; Bhattacharyya, P.; Roychowdhury, S.; Banerjee, R.; Chaudhury, K. Metabolomic Fingerprinting and Systemic Inflammatory Profiling of Asthma COPD Overlap (ACO). Respir. Res. 2020, 21, 126. [Google Scholar] [CrossRef]
- Cruickshank-Quinn, C.I.; Jacobson, S.; Hughes, G.; Powell, R.L.; Petrache, I.; Kechris, K.; Bowler, R.; Reisdorph, N. Metabolomics and Transcriptomics Pathway Approach Reveals Outcome-Specific Perturbations in COPD. Sci. Rep. 2018, 8, 17132. [Google Scholar] [CrossRef]
- Paris, D.; Palomba, L.; Tramice, A.; Motta, L.; Fuschillo, S.; Maniscalco, M.; Motta, A. Identification of Biomarkers in COPD by Metabolomics of Exhaled Breath Condensate and Serum/Plasma. Minerva Med. 2022, 113, 424–435. [Google Scholar] [CrossRef]
- Esther, C.R.; O’Neal, W.K.; Anderson, W.H.; Kesimer, M.; Ceppe, A.; Doerschuk, C.M.; Alexis, N.E.; Hastie, A.T.; Barr, R.G.; Bowler, R.P.; et al. Subpopulations and Intermediate Outcome Measures in COPD Study Identification of Sputum Biomarkers Predictive of Pulmonary Exacerbations in COPD. Chest 2022, 161, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liang, Q.; Frost-Pineda, K.; Muhammad-Kah, R.; Rimmer, L.; Roethig, H.; Mendes, P.; Sarkar, M. Relationship between Biomarkers of Cigarette Smoke Exposure and Biomarkers of Inflammation, Oxidative Stress, and Platelet Activation in Adult Cigarette Smokers. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Kilk, K.; Aug, A.; Ottas, A.; Soomets, U.; Altraja, S.; Altraja, A. Phenotyping of Chronic Obstructive Pulmonary Disease Based on the Integration of Metabolomes and Clinical Characteristics. Int. J. Mol. Sci. 2018, 19, 666. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Kolmert, J.; Yang, M.; Reinke, S.N.; Kamleh, M.A.; Snowden, S.; Heyder, T.; Levänen, B.; Erle, D.J.; Sköld, C.M.; et al. Metabolomics Analysis Identifies Sex-Associated Metabotypes of Oxidative Stress and the Autotaxin-lysoPA Axis in COPD. Eur. Respir. J. 2017, 49, 1602322. [Google Scholar] [CrossRef]
- Ausín, P.; Martínez-Llorens, J.; Sabaté-Bresco, M.; Casadevall, C.; Barreiro, E.; Gea, J. Sex Differences in Function and Structure of the Quadriceps Muscle in Chronic Obstructive Pulmonary Disease Patients. Chron. Respir. Dis. 2017, 14, 127–139. [Google Scholar] [CrossRef]
- Lopez Varela, M.V.; Montes de Oca, M.; Halbert, R.J.; Muiño, A.; Perez-Padilla, R.; Tálamo, C.; Jardim, J.R.B.; Valdivia, G.; Pertuzé, J.; Moreno, D.; et al. PLATINO Team Sex-Related Differences in COPD in Five Latin American Cities: The PLATINO Study. Eur. Respir. J. 2010, 36, 1034–1041. [Google Scholar] [CrossRef]
- Holz, O.; DeLuca, D.S.; Roepcke, S.; Illig, T.; Weinberger, K.M.; Schudt, C.; Hohlfeld, J.M. Smokers with COPD Show a Shift in Energy and Nitrogen Metabolism at Rest and During Exercise. Int. J. Chron. Obstruct. Pulmon. Dis. 2020, 15, 1–13. [Google Scholar] [CrossRef]
- Ambrose, J.A.; Barua, R.S. The Pathophysiology of Cigarette Smoking and Cardiovascular Disease: An Update. J. Am. Coll. Cardiol. 2004, 43, 1731–1737. [Google Scholar] [CrossRef]
- James, R.W.; Leviev, I.; Righetti, A. Smoking Is Associated with Reduced Serum Paraoxonase Activity and Concentration in Patients with Coronary Artery Disease. Circulation 2000, 101, 2252–2257. [Google Scholar] [CrossRef]
- Jiang, Z.; Knudsen, N.H.; Wang, G.; Qiu, W.; Naing, Z.Z.C.; Bai, Y.; Ai, X.; Lee, C.-H.; Zhou, X. Genetic Control of Fatty Acid β-Oxidation in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2017, 56, 738–748. [Google Scholar] [CrossRef]
- Madsen, C.; Nafstad, P.; Eikvar, L.; Schwarze, P.E.; Rønningen, K.S.; Haaheim, L.L. Association between Tobacco Smoke Exposure and Levels of C-Reactive Protein in the Oslo II Study. Eur. J. Epidemiol. 2007, 22, 311–317. [Google Scholar] [CrossRef]
- Vassalle, C.; Petrozzi, L.; Botto, N.; Andreassi, M.G.; Zucchelli, G.C. Oxidative Stress and Its Association with Coronary Artery Disease and Different Atherogenic Risk Factors. J. Intern. Med. 2004, 256, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Yasue, H.; Hirai, N.; Mizuno, Y.; Harada, E.; Itoh, T.; Yoshimura, M.; Kugiyama, K.; Ogawa, H. Low-Grade Inflammation, Thrombogenicity, and Atherogenic Lipid Profile in Cigarette Smokers. Circ. J. 2006, 70, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Gillenwater, L.A.; Pratte, K.A.; Hobbs, B.D.; Cho, M.H.; Zhuang, Y.; Halper-Stromberg, E.; Cruickshank-Quinn, C.; Reisdorph, N.; Petrache, I.; Labaki, W.W.; et al. Plasma Metabolomic Signatures of Chronic Obstructive Pulmonary Disease and the Impact of Genetic Variants on Phenotype-Driven Modules. Netw. Syst. Med. 2020, 3, 159–181. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Barr, D.B.; Ryan, P.B.; Fedirko, V.; Sarnat, J.A.; Gaskins, A.J.; Chang, C.-J.; Tang, Z.; Marsit, C.J.; Corwin, E.J.; et al. High-Resolution Metabolomics of Exposure to Tobacco Smoke during Pregnancy and Adverse Birth Outcomes in the Atlanta African American Maternal-Child Cohort. Environ. Pollut. Barking Essex 1987 2022, 292, 118361. [Google Scholar] [CrossRef]
- Mastej, E.; Gillenwater, L.; Zhuang, Y.; Pratte, K.A.; Bowler, R.P.; Kechris, K. Identifying Protein-Metabolite Networks Associated with COPD Phenotypes. Metabolites 2020, 10, 124. [Google Scholar] [CrossRef]
- Chen, Q.; Deeb, R.S.; Ma, Y.; Staudt, M.R.; Crystal, R.G.; Gross, S.S. Serum Metabolite Biomarkers Discriminate Healthy Smokers from COPD Smokers. PLoS ONE 2015, 10, e0143937. [Google Scholar] [CrossRef]
- Frigerio, G.; Mercadante, R.; Campo, L.; Polledri, E.; Boniardi, L.; Olgiati, L.; Missineo, P.; Nash, W.J.; Dunn, W.B.; Fustinoni, S. Urinary Biomonitoring of Subjects with Different Smoking Habits. Part II: An Untargeted Metabolomic Approach and the Comparison with the Targeted Measurement of Mercapturic Acids. Toxicol. Lett. 2020, 329, 56–66. [Google Scholar] [CrossRef]
- Zhao, M.; Fan, K.; Wang, J.; Wang, J.; Xu, Q.; Wei, D.; Chen, Y.; Zhou, L.; Mao, Z.; Chen, T. Lipidomic Analysis Reveals the Effect of Passive Smoking on Facial Skin Surface Lipid in Females. Chem. Phys. Lipids 2022, 247, 105228. [Google Scholar] [CrossRef]
- Zhu, H.; Abdullah, A.S.; He, J.; Xi, J.; Mao, Y.; Feng, Y.; Xiao, Q.; Zheng, P. Untargeted Urinary Metabolomics and Children’s Exposure to Secondhand Smoke: The Influence of Individual Differences. Int. J. Environ. Res. Public. Health 2021, 18, 710. [Google Scholar] [CrossRef] [PubMed]
- Ohmomo, H.; Harada, S.; Komaki, S.; Ono, K.; Sutoh, Y.; Otomo, R.; Umekage, S.; Hachiya, T.; Katanoda, K.; Takebayashi, T.; et al. DNA Methylation Abnormalities and Altered Whole Transcriptome Profiles after Switching from Combustible Tobacco Smoking to Heated Tobacco Products. Cancer Epidemiol. Biomark. Prev. 2022, 31, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Bradicich, M.; Schuurmans, M.M. Smoking Status and Second-Hand Smoke Biomarkers in COPD, Asthma and Healthy Controls. ERJ Open Res. 2020, 6, 192–2019. [Google Scholar] [CrossRef]
- Behr, C.M.; Koffijberg, H.; Degeling, K.; Vliegenthart, R.; IJzerman, M.J. Can We Increase Efficiency of CT Lung Cancer Screening by Combining with CVD and COPD Screening? Results of an Early Economic Evaluation. Eur. Radiol. 2022, 32, 3067–3075. [Google Scholar] [CrossRef]
- Kotaki, K.; Ikeda, H.; Fukuda, T.; Yuki, F.; Hasuo, K.; Kawano, Y.; Kawasaki, M. Effectiveness of Diagnostic Screening Tests in Mass Screening for COPD Using a Cooperative Regional System in a Region with Heavy Air Pollution: A Cross-Sectional Study. BMJ Open 2017, 7, e012923. [Google Scholar] [CrossRef] [PubMed]
- Llordés, M.; Zurdo, E.; Jaén, Á.; Vázquez, I.; Pastrana, L.; Miravitlles, M. Which Is the Best Screening Strategy for COPD among Smokers in Primary Care? COPD J. Chronic Obstr. Pulm. Dis. 2017, 14, 43–51. [Google Scholar] [CrossRef]
- Tamaki, K.; Sakihara, E.; Miyata, H.; Hirahara, N.; Kirichek, O.; Tawara, R.; Akiyama, S.; Katsumata, M.; Haruya, M.; Ishii, T.; et al. Utility of Self-Administered Questionnaires for Identifying Individuals at Risk of COPD in Japan: The OCEAN (Okinawa COPD casE Finding AssessmeNt) Study. Int. J. Chron. Obstruct. Pulmon. Dis. 2021, 16, 1771–1782. [Google Scholar] [CrossRef]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef]
- Shramko, V.S.; Polonskaya, Y.V.; Kashtanova, E.V.; Stakhneva, E.M.; Ragino, Y.I. The Short Overview on the Relevance of Fatty Acids for Human Cardiovascular Disorders. Biomolecules 2020, 10, 1127. [Google Scholar] [CrossRef]
- Hoffer, L.J.; Taveroff, A.; Robitaille, L.; Mamer, O.A.; Reimer, M.L. Alpha-Keto and Alpha-Hydroxy Branched-Chain Acid Interrelationships in Normal Humans. J. Nutr. 1993, 123, 1513–1521. [Google Scholar] [CrossRef]
- Brosnan, M.E.; Brosnan, J.T. Histidine Metabolism and Function. J. Nutr. 2020, 150, 2570S–2575S. [Google Scholar] [CrossRef]
- Anastassiadis, S.; Morgunov, I.G. Gluconic Acid Production. Recent Pat. Biotechnol. 2007, 1, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.A.; Reece, J.B.; Urry, L.A.; Cain, M.L.; Wasserman, S.A.; Minorsky, P.V.; Jackson, R.B. Biology, 8th ed.; Pearson Benjamin Cummings: San Francisco, CA, USA, 2008; ISBN 9780558230647. [Google Scholar]
- Jimoh, T.A.; Omoarukhe, F.O.; Epelle, E.I.; Okoye, P.U.; Oke Olusola, E.; Mukherjee, A.; Okolie, J.A. Introduction to Carbon Capture by Solvent-based Technologies. In Encyclopedia of Renewable Energy, Sustainability and the Environment; Rahimpour, M.R., Ed.; Elsevier: Oxford, UK, 2024; Volume 4, pp. 511–524. ISBN 978-0-323-93941-6. [Google Scholar]
- MacKay, D.; Hathcock, J.; Guarneri, E. Niacin: Chemical Forms, Bioavailability, and Health Effects. Nutr. Rev. 2012, 70, 357–366. [Google Scholar] [CrossRef]
- Gea, J.; Pascual, S.; Castro-Acosta, A.; Hernández-Carcereny, C.; Castelo, R.; Márquez-Martín, E.; Montón, C.; Palou, A.; Faner, R.; Furlong, L.I.; et al. The BIOMEPOC Project: Personalized Biomarkers and Clinical Profiles in Chronic Obstructive Pulmonary Disease. Arch. Bronconeumol. 2019, 55, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Borràs-Santos, A.; Garcia-Aymerich, J.; Soler-Cataluña, J.J.; Vigil Giménez, L.; Gea Guiral, J.; Rodríguez Chiaradía, D.; Pascual-Guardia, S.; Marcos Rodríguez, P.J.; Alvarez Martinez, C.J.; Casanova Macario, C.; et al. Determinantes de la aparición y progresión de la enfermedad pulmonar obstructiva crónica en adultos jóvenes. Protocolo de un estudio caso-control con seguimiento. Arch. Bronconeumol. 2019, 55, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Global Initiative for Chronic Obstructive Lung Disease Global Strategy for the Diagnosis, Management and Prevention of Chronic Obstructive Pulmonary Disesease; Global Initiative for Chronic Obstructive Lung Disease: Deer Park, IL, USA, 2024; ISBN 979-8-8839-2147-5.
- Mackay, G.M.; Zheng, L.; van den Broek, N.J.F.; Gottlieb, E. Analysis of Cell Metabolism Using LC-MS and Isotope Tracers. Methods Enzymol. 2015, 561, 171–196. [Google Scholar] [CrossRef]
- Ghrayeb, A.; Finney, A.C.; Agranovich, B.; Peled, D.; Anand, S.K.; McKinney, M.P.; Sarji, M.; Yang, D.; Weissman, N.; Drucker, S.; et al. Serine Synthesis via Reversed SHMT2 Activity Drives Glycine Depletion and Acetaminophen Hepatotoxicity in MASLD. Cell Metab. 2024, 36, 116–129.e7. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a Unified Platform for Metabolomics Data Processing, Analysis and Interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Hsieh, S.J.; Ware, L.B.; Eisner, M.D.; Yu, L.; Jacob, P.; Havel, C.; Goniewicz, M.L.; Matthay, M.A.; Benowitz, N.L.; Calfee, C.S. Biomarkers Increase Detection of Active Smoking and Secondhand Smoke Exposure in Critically Ill Patients. Crit. Care Med. 2011, 39, 40–45. [Google Scholar] [CrossRef]
| HC (N = 91) | COPD (N = 91) | |
|---|---|---|
| Age, median (IQR), yr. | 48 (44–61) | 50 (46–66) |
| Male, N (%) | 50 (55) | 46 (51) |
| Body Mass Index (kg/m2), mean ± SD | 26.5 ± 4.4 | 26.1 ± 5.5 |
| Current or former smokers, N (%) | 91 (100) | 91 (100) |
| Pack Years, median (IQR) | 23 (15–32) | 39 (24–60) *** |
| Post-BD FEV1 (% pred.), median (IQR) | 95 (85–104) | 56 (43–76) *** |
| FEV1/FVC ratio, median (IQR) | 82 (74–87) | 56 (40–65) *** |
| DLco, median (IQR) | 85 (79–96) | 56 (43–76) *** |
| GOLD class, N (%) | ||
| GOLD 1 | - | 19 (21) |
| GOLD 2 | - | 37 (41) |
| GOLD 3 | - | 24 (26) |
| GOLD 4 | - | 11 (12) |
| Compound Name | logFC | HMDB | Chemical Taxonomy (Super Class) | Chemical Taxonomy (Sub Class) | KEGG Pathways |
|---|---|---|---|---|---|
| Hexadecanoic acid (Palmitic acid) | −0.057507 | HMDB0000220 | Lipids and lipid-like molecules | Fatty acids and conjugates (LCFA) | Fatty acid Metabolism |
| Glyceric acid | −0.32747 | HMDB0000139 | Organic Oxygen Compounds | Carbohydrates and their conjugates | Pentose phosphate pathway |
| Urocanate | −0.45756 | HMDB0000301 | Organoheterocyclic Compounds | Imidazoles | Histidine metabolism |
| 2-aminonicotinic acid | −0.32725 | HMDB0061680 | Organoheterocyclic Compounds | Pyridinecarboxylic acids and derivatives | |
| 2-Hydroxyisocaproic acid (Leucic acid) | 0.33133 | HMDB0000665 | Lipids and Lipid-like Molecules | Fatty acids and conjugates (MC Hydroxy fatty acid) | Fatty acid Metabolism |
| Diethanolamine | 0.16912 | HMDB0004437 | Organic Nitrogen Compounds | Amines | Glycerophospholipid metabolism |
| 1-Tetradecylamine | 3.0246 | HMDB0258887 | Organic Nitrogen Compounds | Amines | |
| Pentapropylene glycol (PPG n5) | 0.5634 | Organic Oxygen Compounds | Alcohols and polyols | ||
| 2-Naphthalenesulfonic acid | −1.0249 | HMDB0255446 | Benzenoids | Naphthalene sulfonic acids and derivatives | |
| 4-Dodecylbenzenesulfonic acid | −0.85833 | HMDB0059915 | Benzenoids | Benzene sulfonic acids and derivatives |
| Compound Name | logFC | HMDB | Chemical Taxonomy (Super Class) | Chemical Taxonomy (Sub Class) | KEGG Pathways |
|---|---|---|---|---|---|
| Hexadecanoic acid (Palmitic acid) | −0.057507 | HMDB0000220 | Lipids and Lipid-like Molecules | Fatty acids and conjugates (LCFA) | Fatty acid Metabolism |
| Glyceric acid | −0.32747 | HMDB0000139 | Organic Oxygen Compounds | Carbohydrates and Carbohydrate conjugates | Pentose phosphate pathway |
| Urocanate | −0.45756 | HMDB0000301 | Organoheterocyclic Compounds | Imidazoles | Histidine metabolism |
| 2-Aminonicotinic acid | −0.32725 | HMDB0061680 | Organoheterocyclic Compounds | Pyridinecarboxylic acids and derivatives | |
| 2-Hydroxyisocaproic acid (Leucic acid) | 0.33133 | HMDB0000665 | Lipids and Lipid-like Molecules | Fatty acids and conjugates (MC Hydroxy fatty acid) | Fatty acid Metabolism |
| Diethanolamine | 0.16912 | HMDB0004437 | Organic Nitrogen Compounds | Amines | Glycerophospholipid Metabolism |
| Gluconic acid | 0.24119 | HMDB0000625 | Organic Oxygen Compounds | Carbohydrates and their conjugates | Pentose phosphate pathway |
| 2-Hydroxytetradecanoic acid (2-Hydroxymyristic acid) | −0.20222 | HMDB0002261 | Lipids and Lipid-like Molecules | Fatty acids and conjugates (LC Hydroxy fatty acid) | Fatty acid Metabolism |
| 14-Methylhexadecanoic acid | −0.12759 | HMDB0031067 | Lipids and Lipid-like Molecules | Fatty acids and conjugates (LC chain Methyl fatty acid) | Fatty acid Metabolism |
| N-Methylglutamate | 0.14171 | HMDB0062660 | Organic Acids and Derivatives | Amino acids, peptides and analogues | Derivative of Glutamic acid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casadevall, C.; Agranovich, B.; Enríquez-Rodríguez, C.J.; Faner, R.; Pascual-Guàrdia, S.; Castro-Acosta, A.; Camps-Ubach, R.; Garcia-Aymerich, J.; Barreiro, E.; Monsó, E.; et al. Metabolomic Plasma Profile of Chronic Obstructive Pulmonary Disease Patients. Int. J. Mol. Sci. 2025, 26, 4526. https://doi.org/10.3390/ijms26104526
Casadevall C, Agranovich B, Enríquez-Rodríguez CJ, Faner R, Pascual-Guàrdia S, Castro-Acosta A, Camps-Ubach R, Garcia-Aymerich J, Barreiro E, Monsó E, et al. Metabolomic Plasma Profile of Chronic Obstructive Pulmonary Disease Patients. International Journal of Molecular Sciences. 2025; 26(10):4526. https://doi.org/10.3390/ijms26104526
Chicago/Turabian StyleCasadevall, Carme, Bella Agranovich, Cesar Jesse Enríquez-Rodríguez, Rosa Faner, Sergi Pascual-Guàrdia, Ady Castro-Acosta, Ramon Camps-Ubach, Judith Garcia-Aymerich, Esther Barreiro, Eduard Monsó, and et al. 2025. "Metabolomic Plasma Profile of Chronic Obstructive Pulmonary Disease Patients" International Journal of Molecular Sciences 26, no. 10: 4526. https://doi.org/10.3390/ijms26104526
APA StyleCasadevall, C., Agranovich, B., Enríquez-Rodríguez, C. J., Faner, R., Pascual-Guàrdia, S., Castro-Acosta, A., Camps-Ubach, R., Garcia-Aymerich, J., Barreiro, E., Monsó, E., Seijo, L., Soler-Cataluña, J. J., Santos, S., Peces-Barba, G., López-Campos, J. L., Casanova, C., Agustí, A., Cosío, B. G., Abramovich, I., & Gea, J., on behalf of the EARLY COPD and BIOMEPOC Groups. (2025). Metabolomic Plasma Profile of Chronic Obstructive Pulmonary Disease Patients. International Journal of Molecular Sciences, 26(10), 4526. https://doi.org/10.3390/ijms26104526









