Novel FANCI and RAD54B Variants and the Observed Clinical Outcomes in a Hungarian Melanoma Cohort
Abstract
1. Introduction
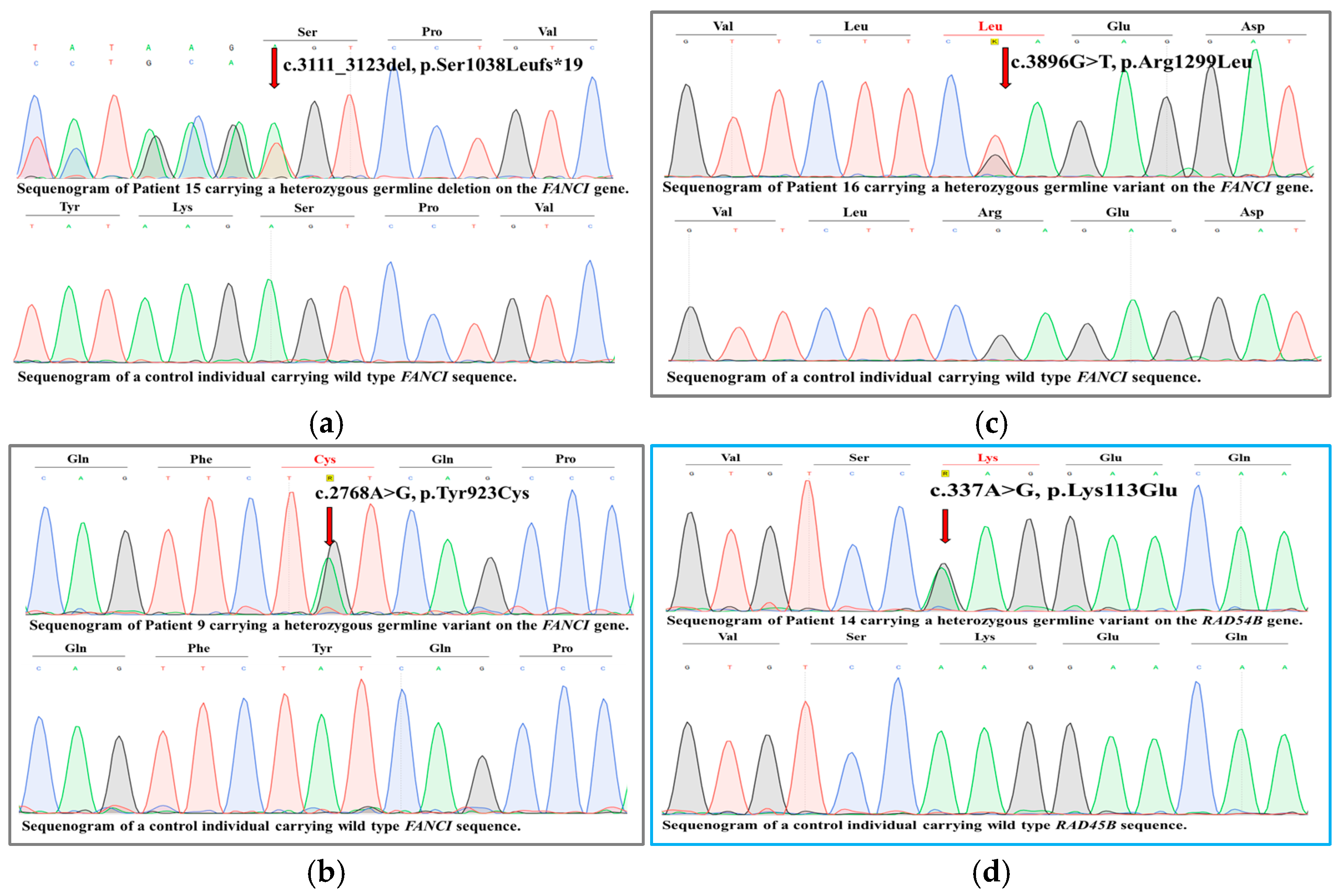
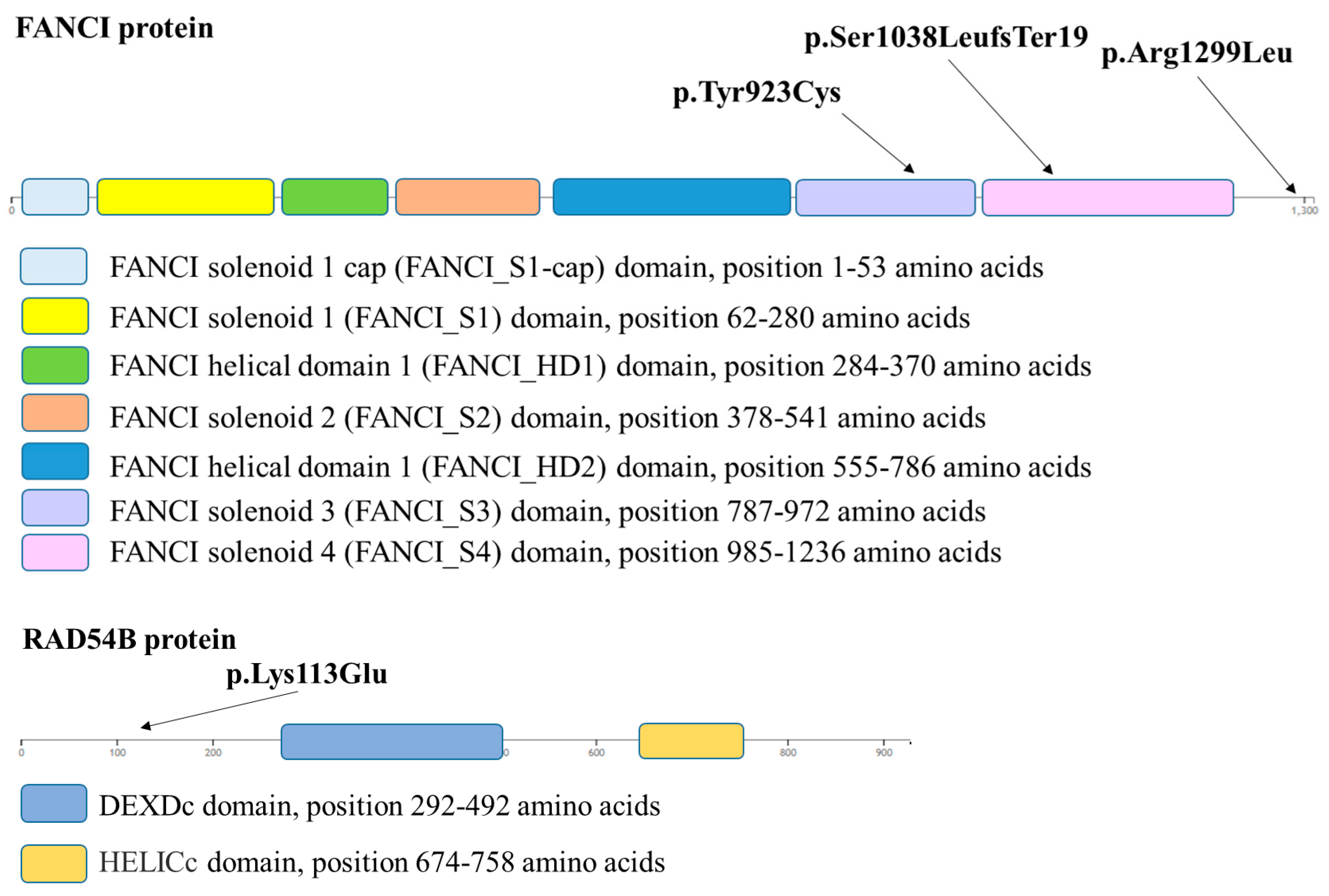
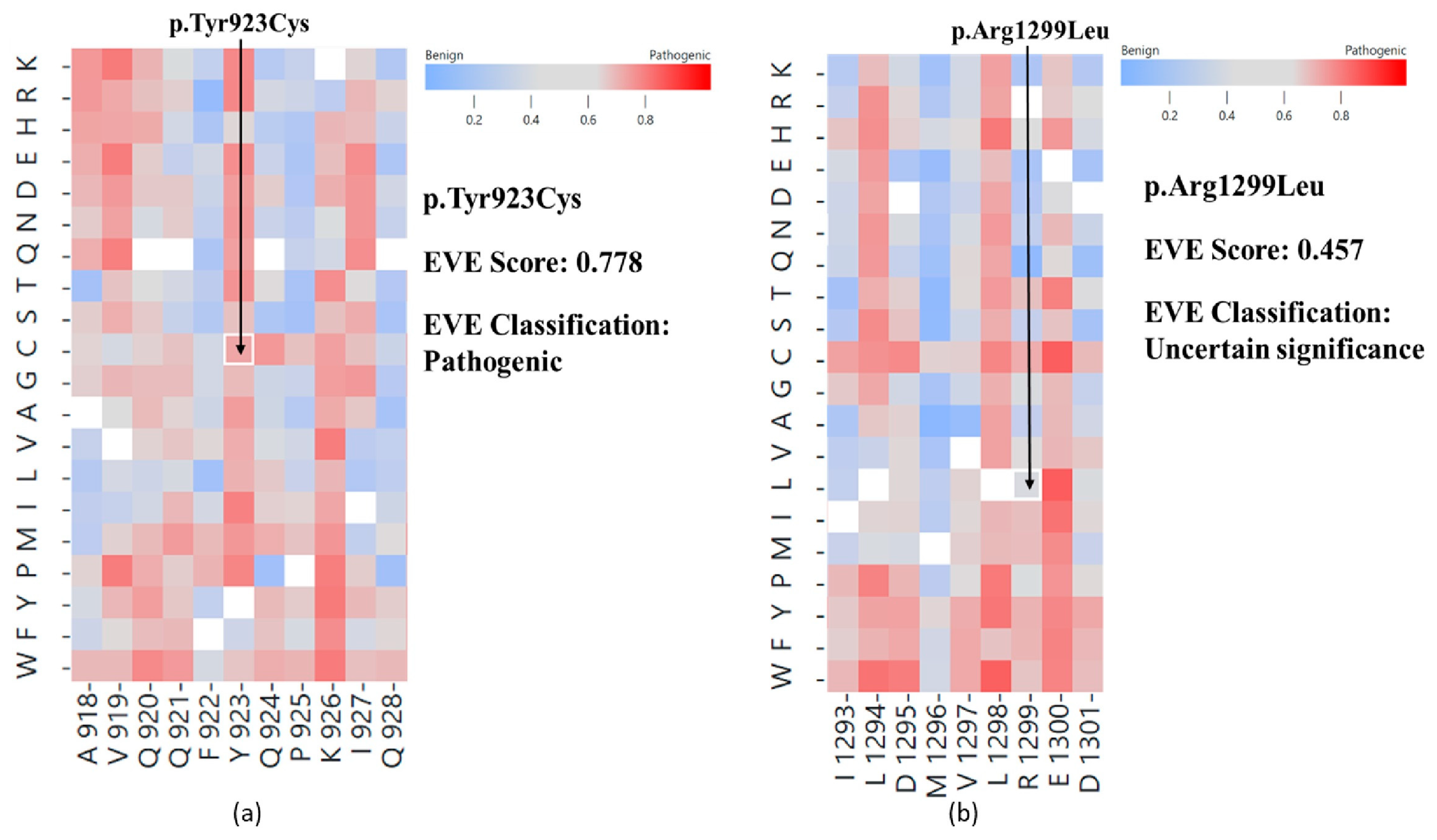
2. Results
2.1. In Silico Variant Analysis
2.2. Clinical Outcomes
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Targeted Next-Generation Sequencing with a Virtual Gene Panel
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Read, J.; Wadt, K.A.; Hayward, N.K. Melanoma genetics. J. Med. Genet. 2016, 53, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Soura, E.; Eliades, P.J.; Shannon, K.; Stratigos, A.J.; Tsao, H. Hereditary melanoma: Update on syndromes and management: Genetics of familial atypical multiple mole melanoma syndrome. J. Am. Acad. Dermatol. 2016, 74, 395–407. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nan, H.; Qureshi, A.A.; Han, J. Melanoma susceptibility variants on chromosome 20q11.22 are associated with pigmentary traits and the risk of nonmelanoma skin cancer. Br. J. Dermatol. 2010, 162, 461–463. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Funchain, P.; Ni, Y.; Heald, B.; Bungo, B.; Arbesman, M.; Behera, T.R.; McCormick, S.; Song, J.M.; Kennedy, L.B.; Nielsen, S.M.; et al. Germline cancer susceptibility in individuals with melanoma. J. Am. Acad. Dermatol. 2024, 91, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Bokor, B.A.; Abdolreza, A.; Kaptás, F.; Pál, M.; Battyani, Z.; Széll, M.; Nagy, N. Novel variants in medium and low penetrance predisposing genes in a Hungarian malignant melanoma cohort with increased risk. Pigment. Cell Melanoma Res. 2024, in press. [Google Scholar] [CrossRef] [PubMed]
- Amaral, T.; Schulze, M.; Sinnberg, T.; Nieser, M.; Martus, P.; Battke, F.; Garbe, C.; Biskup, S.; Forschner, A. Are Pathogenic Germline Variants in Metastatic Melanoma Associated with Resistance to Combined Immunotherapy? Cancers 2020, 12, 1101. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Song, H.; Dicks, E.M.; Tyrer, J.; Intermaggio, M.; Chenevix-Trench, G.; Bowtell, D.D.; Traficante, N.; Group, A.; Brenton, J.; Goranova, T.; et al. Population-based targeted sequencing of 54 candidate genes identifies PALB2 as a susceptibility gene for high-grade serous ovarian cancer. J. Med. Genet. 2021, 58, 305–313. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liede, A.; Karlan, B.Y.; Narod, S.A. Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: A review of the literature. J. Clin. Oncol. 2004, 22, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.; Teule, A.; Caminal, J.M.; Blanco, I.; Piulats, J.M. Uveal melanoma and BRCA1/BRCA2 genes: A relationship that needs further investigation. J. Clin. Oncol. 2011, 29, e827–e829. [Google Scholar] [CrossRef] [PubMed]
- Mersch, J.; Jackson, M.A.; Park, M.; Nebgen, D.; Peterson, S.K.; Singletary, C.; Arun, B.K.; Litton, J.K. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 2015, 121, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.A.; Nathan, V.; Bourke, L.M.; Palmer, J.M.; Zhang, T.; Symmons, J.; Howlie, M.; Patch, A.M.; Read, J.; Holland, E.A.; et al. Evaluation of the contribution of germline variants in BRCA1 and BRCA2 to uveal and cutaneous melanoma. Melanoma Res. 2019, 29, 483–490. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aoude, L.G.; Heitzer, E.; Johansson, P.; Gartside, M.; Wadt, K.; Pritchard, A.L.; Palmer, J.M.; Symmons, J.; Gerdes, A.M.; Montgomery, G.W.; et al. POLE mutations in families predisposed to cutaneous melanoma. Fam. Cancer 2015, 14, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Stolarova, L.; Jelinkova, S.; Storchova, R.; Machackova, E.; Zemankova, P.; Vocka, M.; Kodet, O.; Kral, J.; Cerna, M.; Volkova, Z.; et al. Identification of Germline Mutations in Melanoma Patients with Early Onset, Double Primary Tumors, or Family Cancer History by NGS Analysis of 217 Genes. Biomedicines 2020, 8, 404. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fidalgo, F.; Torrezan, G.T.; Sá, B.C.S.; Barros, B.D.F.; Moredo, L.F.; Valieris, R.; de Souza, S.J.; Duprat, J.P.; Krepischi, A.C.V.; Carraro, D.M. Family-based whole-exome sequencing identifies rare variants potentially related to cutaneous melanoma predisposition in Brazilian melanoma-prone families. PLoS ONE 2022, 17, e0262419. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abdel-Rahman, M.H.; Sample, K.M.; Pilarski, R.; Walsh, T.; Grosel, T.; Kinnamon, D.; Boru, G.; Massengill, J.B.; Schoenfield, L.; Kelly, B.; et al. Whole Exome Sequencing Identifies Candidate Genes Associated with Hereditary Predisposition to Uveal Melanoma. Ophthalmology 2020, 127, 668–678. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vergani, E.; Frigerio, S.; Dugo, M.; Devecchi, A.; Feltrin, E.; De Cecco, L.; Vallacchi, V.; Cossa, M.; Di Guardo, L.; Manoukian, S.; et al. Genetic Variants and Somatic Alterations Associated with MITF-E318K Germline Mutation in Melanoma Patients. Genes. 2021, 12, 1440. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, J.; Cai, X.; Kang, L.; Chen, S.; Liu, H. Identification of novel biomarkers and candidate small-molecule drugs in cutaneous melanoma by comprehensive gene microarrays analysis. J. Cancer 2021, 12, 1307–1317. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cai, Z.; Duan, Y.; Li, W.; Liu, Z.; Gong, Z.; Hong, S.; He, X.; Xuanyuan, X.; Chen, Y.; Bi, X.; et al. FANCI serve as a prognostic biomarker correlated with immune infiltrates in skin cutaneous melanoma. Front. Immunol. 2023, 14, 1295831. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Campos Segura, A.V.; Velásquez Sotomayor, M.B.; Gutiérrez Román, A.I.F.; Ortiz Rojas, C.A.; Murillo Carrasco, A.G. Impact of mini-driver genes in the prognosis and tumor features of colorectal cancer samples: A novel perspective to support current biomarkers. PeerJ 2023, 11, e15410. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Patient No. | Age of Onset | No. of Primary Melanomas | Lymphatic Metastasis | Other Metastasis | Therapy | ||||
|---|---|---|---|---|---|---|---|---|---|
| Excision | Targeted Molecular Therapy | Immuno- Therapy | Radio- Therapy | Chemo-Therapy | |||||
| 1 | 58 | 1 | no | no | yes | no | no | no | no |
| 2 | 76 | 1 | no | no | yes | no | no | no | no |
| 3 | 82 | 1 | no | no | yes | no | no | no | no |
| 4 | 55 | 1 | no | no | yes | no | no | no | no |
| 5 | 26 | 2 | no | no | yes | no | no | no | no |
| 6 | 51 | 3 | yes | no | yes | no | yes | no | no |
| 7 | 44 | 0 | no | no | no | no | no | no | no |
| 8 | 57 | 0 | no | no | no | no | no | no | no |
| 9 | 42 | 1 | no | no | yes | no | no | no | no |
| 10 | 42 | 1 | no | no | yes | no | no | no | no |
| 11 | 51 | 1 | no | no | yes | no | no | no | no |
| 12 | 50 | 1 | no | no | yes | no | yes | no | no |
| 13 | 42 | 1 | no | no | yes | no | no | no | no |
| 14 | 43 | 1 | no | no | yes | no | no | no | no |
| 15 | 31 | 1 | yes | yes, multiple | yes | yes | yes | yes | no |
| 16 | 53 | 0 | no | no | no | no | no | no | no |
| 17 | 40 | 1 | no | no | yes | no | no | no | no |
| Patient No. | pTNM Stage | Breslow Thickness (mm) | Clark Level | Mitotic Rate (mm2) | Presence of Ulceration/ Regression |
|---|---|---|---|---|---|
| 1 | pT2a | 1.28 | III | 1–26 | no |
| 2 | pT1a | 0.5 | II | 0 | regression |
| 3 | NA | NA | NA | NA | NA |
| 4 | pT4a | 6 | IV | 2 | ulceration |
| 5 | pT1a | 0.35 | II | 2 | no |
| pT1a | 0.32 | II | 1 | no | |
| 6 | pT3a | 3.15 | IV | 2–3 | regression |
| pT2a | 2 | III | 2–3 | regression | |
| in situ superficially spreading melanoma | |||||
| 7 | patient is affected by dyspastic naevus syndrome | ||||
| 8 | patient is affected by dysplastic naevus syndrome | ||||
| 9 | pT3a | 0.78 | IV | 2–3 | ulceration |
| 10 | pT1a | 0.75 | III | 2 | no |
| 11 | pT1b | 0.6 | IV | 2 | ulceration |
| 12 | pT2a | 1.9 | IV | 1 | no |
| 13 | pT1a | 0.58 | III | 0 | no |
| 14 | pT4a | 4.7 | III | 3–4 | no |
| 15 | pT1a | 0.8 | II | 0 | no |
| 16 | patient is affected by dyspastic naevus syndrome | ||||
| 17 | pT2a | 1.21 | IV | 2 | no |
| Patient No. | Immune System Disease | Family History of Melanoma | Presence of Any Germline CDKN2A Variant |
|---|---|---|---|
| 1 | unknown | negative | no |
| 2 | unknown | positive | no |
| 3 | unknown | negative | no |
| 4 | unknown | negative | yes, a leaning-pathogenic VUS |
| 5 | unknown | negative | no |
| 6 | unknown | negative | no |
| 7 | unknown | negative | no |
| 8 | ulcerative colitis | negative | no |
| 9 | unknown | positive | no |
| 10 | unknown | negative | no |
| 11 | unknown | negative | no |
| 12 | unknown | negative | no |
| 13 | unknown | negative | no |
| 14 | unknown | negative | no |
| 15 | unknown | negative | no |
| 16 | unknown | positive | no |
| 17 | unknown | negative | no |
| FANCI exon 25 forward primer | TTGTGGGGAGATTACACAACC |
| FANCI exon 25 reverse primer | TCTCAAGTGTCTTCTGGTAGGT |
| FANCI exon 25 forward primer | CAATACCACTTTCTCCTGCTTC |
| FANCI exon 25 reverse primer | CAGCCACTCTTTGTGGTTGA |
| FANCI exon 37 forward primer | GTGCGTGCTTGCTTTAGGTA |
| FANCI exon 37 reverse primer | ATCAAACAAGTCGGGGCAAC |
| RAD54B exon 4 forward primer | TGTGCCTTTTGGTTTTGTTTGAAT |
| RAD54B exon 4 reverse primer | AGATTGTCAGGCTCACTAACCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bokor, B.A.; Abdolreza, A.; Kaptás, F.; Pál, M.; Battyani, Z.; Széll, M.; Nagy, N. Novel FANCI and RAD54B Variants and the Observed Clinical Outcomes in a Hungarian Melanoma Cohort. Int. J. Mol. Sci. 2025, 26, 23. https://doi.org/10.3390/ijms26010023
Bokor BA, Abdolreza A, Kaptás F, Pál M, Battyani Z, Széll M, Nagy N. Novel FANCI and RAD54B Variants and the Observed Clinical Outcomes in a Hungarian Melanoma Cohort. International Journal of Molecular Sciences. 2025; 26(1):23. https://doi.org/10.3390/ijms26010023
Chicago/Turabian StyleBokor, Barbara Anna, Aliasgari Abdolreza, Flóra Kaptás, Margit Pál, Zita Battyani, Márta Széll, and Nikoletta Nagy. 2025. "Novel FANCI and RAD54B Variants and the Observed Clinical Outcomes in a Hungarian Melanoma Cohort" International Journal of Molecular Sciences 26, no. 1: 23. https://doi.org/10.3390/ijms26010023
APA StyleBokor, B. A., Abdolreza, A., Kaptás, F., Pál, M., Battyani, Z., Széll, M., & Nagy, N. (2025). Novel FANCI and RAD54B Variants and the Observed Clinical Outcomes in a Hungarian Melanoma Cohort. International Journal of Molecular Sciences, 26(1), 23. https://doi.org/10.3390/ijms26010023







