AMPK Phosphorylates LMX1b to Regulate a Brainstem Neurogenic Network Important for Control of Breathing in Neonatal Mice
Abstract
1. Introduction
2. Results
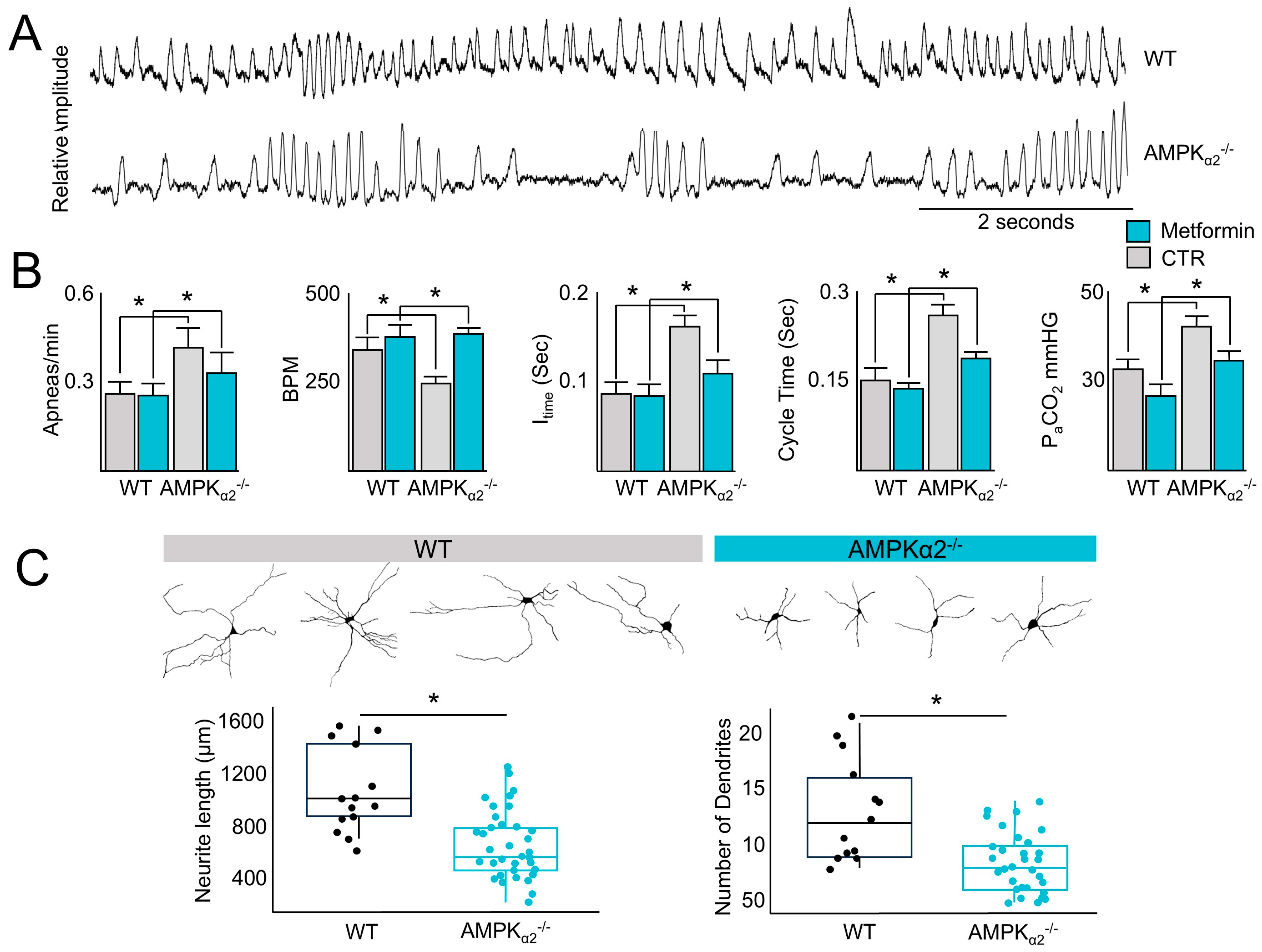
2.1. AMPK Regulates the Control of Ventilation That Can Be Rescued via Metformin Administration
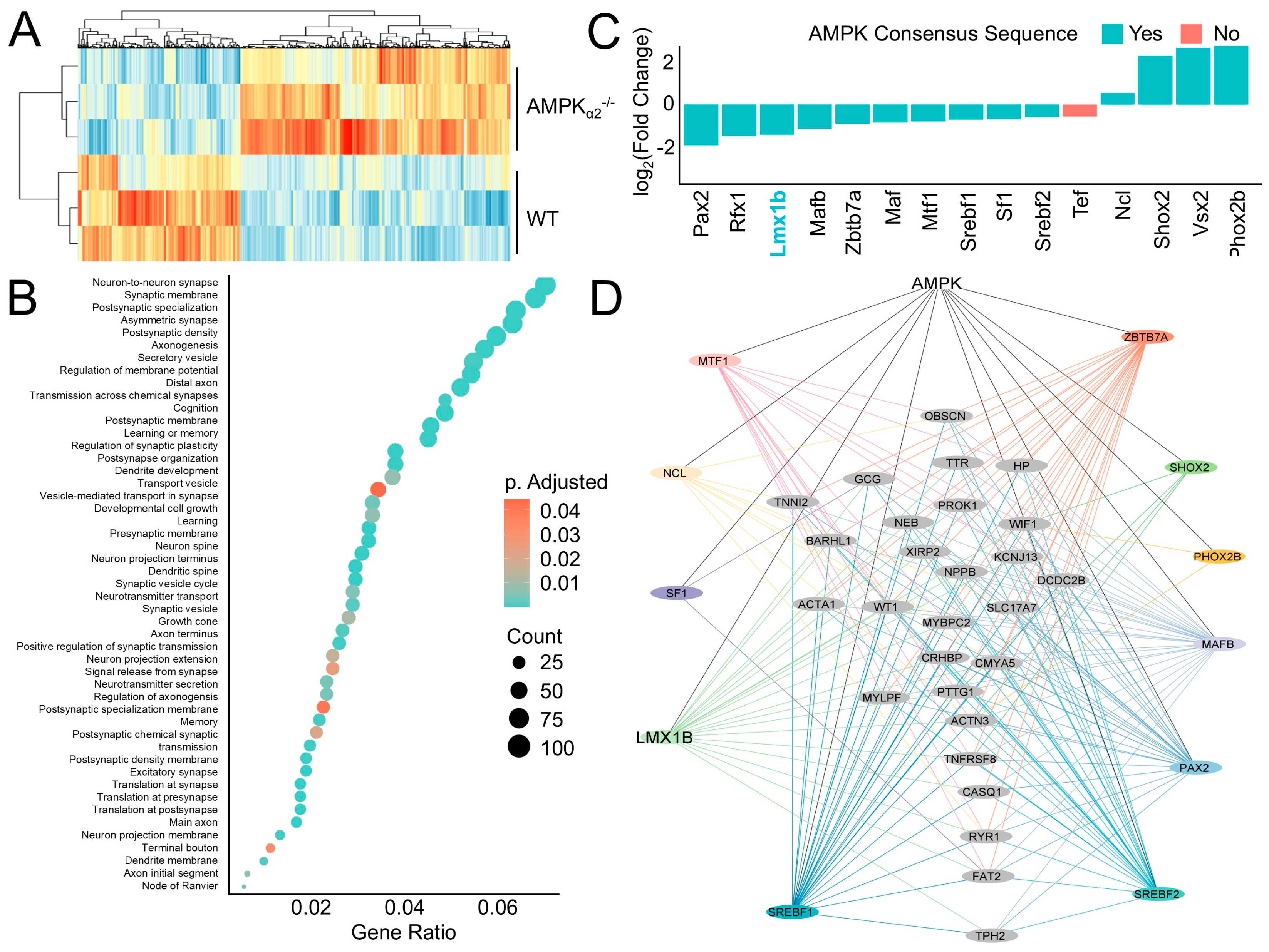
2.2. AMPK Induces a Neurogenic Regulatory Network
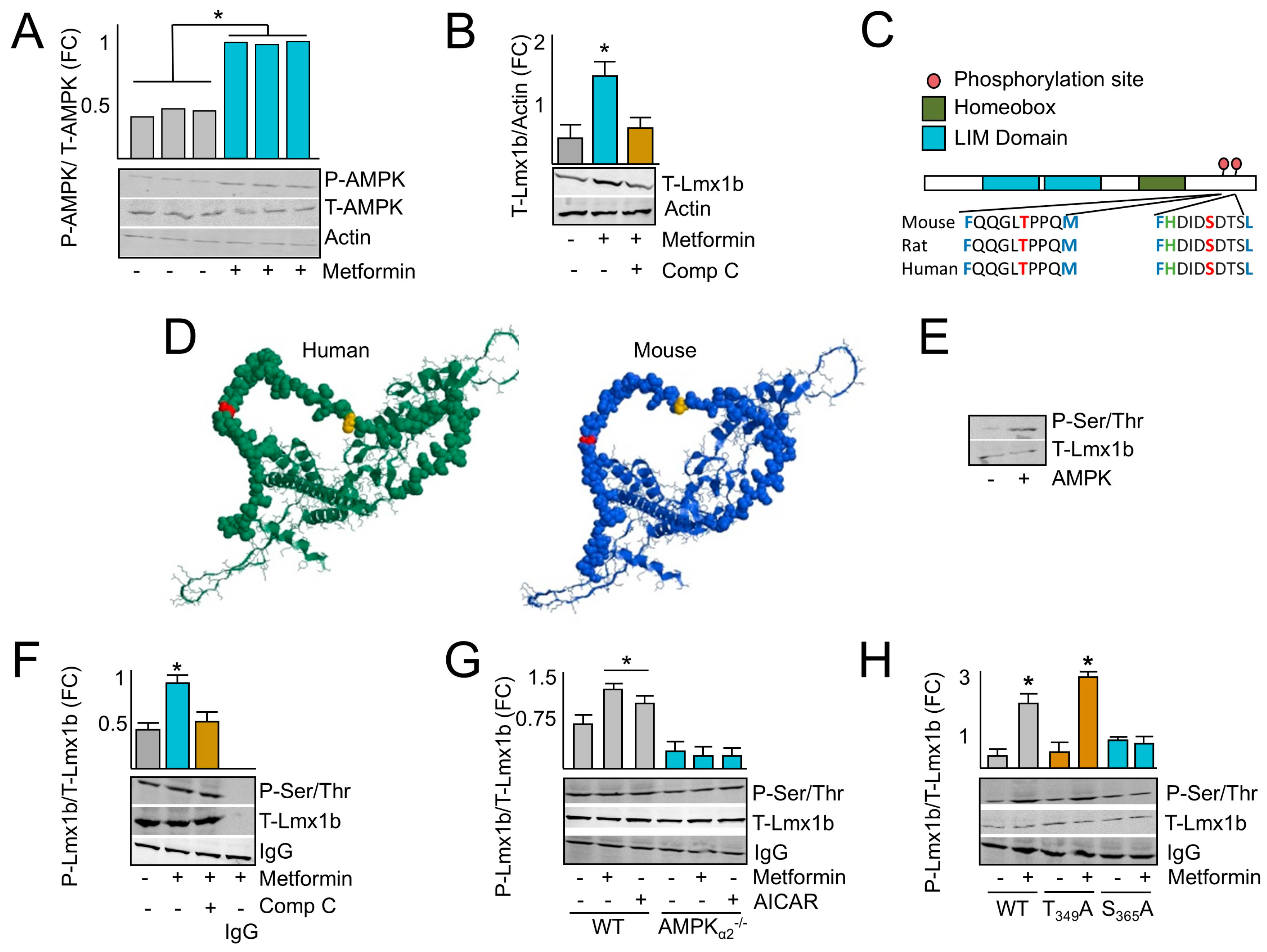
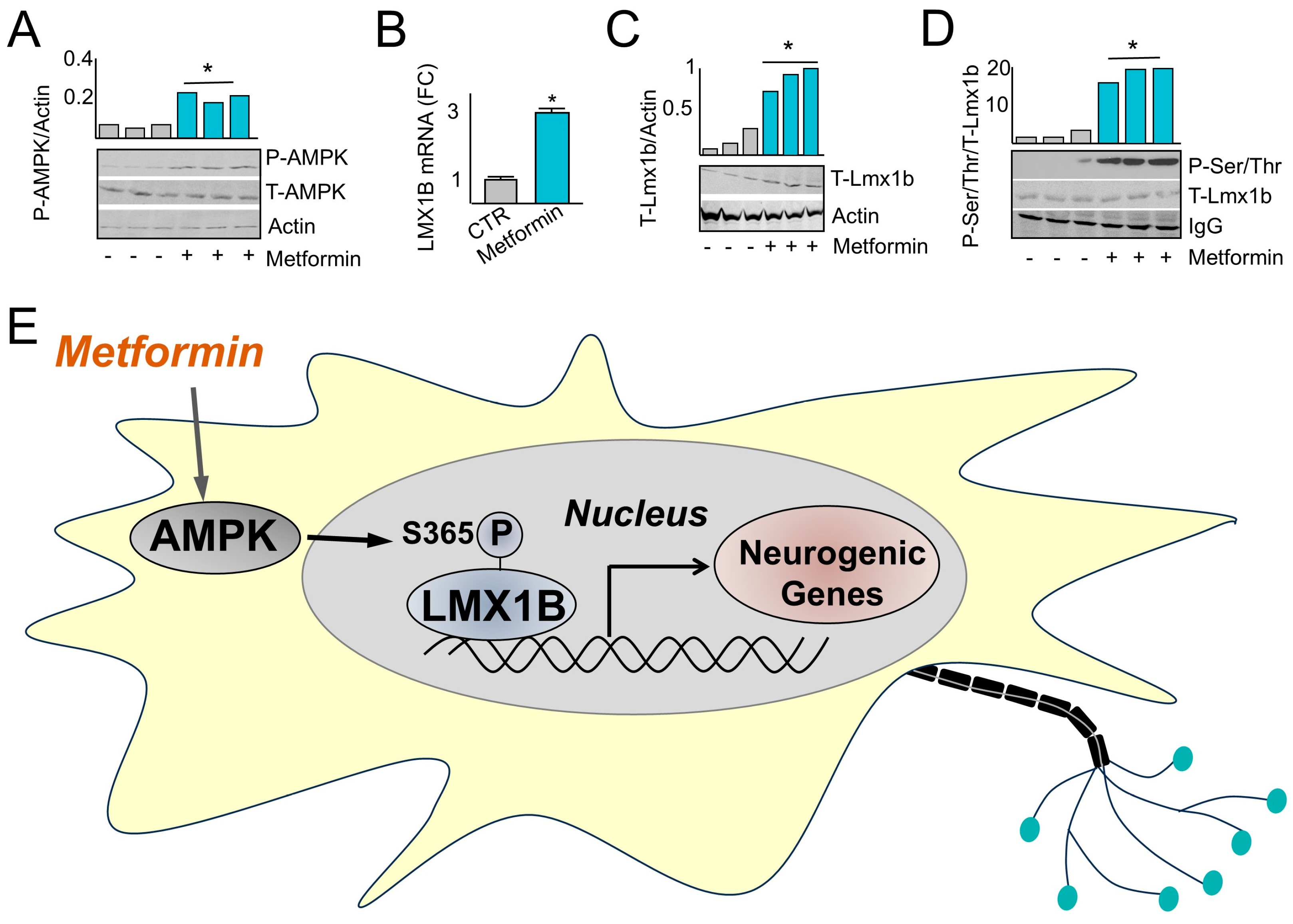
2.3. AMPK Induces LMX1B
2.4. AMPK Phosphorylates LMX1B at Serine 365
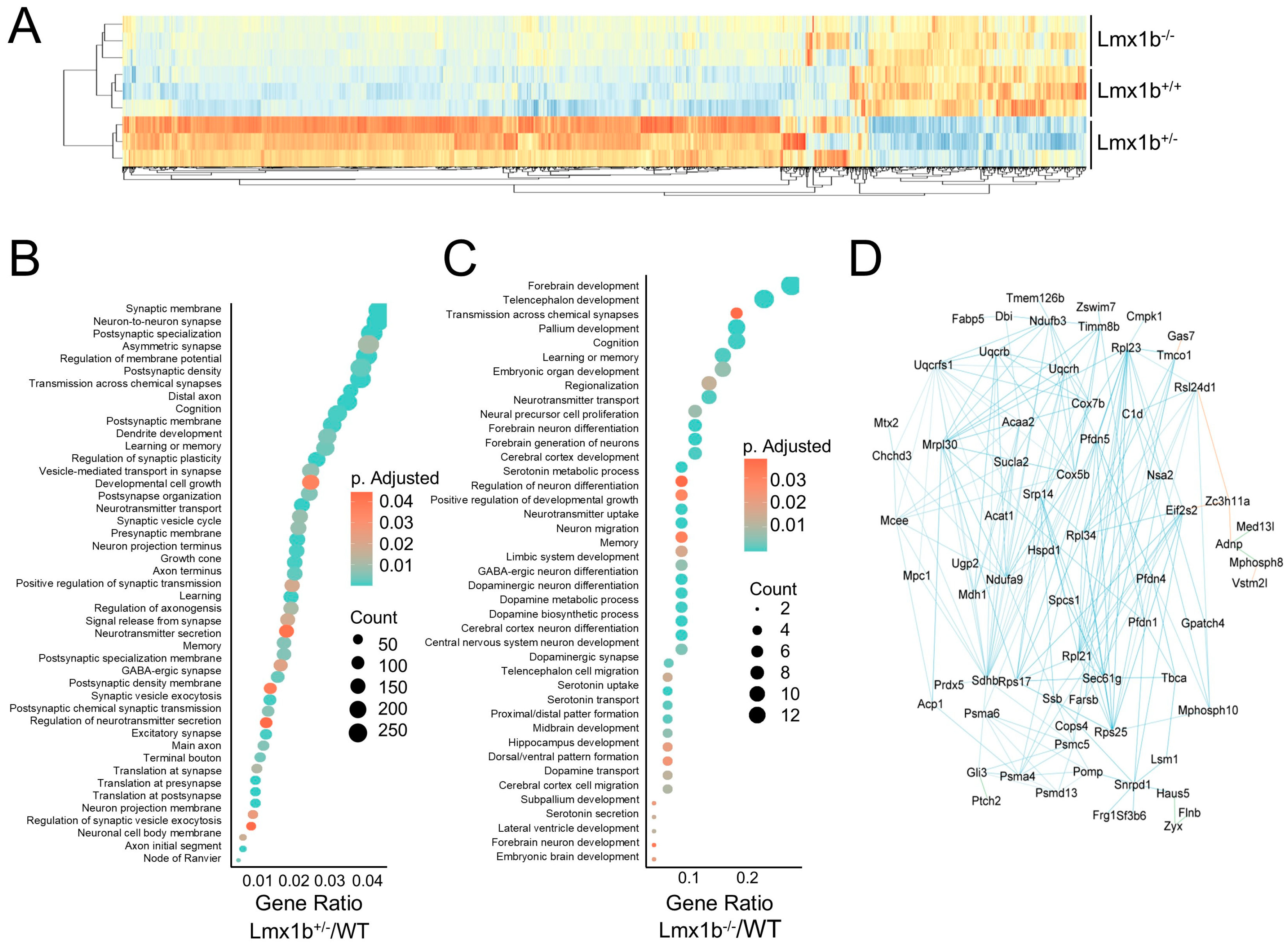
2.5. Lmx1b Governs a Transcriptional Network Important for Ventilatory Center Neuron Maturation
2.6. Metformin Administration Induces Fetal Expression and Phosphorylation of Lmx1b
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohuma, E.O.; Moller, A.B.; Bradley, E.; Chakwera, S.; Hussain-Alkhateeb, L.; Lewin, A.; Okwaraji, Y.B.; Mahanani, W.R.; Johansson, E.W.; Lavin, T.; et al. National, regional, and global estimates of preterm birth in 2020, with trends from 2010: A systematic analysis. Lancet 2023, 402, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Eichenwald, E.C. Committee on Fetus and Newborn; American Academy of Pediatrics. Apnea of Prematurity. Pediatrics 2016, 137, e20153757. [Google Scholar] [CrossRef]
- Williamson, M.; Poorun, R.; Hartley, C. Apnoea of Prematurity and Neurodevelopmental Outcomes: Current Understanding and Future Prospects for Research. Front. Pediatr. 2021, 9, 755677. [Google Scholar] [CrossRef] [PubMed]
- Alheid, G.F.; McCrimmon, D.R. The chemical neuroanatomy of breathing. Respir. Physiol. Neurobiol. 2008, 164, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Erickson, G.; Dobson, N.R.; Hunt, C.E. Immature control of breathing and apnea of prematurity: The known and unknown. J. Perinatol. 2021, 41, 2111–2123. [Google Scholar] [CrossRef]
- Tupal, S.; Huang, W.-H.; Picardo, M.D.; Ling, G.-Y.; Del Negro, C.A.; Zoghbi, H.Y.; Gray, P.A. Atoh1-dependent rhombic lip neurons are required for temporal delay between independent respiratory oscillators in embryonic mice. Elife 2014, 3, e02265. [Google Scholar] [CrossRef] [PubMed]
- Marin, T.L.; Gongol, B.; Zhang, F.; Martin, M.; Johnson, D.A.; Xiao, H.; Wang, Y.; Subramaniam, S.; Chien, S.; Shyy, J.Y. AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Sci. Signal. 2017, 10, eaaf7478. [Google Scholar] [CrossRef] [PubMed]
- MacMillan, S.; Evans, A.M. AMPK facilitates the hypoxic ventilatory response through non-adrenergic mechanisms at the brainstem. Pflugers Arch.-Eur. J. Physiol. 2023, 475, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Viollet, B.; Athea, Y.; Mounier, R.; Guigas, B.; Zarrinpashneh, E.; Horman, S.; Lantier, L.; Hebrard, S.; Devin-Leclerc, J.; Beauloye, C.; et al. AMPK: Lessons from transgenic and knockout animals. Front. Biosci. 2009, 14, 19–44. [Google Scholar] [CrossRef]
- Sharma, S.; Zhang, Y.; Akter, K.A.; Nozohouri, S.; Archie, S.R.; Patel, D.; Villalba, H.; Abbruscato, T. Permeability of Metformin across an In Vitro Blood-Brain Barrier Model during Normoxia and Oxygen-Glucose Deprivation Conditions: Role of Organic Cation Transporters (Octs). Pharmaceutics 2023, 15, 1357. [Google Scholar] [CrossRef] [PubMed]
- Kovo, M.; Haroutiunian, S.; Feldman, N.; Hoffman, A.; Glezerman, M. Determination of metformin transfer across the human placenta using a dually perfused ex vivo placental cotyledon model. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008, 136, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Park, C.S.; Bang, B.R.; Kwon, H.S.; Moon, K.A.; Kim, T.B.; Lee, K.Y.; Moon, H.B.; Cho, Y.S. Metformin reduces airway inflammation and remodeling via activation of AMP-activated protein kinase. Biochem. Pharmacol. 2012, 84, 1660–1670. [Google Scholar] [CrossRef]
- Chen, X.; Walther, F.J.; Sengers, R.M.A.; Laghmani, E.H.; Salam, A.; Folkerts, G.; Pera, T.; Wagenaar, G.T. Metformin attenuates hyperoxia-induced lung injury in neonatal rats by reducing the inflammatory response. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2015, 309, L262–L270. [Google Scholar] [CrossRef] [PubMed]
- Morin-Papunen, L.; Rantala, A.S.; Unkila-Kallio, L.; Tiitinen, A.; Hippeläinen, M.; Perheentupa, A.; Tinkanen, H.; Bloigu, R.; Puukka, K.; Ruokonen, A.; et al. Metformin improves pregnancy and live-birth rates in women with polycystic ovary syndrome (PCOS): A multicenter, double-blind, placebo-controlled randomized trial. J. Clin. Endocrinol. 2012, 97, 1492–1500. [Google Scholar] [CrossRef]
- Tosti, G.; Barberio, A.; Tartaglione, L.; Rizzi, A.; Di Leo, M.; Viti, L.; Sirico, A.; De Carolis, S.; Pontecorvi, A.; Lanzone, A.; et al. Lights and shadows on the use of metformin in pregnancy: From the preconception phase to breastfeeding and beyond. Front. Endocrinol. 2023, 14, 1176623. [Google Scholar] [CrossRef] [PubMed]
- Castilla-Ibeas, A.; Zdral, S.; Oberg, K.C.; Ros, M.A. The limb dorsoventral axis: Lmx1b’s role in development, pathology, evolution, and regeneration. Dev. Dyn. 2024, 253, 798–814. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.D.; Lewis, S.; Juričić, L.; Udoh, U.A.; Hartmann, S.; Jansen, M.A.; Ogunbayo, O.A.; Puggioni, P.; Holmes, A.P.; Kumar, P.; et al. AMP-activated Protein Kinase Deficiency Blocks the Hypoxic Ventilatory Response and Thus Precipitates Hypoventilation and Apnea. Am. J. Respir. Crit. Care Med. 2016, 193, 1032–1043. [Google Scholar] [CrossRef]
- Marin, T.L.; Gongol, B.; Martin, M.; King, S.J.; Smith, L.; Johnson, D.A.; Subramaniam, S.; Chien, S.; Shyy, J.Y. Identification of AMP-activated protein kinase targets by a consensus sequence search of the proteome. BMC Syst. Biol. 2015, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Lv, N.; Wang, Y.; Zhao, M.; Dong, L.; Wei, H. The Role of PAX2 in Neurodevelopment and Disease. Neuropsychiatr. Dis. Treat. 2021, 17, 3559–3567. [Google Scholar] [CrossRef]
- Perry, R.B.; Rishal, I.; Doron-Mandel, E.; Kalinski, A.L.; Medzihradszky, K.F.; Terenzio, M.; Alber, S.; Koley, S.; Lin, A.; Rozenbaum, M.; et al. Nucleolin-Mediated RNA Localization Regulates Neuron Growth and Cycling Cell Size. Cell Rep. 2016, 16, 1664–1676. [Google Scholar] [CrossRef] [PubMed]
- Pai, E.L.; Vogt, D.; Clemente-Perez, A.; McKinsey, G.L.; Cho, F.S.; Hu, J.S.; Wimer, M.; Paul, A.; Fazel Darbandi, S.; Pla, R.; et al. Mafb and c-Maf Have Prenatal Compensatory and Postnatal Antagonistic Roles in Cortical Interneuron Fate and Function. Cell Rep. 2019, 26, 1157–1173. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Xu, W.; Zuo, Z. Knockout of the regulatory factor X1 gene leads to early embryonic lethality. Biochem. Biophys. Res. Commun. 2009, 386, 715–717. [Google Scholar] [CrossRef]
- Kele, J.; Andersson, E.R.; Villaescusa, J.C.; Cajanek, L.; Parish, C.L.; Bonilla, S.; Toledo, E.M.; Bryja, V.; Rubin, J.S.; Shimono, A.; et al. SFRP1 and SFRP2 dose-dependently regulate midbrain dopamine neuron development in vivo and in embryonic stem cells. Stem Cells 2012, 30, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.S.; Pinilla, I.; Saha, J.; Clermont, J.M.; Lien, J.S.; Borys, K.D.; Capowski, E.E.; Phillips, M.J.; Gamm, D.M. VSX2 and ASCL1 Are Indicators of Neurogenic Competence in Human Retinal Progenitor Cultures. PLoS ONE 2015, 10, e0135830. [Google Scholar] [CrossRef] [PubMed]
- Fulton, S.L.; Bendl, J.; Gameiro-Ros, I.; Fullard, J.F.; Al-Kachak, A.; Lepack, A.E.; Stewart, A.F.; Singh, S.; Poller, W.C.; Bastle, R.M.; et al. ZBTB7A regulates MDD-specific chromatin signatures and astrocyte-mediated stress vulnerability in orbitofrontal cortex. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yu, D.; Febbo, I.G.; Maroteaux, M.J.; Wang, H.; Song, Y.; Han, X.; Sun, C.; Meyer, E.E.; Rowe, S.; Chen, Y.; et al. The Transcription Factor Shox2 Shapes Neuron Firing Properties and Suppresses Seizures by Regulation of Key Ion Channels in Thalamocortical Neurons. Cereb. Cortex 2021, 31, 3194–3212. [Google Scholar] [CrossRef] [PubMed]
- Ferlazzo, G.M.; Gambetta, A.M.; Amato, S.; Cannizzaro, N.; Angiolillo, S.; Arboit, M.; Diamante, L.; Carbognin, E.; Romani, P.; La Torre, F.; et al. Genome-wide screening in pluripotent cells identifies Mtf1 as a suppressor of mutant huntingtin toxicity. Nat. Commun. 2023, 14, 3962. [Google Scholar] [CrossRef] [PubMed]
- Fosch, A.; Zagmutt, S.; Casals, N.; Rodríguez-Rodríguez, R. New Insights of SF1 Neurons in Hypothalamic Regulation of Obesity and Diabetes. Int. J. Mol. Sci. 2021, 22, 6186. [Google Scholar] [CrossRef] [PubMed]
- Hua, P.; Liu, W.; Zhao, Y.; Ding, H.; Wang, L.; Xiao, H. Tef polymorphism is associated with sleep disturbances in patients with Parkinson’s disease. Sleep Med. 2012, 13, 297–300. [Google Scholar] [CrossRef]
- Gao, S.P.; Sun, H.F.; Jiang, H.L.; Li, L.D.; Hu, X.; Xu, X.E.; Jin, W. Loss of COX5B inhibits proliferation and promotes senescence via mitochondrial dysfunction in breast cancer. Oncotarget 2015, 6, 43363–43374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Zhang, L.; Xu, J.; Song, M.; Shao, T.; Huang, Z.; Li, Y. RBP EIF2S2 Promotes Tumorigenesis and Progression by Regulating MYC-Mediated Inhibition via FHIT-Related Enhancers. Mol. Ther. 2020, 28, 1105–1118. [Google Scholar] [CrossRef]
- Fan, S.; Chen, Y.; Jiang, Y.; Hu, K.; Li, C. Prefoldin subunit MM1 promotes cell migration via facilitating filopodia formation. Biochem. Biophys. Res. Commun. 2020, 533, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Brajanovski, N.; Chan, K.T.; Xuan, J.; Pearson, R.B.; Sanij, E. Ribosomal proteins and human diseases: Molecular mechanisms and targeted therapy. Signal Transduct. Target. Ther. 2021, 6, 323. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Huang, R.; Lai, Y. Identification and validation of signal recognition particle 14 as a prognostic biomarker predicting overall survival in patients with acute myeloid leukemia. BMC Med. Genom. 2021, 14, 127. [Google Scholar] [CrossRef] [PubMed]
- Kinney, H.C. Brainstem mechanisms underlying the sudden infant death syndrome: Evidence from human pathologic studies. Dev. Psychobiol. J. Int. Soc. Dev. Psychobiol. 2009, 51, 223–233. [Google Scholar] [CrossRef]
- Xu, P.; Zheng, Y.; Liao, J.; Hu, M.; Yang, Y.; Zhang, B.; Kilby, M.D.; Fu, H.; Liu, Y.; Zhang, F.; et al. AMPK regulates homeostasis of invasion and viability in trophoblasts by redirecting glucose metabolism: Implications for pre-eclampsia. Cell Prolif. 2023, 56, e13358. [Google Scholar] [CrossRef]
- Moore, L.G.; Lorca, R.A.; Gumina, D.L.; Wesolowski, S.R.; Reisz, J.A.; Cioffi-Ragan, D.; Houck, J.A.; Banerji, S.; Euser, A.G.; D’Alessandro, A.; et al. Maternal AMPK pathway activation with uterine artery blood flow and fetal growth maintenance during hypoxia. Am. J. Physiol.-Heart Circ. Physiol. 2024, 327, H778–H792. [Google Scholar] [CrossRef]
- Hardie, D.G. AMP-activated proteion kinase: An energy sensor that regulates all aspects of cell function. Genes Dev. 2011, 25, 1895–1908. [Google Scholar] [CrossRef]
- Bataillon, T.; Gauthier, P.; Villesen, P.; Santoni, S.; Thompson, J.D.; Ehlers, B.K. From genotype to phenotype: Genetic redundancy and the maintenance of an adaptive polymorphism in the context of high gene flow. Evol. Lett. 2022, 6, 189–202. [Google Scholar] [CrossRef]
- Rousset, C.I.; Leiper, F.C.; Kichev, A.; Gressens, P.; Carling, D.; Hagberg, H.; Thornton, C. A dual role for AMP-activated protein kinase (AMPK) during neonatal hypoxic-ischaemic brain injury in mice. J. Neurochem. 2015, 133, 242–252. [Google Scholar] [CrossRef]
- Williams, T.; Courchet, J.; Viollet, B.; Brenman, J.E.; Polleux, F. AMP-activated protein kinase (AMPK) activity is not required for neuronal development but regulates axogenesis during metabolic stress. Proc. Natl. Acad. Sci. USA 2011, 108, 5849–5854. [Google Scholar] [CrossRef]
- Ramamurthy, S.; Chang, E.; Cao, Y.; Zhu, J.; Ronnett, G.V. AMPK activation regulates neuronal structure in developing hippocampal neurons. Neuroscience 2014, 259, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Curry, D.W.; Stutz, B.; Andrews, Z.B.; Elsworth, J.D. Targeting AMPK Signaling as a Neuroprotective Strategy in Parkinson’s Disease. J. Parkinson’s Dis. 2018, 8, 161–181. [Google Scholar] [CrossRef]
- Kim, J.; Yang, G.; Kim, Y.; Kim, J.; Ha, J. AMPK activators: Mechanisms of action and physiological activities. Exp. Mol. Med. 2016, 48, e224. [Google Scholar] [CrossRef]
- Krawchuk, D.; Kania, A. Identification of genes controlled by LMX1B in the developing mouse limb bud. Dev. Dyn. 2008, 237, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Spencer, W.C.; Deneris, E.S. Regulatory Mechanisms Controlling Maturation of Serotonin Neuron Identity and Function. Front. Cell. Neurosci. 2017, 11, 215. [Google Scholar] [CrossRef] [PubMed]
- Velyvis, A.; Qin, J. LIM Domain and Its Binding to Target Proteins. In Madame Curie Bioscience Database [Internet]; Landes Bioscience: Austin, TX, USA, 2013. [Google Scholar]
- Kim, K.; Kim, J.H.; Kim, I.; Seong, S.; Han, J.E.; Lee, K.B.; Koh, J.T.; Kim, N. Transcription Factor Lmx1b Negatively Regulates Osteoblast Differentiation and Bone Formation. Int. J. Mol. Sci. 2022, 23, 5225. [Google Scholar] [CrossRef]
- Ninan, K.; Liyanage, S.K.; Murphy, K.E.; Asztalos, E.V.; McDonald, S.D. Evaluation of Long-term Outcomes Associated with Preterm Exposure to Antenatal Corticosteroids: A Systematic Review and Meta-analysis. JAMA Pediatr. 2022, 176, e220483. [Google Scholar] [CrossRef]
- Buchman, A.L. Side effects of corticosteroid therapy. J. Clin. Gastroenterol. 2001, 33, 289–294. [Google Scholar] [CrossRef]
- Siffel, C.; Kistler, K.D.; Sarda, S.P. Global incidence of intraventricular hemorrhage among extremely preterm infants: A systematic literature review. J. Perinat. Med. 2021, 49, 1017–1026. [Google Scholar] [CrossRef]
- Zhong, F.; Liu, L.; Wei, J.L.; Dai, R.P. Step by Step Golgi-Cox Staining for Cryosection. Front. Neuroanat. 2019, 13, 62. [Google Scholar] [CrossRef]
- Zaqout, S.; Kaindl, A.M. Golgi-Cox Staining Step by Step. Front Neuroanat 2016, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Balan, K.V.; Kc, P.; Mayer, C.A.; Wilson, C.G.; Belkadi, A.; Martin, R.J. Intrapulmonary lipopolysaccharide exposure upregulates cytokine expression in the neonatal brainstem. Acta Paediatr. 2012, 101, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Bruggink, S.; Kentch, K.; Kronenfeld, J.; Renquist, B.J. A leak-free head-out plethysmography system to accurately assess lung function in mice. J. Appl. Physiol. 2022, 133, 104–118. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marin, T.L.; Wilson, C.G.; Ramirez, M.L.; Sun, W.; Malhotra, A.; Gongol, B. AMPK Phosphorylates LMX1b to Regulate a Brainstem Neurogenic Network Important for Control of Breathing in Neonatal Mice. Int. J. Mol. Sci. 2025, 26, 213. https://doi.org/10.3390/ijms26010213
Marin TL, Wilson CG, Ramirez ML, Sun W, Malhotra A, Gongol B. AMPK Phosphorylates LMX1b to Regulate a Brainstem Neurogenic Network Important for Control of Breathing in Neonatal Mice. International Journal of Molecular Sciences. 2025; 26(1):213. https://doi.org/10.3390/ijms26010213
Chicago/Turabian StyleMarin, Traci L., Christopher G. Wilson, Miguel Lopez Ramirez, Wei Sun, Atul Malhotra, and Brendan Gongol. 2025. "AMPK Phosphorylates LMX1b to Regulate a Brainstem Neurogenic Network Important for Control of Breathing in Neonatal Mice" International Journal of Molecular Sciences 26, no. 1: 213. https://doi.org/10.3390/ijms26010213
APA StyleMarin, T. L., Wilson, C. G., Ramirez, M. L., Sun, W., Malhotra, A., & Gongol, B. (2025). AMPK Phosphorylates LMX1b to Regulate a Brainstem Neurogenic Network Important for Control of Breathing in Neonatal Mice. International Journal of Molecular Sciences, 26(1), 213. https://doi.org/10.3390/ijms26010213





