Tumor Antigens beyond the Human Exome
Abstract
1. Introduction
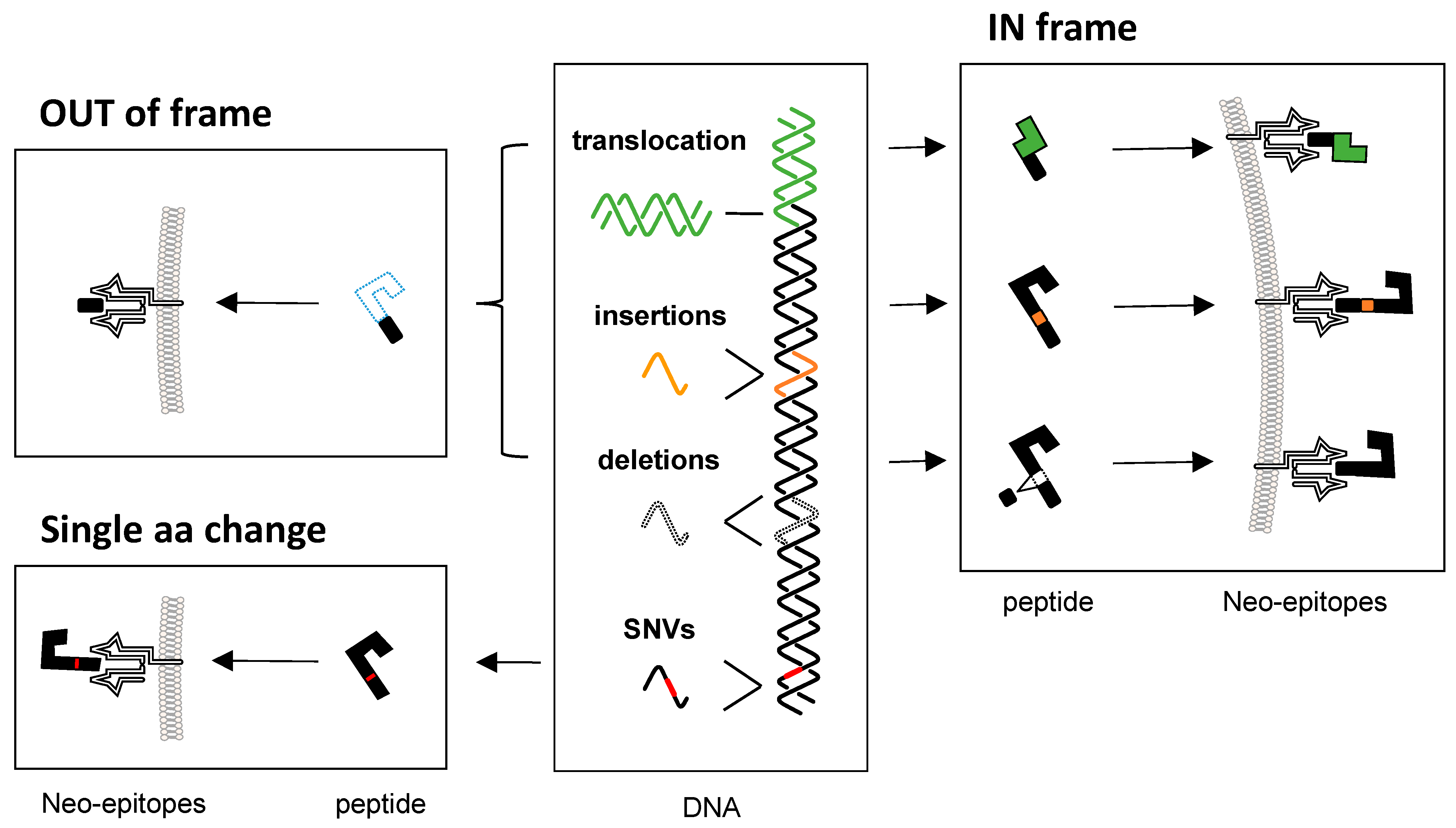
2. Canonical Neo-Epitopes
2.1. Single Nucleotide Variants
2.2. Insertions and Deletions
2.3. Gene Fusions
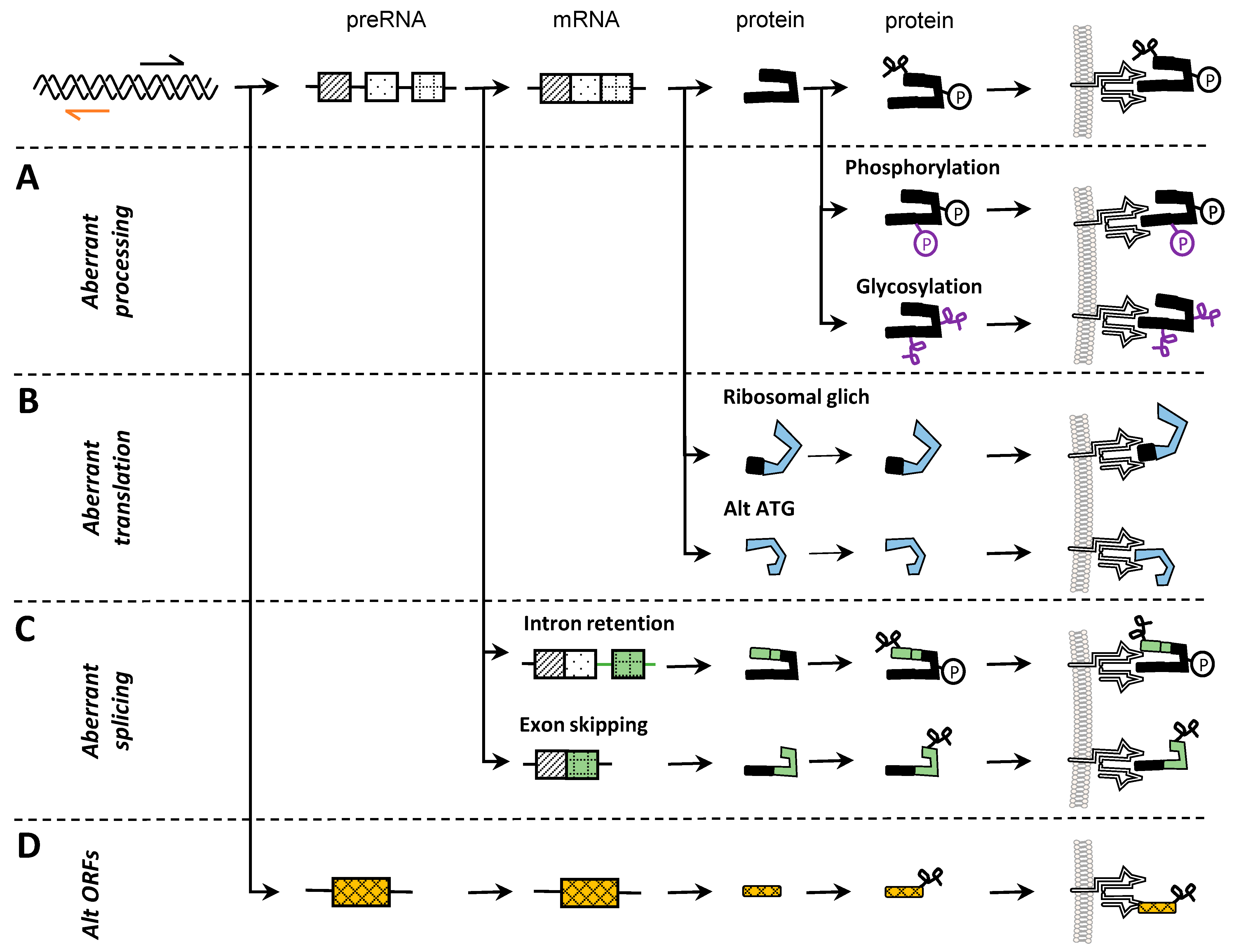
3. Noncanonical Neo-Epitopes
3.1. Expression of Noncoding DNA Regions
3.2. Splicing
3.2.1. Cis-Acting Mutations
3.2.2. Trans-Acting Mutations
3.3. Translation
3.3.1. Initiation of mRNA Translation
3.3.2. Elongation of mRNA Translation
3.3.3. Termination of mRNA Translation
3.4. Post-Translational Modifications
3.4.1. Phosphorylation
3.4.2. Glycosylation
3.4.3. Other Post-Translational Modifications
4. Viral Neo-Epitopes
4.1. Human Papillomavirus
4.2. Human T-Cell Leukemia Virus
4.3. Epstein–Barr Virus
4.4. Cytomegalovirus
4.5. Merkel Cell Polyoma Virus
4.6. Kaposi’s Sarcoma Assoiated Herpesvirus
4.7. Hepatitis B and C Virus
4.8. Human Endogenous Retroviruses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Mo, Y.; Wang, Y.; Wu, P.; Zhang, Y.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; Li, X.; et al. Neo-antigen vaccine: An emerging tumor immunotherapy. Mol. Cancer 2019, 18, 128. [Google Scholar] [CrossRef] [PubMed]
- Lang, F.; Schrors, B.; Lower, M.; Tureci, O.; Sahin, U. Identification of neo-antigens for individualized therapeutic cancer vaccines. Nat. Rev. Drug Discov. 2022, 21, 261–282. [Google Scholar] [CrossRef] [PubMed]
- Vormehr, M.; Tureci, O.; Sahin, U. Harnessing Tumor Mutations for Truly Individualized Cancer Vaccines. Annu. Rev. Med. 2019, 70, 395–407. [Google Scholar] [CrossRef]
- Germano, G.; Lamba, S.; Rospo, G.; Barault, L.; Magri, A.; Maione, F.; Russo, M.; Crisafulli, G.; Bartolini, A.; Lerda, G.; et al. Inactivation of DNA repair triggers neo-antigen generation and impairs tumour growth. Nature 2017, 552, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D.H.; Shankaran, V.; Dighe, A.S.; Stockert, E.; Aguet, M.; Old, L.J.; Schreiber, R.D. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. USA 1998, 95, 7556–7561. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, V.P.; Luksza, M.; Zhao, J.N.; Makarov, V.; Moral, J.A.; Remark, R.; Herbst, B.; Askan, G.; Bhanot, U.; Senbabaoglu, Y.; et al. Identification of unique neo-antigen qualities in long-term survivors of pancreatic cancer. Nature 2017, 551, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Blass, E.; Ott, P.A. Advances in the development of personalized neo-antigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 2021, 18, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Strickland, K.C.; Howitt, B.E.; Shukla, S.A.; Rodig, S.; Ritterhouse, L.L.; Liu, J.F.; Garber, J.E.; Chowdhury, D.; Wu, C.J.; D’Andrea, A.D.; et al. Association and prognostic significance of BRCA1/2-mutation status with neo-antigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016, 7, 13587–13598. [Google Scholar] [CrossRef]
- Nagel, R.; Pataskar, A.; Champagne, J.; Agami, R. Boosting Antitumor Immunity with an Expanded Neo-epitope Landscape. Cancer Res. 2022, 82, 3637–3649. [Google Scholar] [CrossRef]
- Finn, O.J.; Rammensee, H.G. Is It Possible to Develop Cancer Vaccines to Neo-antigens, What Are the Major Challenges, and How Can These Be Overcome? Neo-antigens: Nothing New in Spite of the Name. Cold Spring Harb. Perspect. Biol. 2018, 10, a028829. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neo-antigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, N.P.; Heeke, C.; Tvingsholm, S.A.; Borch, A.; Draghi, A.; Crowther, M.D.; Carri, I.; Munk, K.K.; Holm, J.S.; Bjerregaard, A.M.; et al. Neo-antigen-reactive CD8+ T cells affect clinical outcome of adoptive cell therapy with tumor-infiltrating lymphocytes in melanoma. J. Clin. Investig. 2022, 132, JCI150535. [Google Scholar] [CrossRef] [PubMed]
- Overwijk, W.W.; Wang, E.; Marincola, F.M.; Rammensee, H.G.; Restifo, N.P. Mining the mutanome: Developing highly personalized Immunotherapies based on mutational analysis of tumors. J. Immunother. Cancer 2013, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Tureci, O. Personalized vaccines for cancer immunotherapy. Science 2018, 359, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Laumont, C.M.; Vincent, K.; Hesnard, L.; Audemard, E.; Bonneil, E.; Laverdure, J.P.; Gendron, P.; Courcelles, M.; Hardy, M.P.; Cote, C.; et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci. Transl. Med. 2018, 10, aau5516. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Zhou, C.; Zhang, Z.; Guan, M.; Zhang, C.; Liu, Z.; Liu, Q. The Landscape of Tumor Fusion Neo-antigens: A Pan-Cancer Analysis. iScience 2019, 21, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wu, J.; Chen, S.; Zhou, Z. Shared neo-antigens: Ideal targets for off-the-shelf cancer immunotherapy. Pharmacogenomics 2020, 21, 637–645. [Google Scholar] [CrossRef]
- Hahn, C.N.; Venugopal, P.; Scott, H.S.; Hiwase, D.K. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol. Rev. 2015, 263, 257–278. [Google Scholar] [CrossRef]
- Chen, P.; Fang, Q.X.; Chen, D.B.; Chen, H.S. Neo-antigen vaccine: An emerging immunotherapy for hepatocellular carcinoma. World J. Gastrointest. Oncol. 2021, 13, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Nejo, T.; Yamamichi, A.; Almeida, N.D.; Goretsky, Y.E.; Okada, H. Tumor antigens in glioma. Semin. Immunol. 2020, 47, 101385. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Cha, H.; Kim, K.; Sung, C.; An, J.; Bang, H.; Kim, H.; Yang, J.O.; Chang, S.; Shin, I.; et al. MHC II immunogenicity shapes the neo-epitope landscape in human tumors. Nat. Genet. 2023, 55, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Chen, F.; Meng, F.; Wei, J.; Liu, B. MHC class II restricted neo-antigen: A promising target in tumor immunotherapy. Cancer Lett. 2017, 392, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Capietto, A.H.; Hoshyar, R.; Delamarre, L. Sources of Cancer Neo-antigens beyond Single-Nucleotide Variants. Int. J. Mol. Sci. 2022, 23, 10131. [Google Scholar] [CrossRef] [PubMed]
- Hansen, U.K.; Ramskov, S.; Bjerregaard, A.M.; Borch, A.; Andersen, R.; Draghi, A.; Donia, M.; Bentzen, A.K.; Marquard, A.M.; Szallasi, Z.; et al. Tumor-Infiltrating T Cells from Clear Cell Renal Cell Carcinoma Patients Recognize Neo-epitopes Derived From Point and Frameshift Mutations. Front. Immunol. 2020, 11, 373. [Google Scholar] [CrossRef] [PubMed]
- Vormehr, M.; Diken, M.; Boegel, S.; Kreiter, S.; Tureci, O.; Sahin, U. Mutanome directed cancer immunotherapy. Curr. Opin. Immunol. 2016, 39, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, M.G.; Stanfield, R.L.; Wilson, I.A. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006, 24, 419–466. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.; Duitama, J.; Al Seesi, S.; Ayres, C.M.; Corcelli, S.A.; Pawashe, A.P.; Blanchard, T.; McMahon, D.; Sidney, J.; Sette, A.; et al. Genomic and bioinformatic profiling of mutational neo-epitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med. 2014, 211, 2231–2248. [Google Scholar] [CrossRef]
- Krawczak, M.; Reiss, J.; Cooper, D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum. Genet. 1992, 90, 41–54. [Google Scholar] [CrossRef]
- Reimand, J.; Wagih, O.; Bader, G.D. The mutational landscape of phosphorylation signaling in cancer. Sci. Rep. 2013, 3, 2651. [Google Scholar] [CrossRef] [PubMed]
- Hinrichs, C.S.; Rosenberg, S.A. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol. Rev. 2014, 257, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neo-antigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef] [PubMed]
- Carreno, B.M.; Magrini, V.; Becker-Hapak, M.; Kaabinejadian, S.; Hundal, J.; Petti, A.A.; Ly, A.; Lie, W.R.; Hildebrand, W.H.; Mardis, E.R.; et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neo-antigen-specific T cells. Science 2015, 348, 803–808. [Google Scholar] [CrossRef]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neo-antigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Lower, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrors, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Huang, T.; Liu, L.; Lv, Z.; Zhao, K.; Yi, Q.; Zhang, J. Recent Advances in DNA Vaccines against Lung Cancer: A Mini Review. Vaccines 2022, 10, 1586. [Google Scholar] [CrossRef]
- Niemi, J.V.L.; Sokolov, A.V.; Schioth, H.B. Neo-antigen Vaccines; Clinical Trials, Classes, Indications, Adjuvants and Combinatorial Treatments. Cancers 2022, 14, 5163. [Google Scholar] [CrossRef]
- Ryan, C. FDA Grants Breakthrough Therapy Designation to mRNA-4157/V940 Plus Pembrolizumab in High-Risk Melanoma. 23.02.2023. 2023. Available online: https://www.onclive.com/view/fda-grants-breakthrough-therapy-designation-to-mrna-4157-v940-plus-pembrolizumab-in-high-risk-melanoma (accessed on 31 March 2023).
- Smith, C.C.; Selitsky, S.R.; Chai, S.; Armistead, P.M.; Vincent, B.G.; Serody, J.S. Alternative tumour-specific antigens. Nat. Rev. Cancer 2019, 19, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Timar, J.; Kashofer, K. Molecular epidemiology and diagnostics of KRAS mutations in human cancer. Cancer Metastasis Rev. 2020, 39, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.; Karuppiah, V.; Hartt, A.; Haidar, J.N.; Moureau, S.; Dobrzycki, T.; Hayes, C.; Rowley, C.; Dias, J.; Harper, S.; et al. Therapeutic high affinity T cell receptor targeting a KRAS(G12D) cancer neo-antigen. Nat. Commun. 2022, 13, 5333. [Google Scholar] [CrossRef]
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020, 80, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Tran, E.; Robbins, P.F.; Lu, Y.C.; Prickett, T.D.; Gartner, J.J.; Jia, L.; Pasetto, A.; Zheng, Z.; Ray, S.; Groh, E.M.; et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N. Engl. J. Med. 2016, 375, 2255–2262. [Google Scholar] [CrossRef] [PubMed]
- Sim, M.J.W.; Lu, J.; Spencer, M.; Hopkins, F.; Tran, E.; Rosenberg, S.A.; Long, E.O.; Sun, P.D. High-affinity oligoclonal TCRs define effective adoptive T cell therapy targeting mutant KRAS-G12D. Proc. Natl. Acad. Sci. USA 2020, 117, 12826–12835. [Google Scholar] [CrossRef] [PubMed]
- Leidner, R.; Sanjuan Silva, N.; Huang, H.; Sprott, D.; Zheng, C.; Shih, Y.P.; Leung, A.; Payne, R.; Sutcliffe, K.; Cramer, J.; et al. Neo-antigen T-Cell Receptor Gene Therapy in Pancreatic Cancer. N. Engl. J. Med. 2022, 386, 2112–2119. [Google Scholar] [CrossRef]
- Maoz, A.; Rennert, G.; Gruber, S.B. T-Cell Transfer Therapy Targeting Mutant KRAS. N. Engl. J. Med. 2017, 376, e11. [Google Scholar] [CrossRef]
- Bai, P.; Zhou, Q.; Wei, P.; Bai, H.; Chan, S.K.; Kappler, J.W.; Marrack, P.; Yin, L. Rational discovery of a cancer neo-epitope harboring the KRAS G12D driver mutation. Sci. China Life Sci. 2021, 64, 2144–2152. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, J.A.; Zhang, H.X.; Jiang, Y.N.; Luo, W.H. Cancer vaccines: Targeting KRAS-driven cancers. Expert Rev. Vaccines 2020, 19, 163–173. [Google Scholar] [CrossRef]
- Pearlman, A.H.; Hwang, M.S.; Konig, M.F.; Hsiue, E.H.; Douglass, J.; DiNapoli, S.R.; Mog, B.J.; Bettegowda, C.; Pardoll, D.M.; Gabelli, S.B.; et al. Targeting public neo-antigens for cancer immunotherapy. Nat. Cancer 2021, 2, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.J.; Mason, C.E. The Newly Emerging View of the Genome. In Genomics, Circuits, and Pathways in Clinical Neuropsychiatry; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Roudko, V.; Cimen Bozkus, C.; Greenbaum, B.; Lucas, A.; Samstein, R.; Bhardwaj, N. Lynch Syndrome and MSI-H Cancers: From Mechanisms to “Off-The-Shelf” Cancer Vaccines. Front. Immunol. 2021, 12, 757804. [Google Scholar] [CrossRef]
- Litchfield, K.; Reading, J.L.; Lim, E.L.; Xu, H.; Liu, P.; Al-Bakir, M.; Wong, Y.N.S.; Rowan, A.; Funt, S.A.; Merghoub, T.; et al. Escape from nonsense-mediated decay associates with anti-tumor immunogenicity. Nat. Commun. 2020, 11, 3800. [Google Scholar] [CrossRef]
- Turajlic, S.; Litchfield, K.; Xu, H.; Rosenthal, R.; McGranahan, N.; Reading, J.L.; Wong, Y.N.S.; Rowan, A.; Kanu, N.; Al Bakir, M.; et al. Insertion-and-deletion-derived tumour-specific neo-antigens and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol. 2017, 18, 1009–1021. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; Jian, X.; Tan, X.; Lu, M.; Ouyang, J.; Liu, Z.; Li, Y.; Xu, L.; Chen, L.; et al. ProGeo-Neo v2.0: A One-Stop Software for Neo-antigen Prediction and Filtering Based on the Proteogenomics Strategy. Genes 2022, 13, 783. [Google Scholar] [CrossRef] [PubMed]
- Roudko, V.; Bozkus, C.C.; Orfanelli, T.; McClain, C.B.; Carr, C.; O’Donnell, T.; Chakraborty, L.; Samstein, R.; Huang, K.L.; Blank, S.V.; et al. Shared Immunogenic Poly-Epitope Frameshift Mutations in Microsatellite Unstable Tumors. Cell 2020, 183, 1634–1649.e17. [Google Scholar] [CrossRef] [PubMed]
- Baretti, M.; Le, D.T. DNA mismatch repair in cancer. Pharmacol. Ther. 2018, 189, 45–62. [Google Scholar] [CrossRef]
- Gebert, J.; Gelincik, O.; Oezcan-Wahlbrink, M.; Marshall, J.D.; Hernandez-Sanchez, A.; Urban, K.; Long, M.; Cortes, E.; Tosti, E.; Katzenmaier, E.M.; et al. Recurrent Frameshift Neo-antigen Vaccine Elicits Protective Immunity with Reduced Tumor Burden and Improved Overall Survival in a Lynch Syndrome Mouse Model. Gastroenterology 2021, 161, 1288–1302.e13. [Google Scholar] [CrossRef]
- Kloor, M.; Reuschenbach, M.; Pauligk, C.; Karbach, J.; Rafiyan, M.R.; Al-Batran, S.E.; Tariverdian, M.; Jager, E.; von Knebel Doeberitz, M. A Frameshift Peptide Neo-antigen-Based Vaccine for Mismatch Repair-Deficient Cancers: A Phase I/IIa Clinical Trial. Clin. Cancer Res. 2020, 26, 4503–4510. [Google Scholar] [CrossRef]
- Espanol-Rego, M.; Fernandez-Martos, C.; Elez, E.; Foguet, C.; Pedrosa, L.; Rodriguez, N.; Ruiz-Casado, A.; Pineda, E.; Cid, J.; Cabezon, R.; et al. A Phase I-II multicenter trial with Avelumab plus autologous dendritic cell vaccine in pre-treated mismatch repair-proficient (MSS) metastatic colorectal cancer patients; GEMCAD 1602 study. Cancer Immunol. Immunother. 2022, 72, 827–840. [Google Scholar] [CrossRef]
- Mitelman, F.; Johansson, B.; Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 2007, 7, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Bruford, E.A.; Antonescu, C.R.; Carroll, A.J.; Chinnaiyan, A.; Cree, I.A.; Cross, N.C.P.; Dalgleish, R.; Gale, R.P.; Harrison, C.J.; Hastings, R.J.; et al. HUGO Gene Nomenclature Committee (HGNC) recommendations for the designation of gene fusions. Leukemia 2021, 35, 3040–3043. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Liang, W.W.; Foltz, S.M.; Mutharasu, G.; Jayasinghe, R.G.; Cao, S.; Liao, W.W.; Reynolds, S.M.; Wyczalkowski, M.A.; Yao, L.; et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 2018, 23, 227–238.e3. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, T.; Song, X.; Liu, B.; Wei, J. Gene fusion neo-antigens: Emerging targets for cancer immunotherapy. Cancer Lett. 2021, 506, 45–54. [Google Scholar] [CrossRef]
- Mertens, F.; Johansson, B.; Fioretos, T.; Mitelman, F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer 2015, 15, 371–381. [Google Scholar] [CrossRef]
- Weckselblatt, B.; Rudd, M.K. Human Structural Variation: Mechanisms of Chromosome Rearrangements. Trends Genet. 2015, 31, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Leko, V.; Rosenberg, S.A. Identifying and Targeting Human Tumor Antigens for T Cell-Based Immunotherapy of Solid Tumors. Cancer Cell 2020, 38, 454–472. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neo-antigens: Promising targets for cancer therapy. Signal Transduct. Target Ther. 2023, 8, 9. [Google Scholar] [CrossRef]
- Sato, Y.; Nabeta, Y.; Tsukahara, T.; Hirohashi, Y.; Syunsui, R.; Maeda, A.; Sahara, H.; Ikeda, H.; Torigoe, T.; Ichimiya, S.; et al. Detection and induction of CTLs specific for SYT-SSX-derived peptides in HLA-A24+ patients with synovial sarcoma. J. Immunol. 2002, 169, 1611–1618. [Google Scholar] [CrossRef]
- Yotnda, P.; Firat, H.; Garcia-Pons, F.; Garcia, Z.; Gourru, G.; Vernant, J.P.; Lemonnier, F.A.; Leblond, V.; Langlade-Demoyen, P. Cytotoxic T cell response against the chimeric p210 BCR-ABL protein in patients with chronic myelogenous leukemia. J. Clin. Investig. 1998, 101, 2290–2296. [Google Scholar] [CrossRef] [PubMed]
- Dagher, R.; Long, L.M.; Read, E.J.; Leitman, S.F.; Carter, C.S.; Tsokos, M.; Goletz, T.J.; Avila, N.; Berzofsky, J.A.; Helman, L.J.; et al. Pilot trial of tumor-specific peptide vaccination and continuous infusion interleukin-2 in patients with recurrent Ewing sarcoma and alveolar rhabdomyosarcoma: An inter-institute NIH study. Med. Pediatr. Oncol. 2002, 38, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Pinilla-Ibarz, J.; Cathcart, K.; Korontsvit, T.; Soignet, S.; Bocchia, M.; Caggiano, J.; Lai, L.; Jimenez, J.; Kolitz, J.; Scheinberg, D.A. Vaccination of patients with chronic myelogenous leukemia with bcr-abl oncogene breakpoint fusion peptides generates specific immune responses. Blood 2000, 95, 1781–1787. [Google Scholar] [CrossRef]
- Payer, L.M.; Burns, K.H. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019, 20, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Misiak, B.; Ricceri, L.; Sasiadek, M.M. Transposable Elements and Their Epigenetic Regulation in Mental Disorders: Current Evidence in the Field. Front. Genet. 2019, 10, 580. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Beckermann, K.E.; Bortone, D.S.; De Cubas, A.A.; Bixby, L.M.; Lee, S.J.; Panda, A.; Ganesan, S.; Bhanot, G.; Wallen, E.M.; et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J. Clin. Investig. 2018, 128, 4804–4820. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Rose, C.M.; Cass, A.A.; Williams, A.G.; Darwish, M.; Lianoglou, S.; Haverty, P.M.; Tong, A.J.; Blanchette, C.; Albert, M.L.; et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat. Commun. 2019, 10, 5228. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, M.; Kolenda, T.; Kozlowska-Maslon, J.; Sobocinska, J.; Poter, P.; Guglas, K.; Paszkowska, A.; Blizniak, R.; Teresiak, A.; Kazimierczak, U.; et al. The World of Pseudogenes: New Diagnostic and Therapeutic Targets in Cancers or Still Mystery Molecules? Life 2021, 11, 1354. [Google Scholar] [CrossRef] [PubMed]
- Irajizad, E.; Fahrmann, J.F.; Long, J.P.; Vykoukal, J.; Kobayashi, M.; Capello, M.; Yu, C.Y.; Cai, Y.; Hsiao, F.C.; Patel, N.; et al. A Comprehensive Search of Non-Canonical Proteins in Non-Small Cell Lung Cancer and Their Impact on the Immune Response. Int. J. Mol. Sci. 2022, 23, 8933. [Google Scholar] [CrossRef]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Tokita, S.; Hirama, T.; Kochin, V.; Nakatsugawa, M.; Shinkawa, T.; Hirohashi, Y.; Tsukahara, T.; Hata, F.; Takemasa, I.; et al. CD8+ T-cell Immune Surveillance against a Tumor Antigen Encoded by the Oncogenic Long Noncoding RNA PVT1. Cancer Immunol. Res. 2021, 9, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Baralle, D.; Baralle, M. Splicing in action: Assessing disease causing sequence changes. J. Med. Genet. 2005, 42, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Burge, C.B. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 2008, 14, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Frankiw, L.; Baltimore, D.; Li, G. Alternative mRNA splicing in cancer immunotherapy. Nat. Rev. Immunol. 2019, 19, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef] [PubMed]
- De Kesel, J.; Fijalkowski, I.; Taylor, J.; Ntziachristos, P. Splicing dysregulation in human hematologic malignancies: Beyond splicing mutations. Trends Immunol. 2022, 43, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Kahles, A.; Lehmann, K.V.; Toussaint, N.C.; Huser, M.; Stark, S.G.; Sachsenberg, T.; Stegle, O.; Kohlbacher, O.; Sander, C.; Cancer Genome Atlas Research, N.; et al. Comprehensive Analysis of Alternative Splicing across Tumors from 8705 Patients. Cancer Cell 2018, 34, 211–224.e6. [Google Scholar] [CrossRef]
- Bonnal, S.C.; Lopez-Oreja, I.; Valcarcel, J. Roles and mechanisms of alternative splicing in cancer—Implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474. [Google Scholar] [CrossRef]
- Sciarrillo, R.; Wojtuszkiewicz, A.; Assaraf, Y.G.; Jansen, G.; Kaspers, G.J.L.; Giovannetti, E.; Cloos, J. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Resist. Updates 2020, 53, 100728. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Xu, L.; Suzuki, T.; Yoshikawa, T.; Sakamoto, H.; Uemura, H.; Yoshizawa, A.C.; Suzuki, Y.; Nakatsura, T.; Ishihama, Y.; et al. Aberrant splicing isoforms detected by full-length transcriptome sequencing as transcripts of potential neo-antigens in non-small cell lung cancer. Genome Biol. 2021, 22, 9. [Google Scholar] [CrossRef] [PubMed]
- Smart, A.C.; Margolis, C.A.; Pimentel, H.; He, M.X.; Miao, D.; Adeegbe, D.; Fugmann, T.; Wong, K.K.; Van Allen, E.M. Intron retention is a source of neo-epitopes in cancer. Nat. Biotechnol. 2018, 36, 1056–1058. [Google Scholar] [CrossRef] [PubMed]
- Coulie, P.G.; Lehmann, F.; Lethe, B.; Herman, J.; Lurquin, C.; Andrawiss, M.; Boon, T. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl. Acad. Sci. USA 1995, 92, 7976–7980. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.F.; El-Gamil, M.; Li, Y.F.; Fitzgerald, E.B.; Kawakami, Y.; Rosenberg, S.A. The intronic region of an incompletely spliced gp100 gene transcript encodes an epitope recognized by melanoma-reactive tumor-infiltrating lymphocytes. J. Immunol. 1997, 159, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghe, R.G.; Cao, S.; Gao, Q.; Wendl, M.C.; Vo, N.S.; Reynolds, S.M.; Zhao, Y.; Climente-Gonzalez, H.; Chai, S.; Wang, F.; et al. Systematic Analysis of Splice-Site-Creating Mutations in Cancer. Cell Rep. 2018, 23, 270–281.e3. [Google Scholar] [CrossRef] [PubMed]
- Ohno, K.; Takeda, J.I.; Masuda, A. Rules and tools to predict the splicing effects of exonic and intronic mutations. Wiley Interdiscip. Rev. RNA 2018, 9, e1451. [Google Scholar] [CrossRef] [PubMed]
- Roca, X.; Sachidanandam, R.; Krainer, A.R. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003, 31, 6321–6333. [Google Scholar] [CrossRef] [PubMed]
- Voith von Voithenberg, L.; Sanchez-Rico, C.; Kang, H.S.; Madl, T.; Zanier, K.; Barth, A.; Warner, L.R.; Sattler, M.; Lamb, D.C. Recognition of the 3′ splice site RNA by the U2AF heterodimer involves a dynamic population shift. Proc. Natl. Acad. Sci. USA 2016, 113, E7169–E7175. [Google Scholar] [CrossRef]
- Lewandowska, M.A. The missing puzzle piece: Splicing mutations. Int. J. Clin. Exp. Pathol. 2013, 6, 2675–2682. [Google Scholar]
- Abramowicz, A.; Gos, M. Correction to: Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J. Appl. Genet. 2019, 60, 231. [Google Scholar] [CrossRef] [PubMed]
- Berget, S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995, 270, 2411–2414. [Google Scholar] [CrossRef] [PubMed]
- Lappin, K.M.; Barros, E.M.; Jhujh, S.S.; Irwin, G.W.; McMillan, H.; Liberante, F.G.; Latimer, C.; La Bonte, M.J.; Mills, K.I.; Harkin, D.P.; et al. Cancer-Associated SF3B1 Mutations Confer a BRCA-Like Cellular Phenotype and Synthetic Lethality to PARP Inhibitors. Cancer Res. 2022, 82, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Shukla, G.C.; Singh, J. Mutations of RNA splicing factors in hematological malignancies. Cancer Lett. 2017, 409, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; Roberson, E.D.; Anbunathan, H.; Onken, M.D.; Worley, L.A.; Bowcock, A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013, 45, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Furney, S.J.; Pedersen, M.; Gentien, D.; Dumont, A.G.; Rapinat, A.; Desjardins, L.; Turajlic, S.; Piperno-Neumann, S.; de la Grange, P.; Roman-Roman, S.; et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013, 3, 1122–1129. [Google Scholar] [CrossRef]
- Quesada, V.; Conde, L.; Villamor, N.; Ordonez, G.R.; Jares, P.; Bassaganyas, L.; Ramsay, A.J.; Bea, S.; Pinyol, M.; Martinez-Trillos, A.; et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011, 44, 47–52. [Google Scholar] [CrossRef]
- Darman, R.B.; Seiler, M.; Agrawal, A.A.; Lim, K.H.; Peng, S.; Aird, D.; Bailey, S.L.; Bhavsar, E.B.; Chan, B.; Colla, S.; et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015, 13, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Gozani, O.; Potashkin, J.; Reed, R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell Biol. 1998, 18, 4752–4760. [Google Scholar] [CrossRef]
- Golas, M.M.; Sander, B.; Will, C.L.; Luhrmann, R.; Stark, H. Molecular architecture of the multiprotein splicing factor SF3b. Science 2003, 300, 980–984. [Google Scholar] [CrossRef]
- Schischlik, F.; Jager, R.; Rosebrock, F.; Hug, E.; Schuster, M.; Holly, R.; Fuchs, E.; Milosevic Feenstra, J.D.; Bogner, E.; Gisslinger, B.; et al. Mutational landscape of the transcriptome offers putative targets for immunotherapy of myeloproliferative neoplasms. Blood 2019, 134, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Sette, C.; Paronetto, M.P. Somatic Mutations in Core Spliceosome Components Promote Tumorigenesis and Generate an Exploitable Vulnerability in Human Cancer. Cancers 2022, 14, 1827. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; De Neef, E.; Thomas, J.D.; Sabio, E.; Rousseau, B.; Gigoux, M.; Knorr, D.A.; Greenbaum, B.; Elhanati, Y.; Hogg, S.J.; et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell 2021, 184, 4032–4047.e31. [Google Scholar] [CrossRef] [PubMed]
- Assi, R.; Kantarjian, H.M.; Kadia, T.M.; Pemmaraju, N.; Jabbour, E.; Jain, N.; Daver, N.; Estrov, Z.; Uehara, T.; Owa, T.; et al. Final results of a phase 2, open-label study of indisulam, idarubicin, and cytarabine in patients with relapsed or refractory acute myeloid leukemia and high-risk myelodysplastic syndrome. Cancer 2018, 124, 2758–2765. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Minoshima, Y.; Sagane, K.; Sugi, N.H.; Mitsuhashi, K.O.; Yamamoto, N.; Kamiyama, H.; Takahashi, K.; Kotake, Y.; Uesugi, M.; et al. Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat. Chem. Biol. 2017, 13, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Sriram, A.; Bohlen, J.; Teleman, A.A. Translation acrobatics: How cancer cells exploit alternate modes of translational initiation. EMBO Rep. 2018, 19, e45947. [Google Scholar] [CrossRef] [PubMed]
- Minati, R.; Perreault, C.; Thibault, P. A Roadmap Toward the Definition of Actionable Tumor-Specific Antigens. Front. Immunol. 2020, 11, 583287. [Google Scholar] [CrossRef]
- Malarkannan, S.; Horng, T.; Shih, P.P.; Schwab, S.; Shastri, N. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity 1999, 10, 681–690. [Google Scholar] [CrossRef]
- Dersh, D.; Holly, J.; Yewdell, J.W. A few good peptides: MHC class I-based cancer immunosurveillance and immunoevasion. Nat. Rev. Immunol. 2021, 21, 116–128. [Google Scholar] [CrossRef]
- Starck, S.R.; Tsai, J.C.; Chen, K.; Shodiya, M.; Wang, L.; Yahiro, K.; Martins-Green, M.; Shastri, N.; Walter, P. Translation from the 5′ untranslated region shapes the integrated stress response. Science 2016, 351, aad3867. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Kishton, R.J.; Angel, M.; Conn, C.S.; Dalla-Venezia, N.; Marcel, V.; Vincent, A.; Catez, F.; Ferre, S.; Ayadi, L.; et al. Ribosomal Proteins Regulate MHC Class I Peptide Generation for Immunosurveillance. Mol. Cell 2019, 73, 1162–1173.e5. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, K.M.; Lindstrom, M.S. Role of ribosomal protein mutations in tumor development (Review). Int. J. Oncol. 2016, 48, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Apcher, S.; Tovar-Fernadez, M.; Ducellier, S.; Thermou, A.; Nascimento, M.; Sroka, E.; Fahraeus, R. mRNA translation from an antigen presentation perspective: A tribute to the works of Nilabh Shastri. Mol. Immunol. 2022, 141, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Bullock, T.N.; Eisenlohr, L.C. Ribosomal scanning past the primary initiation codon as a mechanism for expression of CTL epitopes encoded in alternative reading frames. J. Exp. Med. 1996, 184, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Bullock, T.N.; Patterson, A.E.; Franlin, L.L.; Notidis, E.; Eisenlohr, L.C. Initiation codon scanthrough versus termination codon readthrough demonstrates strong potential for major histocompatibility complex class I-restricted cryptic epitope expression. J. Exp. Med. 1997, 186, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Weinzierl, A.O.; Maurer, D.; Altenberend, F.; Schneiderhan-Marra, N.; Klingel, K.; Schoor, O.; Wernet, D.; Joos, T.; Rammensee, H.G.; Stevanovic, S. A cryptic vascular endothelial growth factor T-cell epitope: Identification and characterization by mass spectrometry and T-cell assays. Cancer Res. 2008, 68, 2447–2454. [Google Scholar] [CrossRef]
- Champagne, J.; Mordente, K.; Nagel, R.; Agami, R. Slippy-Sloppy translation: A tale of programmed and induced-ribosomal frameshifting. Trends Genet. 2022, 38, 1123–1133. [Google Scholar] [CrossRef]
- Ketteler, R. On programmed ribosomal frameshifting: The alternative proteomes. Front. Genet. 2012, 3, 242. [Google Scholar] [CrossRef]
- Champagne, J.; Pataskar, A.; Blommaert, N.; Nagel, R.; Wernaart, D.; Ramalho, S.; Kenski, J.; Bleijerveld, O.B.; Zaal, E.A.; Berkers, C.R.; et al. Oncogene-dependent sloppiness in mRNA translation. Mol. Cell 2021, 81, 4709–4721.e9. [Google Scholar] [CrossRef] [PubMed]
- Bartok, O.; Pataskar, A.; Nagel, R.; Laos, M.; Goldfarb, E.; Hayoun, D.; Levy, R.; Korner, P.R.; Kreuger, I.Z.M.; Champagne, J.; et al. Anti-tumour immunity induces aberrant peptide presentation in melanoma. Nature 2021, 590, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Pataskar, A.; Champagne, J.; Nagel, R.; Kenski, J.; Laos, M.; Michaux, J.; Pak, H.S.; Bleijerveld, O.B.; Mordente, K.; Navarro, J.M.; et al. Tryptophan depletion results in tryptophan-to-phenylalanine substitutants. Nature 2022, 603, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Nollen, E.A.A.; Rohrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.L.; Philips, M.R. Post-translational modification of RAS proteins. Curr. Opin. Struct. Biol. 2021, 71, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, S.; Tao, Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target Ther. 2020, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Krueger, K.E.; Srivastava, S. Posttranslational protein modifications: Current implications for cancer detection, prevention, and therapeutics. Mol. Cell Proteom. 2006, 5, 1799–1810. [Google Scholar] [CrossRef]
- Witze, E.S.; Old, W.M.; Resing, K.A.; Ahn, N.G. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 2007, 4, 798–806. [Google Scholar] [CrossRef]
- Xu, H.; Wang, Y.; Lin, S.; Deng, W.; Peng, D.; Cui, Q.; Xue, Y. PTMD: A Database of Human Disease-associated Post-translational Modifications. Genom. Proteom. Bioinform. 2018, 16, 244–251. [Google Scholar] [CrossRef]
- Mohammed, F.; Cobbold, M.; Zarling, A.L.; Salim, M.; Barrett-Wilt, G.A.; Shabanowitz, J.; Hunt, D.F.; Engelhard, V.H.; Willcox, B.E. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: A molecular basis for the presentation of transformed self. Nat. Immunol. 2008, 9, 1236–1243. [Google Scholar] [CrossRef]
- Petersen, J.; Wurzbacher, S.J.; Williamson, N.A.; Ramarathinam, S.H.; Reid, H.H.; Nair, A.K.; Zhao, A.Y.; Nastovska, R.; Rudge, G.; Rossjohn, J.; et al. Phosphorylated self-peptides alter human leukocyte antigen class I-restricted antigen presentation and generate tumor-specific epitopes. Proc. Natl. Acad. Sci. USA 2009, 106, 2776–2781. [Google Scholar] [CrossRef] [PubMed]
- Zarling, A.L.; Ficarro, S.B.; White, F.M.; Shabanowitz, J.; Hunt, D.F.; Engelhard, V.H. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J. Exp. Med. 2000, 192, 1755–1762. [Google Scholar] [CrossRef]
- Meyer, V.S.; Drews, O.; Gunder, M.; Hennenlotter, J.; Rammensee, H.G.; Stevanovic, S. Identification of natural MHC class II presented phosphopeptides and tumor-derived MHC class I phospholigands. J. Proteome Res. 2009, 8, 3666–3674. [Google Scholar] [CrossRef] [PubMed]
- Zarling, A.L.; Polefrone, J.M.; Evans, A.M.; Mikesh, L.M.; Shabanowitz, J.; Lewis, S.T.; Engelhard, V.H.; Hunt, D.F. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl. Acad. Sci. USA 2006, 103, 14889–14894. [Google Scholar] [CrossRef] [PubMed]
- Engelhard, V.H.; Obeng, R.C.; Cummings, K.L.; Petroni, G.R.; Ambakhutwala, A.L.; Chianese-Bullock, K.A.; Smith, K.T.; Lulu, A.; Varhegyi, N.; Smolkin, M.E.; et al. MHC-restricted phosphopeptide antigens: Preclinical validation and first-in-humans clinical trial in participants with high-risk melanoma. J. Immunother. Cancer 2020, 8, e000262. [Google Scholar] [CrossRef]
- Ferreira, J.A.; Relvas-Santos, M.; Peixoto, A.; Silva, A.M.; Lara Santos, L. Glycoproteogenomics: Setting the Course for Next-generation Cancer Neo-antigen Discovery for Cancer Vaccines. Genom. Proteom. Bioinform. 2021, 19, 25–43. [Google Scholar] [CrossRef]
- RodrIguez, E.; Schetters, S.T.T.; van Kooyk, Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 2018, 18, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Pinzon Martin, S.; Seeberger, P.H.; Varon Silva, D. Mucins and Pathogenic Mucin-Like Molecules Are Immunomodulators During Infection and Targets for Diagnostics and Vaccines. Front. Chem. 2019, 7, 710. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, D.; Guo, T.; Lin, M. Advances in MUC1-Mediated Breast Cancer Immunotherapy. Biomolecules 2022, 12, 952. [Google Scholar] [CrossRef]
- Holmberg, L.A.; Sandmaier, B.M. Theratope vaccine (STn-KLH). Expert Opin. Biol. Ther. 2001, 1, 881–891. [Google Scholar] [CrossRef]
- Miles, D.; Roche, H.; Martin, M.; Perren, T.J.; Cameron, D.A.; Glaspy, J.; Dodwell, D.; Parker, J.; Mayordomo, J.; Tres, A.; et al. Phase III multicenter clinical trial of the sialyl-TN (STn)-keyhole limpet hemocyanin (KLH) vaccine for metastatic breast cancer. Oncologist 2011, 16, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Gilewski, T.A.; Ragupathi, G.; Dickler, M.; Powell, S.; Bhuta, S.; Panageas, K.; Koganty, R.R.; Chin-Eng, J.; Hudis, C.; Norton, L.; et al. Immunization of high-risk breast cancer patients with clustered sTn-KLH conjugate plus the immunologic adjuvant QS-21. Clin. Cancer Res. 2007, 13, 2977–2985. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, S.; Tachikawa, T.; Shin, S.; Sato, E. Sialosyl-Tn antigen. Its distribution in normal human tissues and expression in adenocarcinomas. Am. J. Clin. Pathol. 1992, 98, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Hauselmann, I.; Borsig, L. Altered tumor-cell glycosylation promotes metastasis. Front. Oncol. 2014, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Sandmaier, B.M.; Oparin, D.V.; Holmberg, L.A.; Reddish, M.A.; MacLean, G.D.; Longenecker, B.M. Evidence of a cellular immune response against sialyl-Tn in breast and ovarian cancer patients after high-dose chemotherapy, stem cell rescue, and immunization with Theratope STn-KLH cancer vaccine. J. Immunother. 1999, 22, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, E.; Sores, J.; Cotton, S.; Peixoto, A.; Ferreira, D.; Freitas, R.; Reis, C.A.; Santos, L.L.; Ferreira, J.A. Esophageal, gastric and colorectal cancers: Looking beyond classical serological biomarkers towards glycoproteomics-assisted precision oncology. Theranostics 2020, 10, 4903–4928. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, V.; Yuriev, E.; Ramsland, P.A.; Halton, J.; Osinski, C.; Li, W.; Plebanski, M.; Paulsen, H.; McKenzie, I.F. A glycopeptide in complex with MHC class I uses the GalNAc residue as an anchor. Proc. Natl. Acad. Sci. USA 2003, 100, 15029–15034. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.K.; Kumar, S.; Dam, V.; Ghersi, D.; Jain, M.; Batra, S.K. MUCIN-4 (MUC4) is a novel tumor antigen in pancreatic cancer immunotherapy. Semin. Immunol. 2020, 47, 101391. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Zhang, X.; Liang, T.; Bai, X. O-GlcNAcylation: An important post-translational modification and a potential therapeutic target for cancer therapy. Mol. Med. 2022, 28, 115. [Google Scholar] [CrossRef]
- Malaker, S.A.; Penny, S.A.; Steadman, L.G.; Myers, P.T.; Loke, J.C.; Raghavan, M.; Bai, D.L.; Shabanowitz, J.; Hunt, D.F.; Cobbold, M. Identification of Glycopeptides as Posttranslationally Modified Neo-antigens in Leukemia. Cancer Immunol. Res. 2017, 5, 376–384. [Google Scholar] [CrossRef]
- Skipper, J.C.; Hendrickson, R.C.; Gulden, P.H.; Brichard, V.; Van, P.A.; Chen, Y.; Shabanowitz, J.; Wolfel, T.; Slingluff, C.L., Jr.; Boon, T.; et al. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 1996, 183, 527–534. [Google Scholar] [CrossRef]
- Blander, J.M. Different routes of MHC-I delivery to phagosomes and their consequences to CD8 T cell immunity. Semin. Immunol. 2023, 66, 101713. [Google Scholar] [CrossRef] [PubMed]
- Chambers, B.; Grufman, P.; Fredriksson, V.; Andersson, K.; Roseboom, M.; Laban, S.; Camps, M.; Wolpert, E.Z.; Wiertz, E.J.; Offringa, R.; et al. Induction of protective CTL immunity against peptide transporter TAP-deficient tumors through dendritic cell vaccination. Cancer Res. 2007, 67, 8450–8455. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.C.; van Hall, T. Alternative Antigen Processing for MHC Class I: Multiple Roads Lead to Rome. Front. Immunol. 2015, 6, 298. [Google Scholar] [CrossRef]
- Oliveira, C.C.; van Hall, T. Importance of TAP-independent processing pathways. Mol. Immunol. 2013, 55, 113–116. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H. Papillomaviruses in human cancer. Cancer 1987, 59, 1692–1696. [Google Scholar] [CrossRef]
- Smalley Rumfield, C.; Roller, N.; Pellom, S.T.; Schlom, J.; Jochems, C. Therapeutic Vaccines for HPV-Associated Malignancies. Immunotargets Ther. 2020, 9, 167–200. [Google Scholar] [CrossRef]
- zur Hausen, H. Papillomavirus infections—A major cause of human cancers. Biochim. Biophys. Acta 1996, 1288, F55–F78. [Google Scholar] [CrossRef]
- Mirabello, L.; Yeager, M.; Yu, K.; Clifford, G.M.; Xiao, Y.; Zhu, B.; Cullen, M.; Boland, J.F.; Wentzensen, N.; Nelson, C.W.; et al. HPV16 E7 Genetic Conservation Is Critical to Carcinogenesis. Cell 2017, 170, 1164–1174.e6. [Google Scholar] [CrossRef]
- Ye, R.; Wang, A.; Bu, B.; Luo, P.; Deng, W.; Zhang, X.; Yin, S. Viral oncogenes, viruses, and cancer: A third-generation sequencing perspective on viral integration into the human genome. Front. Oncol. 2023, 13, 1333812. [Google Scholar] [CrossRef]
- Munger, K.; Baldwin, A.; Edwards, K.M.; Hayakawa, H.; Nguyen, C.L.; Owens, M.; Grace, M.; Huh, K. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 2004, 78, 11451–11460. [Google Scholar] [CrossRef] [PubMed]
- Gruener, M.; Bravo, I.G.; Momburg, F.; Alonso, A.; Tomakidi, P. The E5 protein of the human papillomavirus type 16 down-regulates HLA-I surface expression in calnexin-expressing but not in calnexin-deficient cells. Virol. J. 2007, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Lundine, D.; Leeman, J.E.; Higginson, D.S. Genomic Signatures in HPV-Associated Tumors. Viruses 2021, 13, 1998. [Google Scholar] [CrossRef] [PubMed]
- Cooper, B.; Schneider, S.; Bohl, J.; Jiang, Y.; Beaudet, A.; Vande Pol, S. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology 2003, 306, 87–99. [Google Scholar] [CrossRef]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Vande Pol, S.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef]
- Emeny, R.T.; Wheeler, C.M.; Jansen, K.U.; Hunt, W.C.; Fu, T.M.; Smith, J.F.; MacMullen, S.; Esser, M.T.; Paliard, X. Priming of human papillomavirus type 11-specific humoral and cellular immune responses in college-aged women with a virus-like particle vaccine. J. Virol. 2002, 76, 7832–7842. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Zhai, L.; Tumban, E. Virus-like Particle-Based L2 Vaccines against HPVs: Where Are We Today? Viruses 2019, 12, 18. [Google Scholar] [CrossRef]
- Fallon, J.; Tighe, R.; Kradjian, G.; Guzman, W.; Bernhardt, A.; Neuteboom, B.; Lan, Y.; Sabzevari, H.; Schlom, J.; Greiner, J.W. The immunocytokine NHS-IL12 as a potential cancer therapeutic. Oncotarget 2014, 5, 1869–1884. [Google Scholar] [CrossRef]
- Smalley Rumfield, C.; Pellom, S.T.; Morillon Ii, Y.M.; Schlom, J.; Jochems, C. Immunomodulation to enhance the efficacy of an HPV therapeutic vaccine. J. Immunother. Cancer 2020, 8, e000612. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ (accessed on 25 March 2024).
- Solares, A.M.; Baladron, I.; Ramos, T.; Valenzuela, C.; Borbon, Z.; Fanjull, S.; Gonzalez, L.; Castillo, D.; Esmir, J.; Granadillo, M.; et al. Safety and Immunogenicity of a Human Papillomavirus Peptide Vaccine (CIGB-228) in Women with High-Grade Cervical Intraepithelial Neoplasia: First-in-Human, Proof-of-Concept Trial. ISRN Obstet. Gynecol. 2011, 2011, 292951. [Google Scholar] [CrossRef]
- Voskens, C.J.; Sewell, D.; Hertzano, R.; DeSanto, J.; Rollins, S.; Lee, M.; Taylor, R.; Wolf, J.; Suntharalingam, M.; Gastman, B.; et al. Induction of MAGE-A3 and HPV-16 immunity by Trojan vaccines in patients with head and neck carcinoma. Head Neck 2012, 34, 1734–1746. [Google Scholar] [CrossRef] [PubMed]
- Zandberg, D.P.; Rollins, S.; Goloubeva, O.; Morales, R.E.; Tan, M.; Taylor, R.; Wolf, J.S.; Schumaker, L.M.; Cullen, K.J.; Zimrin, A.; et al. A phase I dose escalation trial of MAGE-A3- and HPV16-specific peptide immunomodulatory vaccines in patients with recurrent/metastatic (RM) squamous cell carcinoma of the head and neck (SCCHN). Cancer Immunol. Immunother. 2015, 64, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Yang, M.C.; Knoff, J.; Wu, T.C.; Hung, C.F. Cancer immunotherapy employing an innovative strategy to enhance CD4+ T cell help in the tumor microenvironment. PLoS ONE 2014, 9, e115711. [Google Scholar] [CrossRef] [PubMed]
- Karkada, M.; Quinton, T.; Blackman, R.; Mansour, M. Tumor Inhibition by DepoVax-Based Cancer Vaccine Is Accompanied by Reduced Regulatory/Suppressor Cell Proliferation and Tumor Infiltration. ISRN Oncol. 2013, 2013, 753427. [Google Scholar] [CrossRef] [PubMed]
- Reuschenbach, M.; Pauligk, C.; Karbach, J.; Rafiyan, M.R.; Kloor, M.; Prigge, E.S.; Sauer, M.; Al-Batran, S.E.; Kaufmann, A.M.; Schneider, A.; et al. A phase 1/2a study to test the safety and immunogenicity of a p16(INK4a) peptide vaccine in patients with advanced human papillomavirus-associated cancers. Cancer 2016, 122, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- Kenter, G.G.; Welters, M.J.; Valentijn, A.R.; Lowik, M.J.; Berends-van der Meer, D.M.; Vloon, A.P.; Drijfhout, J.W.; Wafelman, A.R.; Oostendorp, J.; Fleuren, G.J.; et al. Phase I immunotherapeutic trial with long peptides spanning the E6 and E7 sequences of high-risk human papillomavirus 16 in end-stage cervical cancer patients shows low toxicity and robust immunogenicity. Clin. Cancer Res. 2008, 14, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Kenter, G.G.; Welters, M.J.; Valentijn, A.R.; Lowik, M.J.; Berends-van der Meer, D.M.; Vloon, A.P.; Essahsah, F.; Fathers, L.M.; Offringa, R.; Drijfhout, J.W.; et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N. Engl. J. Med. 2009, 361, 1838–1847. [Google Scholar] [CrossRef]
- van Poelgeest, M.I.; Welters, M.J.; van Esch, E.M.; Stynenbosch, L.F.; Kerpershoek, G.; van Persijn van Meerten, E.L.; van den Hende, M.; Lowik, M.J.; Berends-van der Meer, D.M.; Fathers, L.M.; et al. HPV16 synthetic long peptide (HPV16-SLP) vaccination therapy of patients with advanced or recurrent HPV16-induced gynecological carcinoma, a phase II trial. J. Transl. Med. 2013, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, W.W.; Stratton, S.L.; Myrick, R.S.; Vaughn, R.; Donnalley, L.M.; Coleman, H.N.; Mercado, M.; Moerman-Herzog, A.M.; Spencer, H.J.; Andrews-Collins, N.R.; et al. A phase I dose-escalation clinical trial of a peptide-based human papillomavirus therapeutic vaccine with Candida skin test reagent as a novel vaccine adjuvant for treating women with biopsy-proven cervical intraepithelial neoplasia 2/3. Oncoimmunology 2015, 4, e1031439. [Google Scholar] [CrossRef]
- Coleman, H.N.; Greenfield, W.W.; Stratton, S.L.; Vaughn, R.; Kieber, A.; Moerman-Herzog, A.M.; Spencer, H.J.; Hitt, W.C.; Quick, C.M.; Hutchins, L.F.; et al. Human papillomavirus type 16 viral load is decreased following a therapeutic vaccination. Cancer Immunol. Immunother. 2016, 65, 563–573. [Google Scholar] [CrossRef]
- Wang, X.; Coleman, H.N.; Nagarajan, U.; Spencer, H.J.; Nakagawa, M. Candida skin test reagent as a novel adjuvant for a human papillomavirus peptide-based therapeutic vaccine. Vaccine 2013, 31, 5806–5813. [Google Scholar] [CrossRef]
- Zom, G.G.; Willems, M.; Khan, S.; van der Sluis, T.C.; Kleinovink, J.W.; Camps, M.G.M.; van der Marel, G.A.; Filippov, D.V.; Melief, C.J.M.; Ossendorp, F. Novel TLR2-binding adjuvant induces enhanced T cell responses and tumor eradication. J. Immunother. Cancer 2018, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Slingerland, M.; Speetjens, F.; Welters, M.; Gelderblom, H.; Roozen, I.; van der Velden, L.-A.; Melief, C.J.; Zandvliet, M.; van der Burg, S.; Ossendorp, F. A phase I study in patients with a human papillomavirus type 16 positive oropharyngeal tumor treated with second generation synthetic long peptide vaccine conjugated to a defined adjuvant. J. Clin. Oncol. 2016, 34, TPS3113. [Google Scholar] [CrossRef]
- Maynard, S.K.; Marshall, J.D.; MacGill, R.S.; Yu, L.; Cann, J.A.; Cheng, L.I.; McCarthy, M.P.; Cayatte, C.; Robbins, S.H. Vaccination with synthetic long peptide formulated with CpG in an oil-in-water emulsion induces robust E7-specific CD8 T cell responses and TC-1 tumor eradication. BMC Cancer 2019, 19, 540. [Google Scholar] [CrossRef] [PubMed]
- Galliverti, G.; Tichet, M.; Domingos-Pereira, S.; Hauert, S.; Nardelli-Haefliger, D.; Swartz, M.A.; Hanahan, D.; Wullschleger, S. Nanoparticle Conjugation of Human Papillomavirus 16 E7-long Peptides Enhances Therapeutic Vaccine Efficacy against Solid Tumors in Mice. Cancer Immunol. Res. 2018, 6, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Conarty, J.P.; Wieland, A. The Tumor-Specific Immune Landscape in HPV+ Head and Neck Cancer. Viruses 2023, 15, 1296. [Google Scholar] [CrossRef] [PubMed]
- Stevanović, S.; Draper, L.M.; Langhan, M.M.; Campbell, T.E.; Kwong, M.L.; Wunderlich, J.R.; Dudley, M.E.; Yang, J.C.; Sherry, R.M.; Kammula, U.S.; et al. Complete Regression of Metastatic Cervical Cancer After Treatment with Human Papillomavirus–Targeted Tumor-Infiltrating T Cells. J. Clin. Oncol. 2015, 33, 1543–1550. [Google Scholar] [CrossRef]
- Stevanovic, S.; Helman, S.R.; Wunderlich, J.R.; Langhan, M.M.; Doran, S.L.; Kwong, M.L.M.; Somerville, R.P.T.; Klebanoff, C.A.; Kammula, U.S.; Sherry, R.M.; et al. A Phase II Study of Tumor-infiltrating Lymphocyte Therapy for Human Papillomavirus-associated Epithelial Cancers. Clin. Cancer Res. 2019, 25, 1486–1493. [Google Scholar] [CrossRef]
- Doran, S.L.; Stevanovic, S.; Adhikary, S.; Gartner, J.J.; Jia, L.; Kwong, M.L.M.; Faquin, W.C.; Hewitt, S.M.; Sherry, R.M.; Yang, J.C.; et al. T-Cell Receptor Gene Therapy for Human Papillomavirus-Associated Epithelial Cancers: A First-in-Human, Phase I/II Study. J. Clin. Oncol. 2019, 37, 2759–2768. [Google Scholar] [CrossRef]
- Nagarsheth, N.B.; Norberg, S.M.; Sinkoe, A.L.; Adhikary, S.; Meyer, T.J.; Lack, J.B.; Warner, A.C.; Schweitzer, C.; Doran, S.L.; Korrapati, S.; et al. TCR-engineered T cells targeting E7 for patients with metastatic HPV-associated epithelial cancers. Nat. Med. 2021, 27, 419–425. [Google Scholar] [CrossRef]
- Eberhardt, C.S.; Kissick, H.T.; Patel, M.R.; Cardenas, M.A.; Prokhnevska, N.; Obeng, R.C.; Nasti, T.H.; Griffith, C.C.; Im, S.J.; Wang, X.; et al. Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature 2021, 597, 279–284. [Google Scholar] [CrossRef]
- Iwanaga, M.; Watanabe, T.; Yamaguchi, K. Adult T-cell leukemia: A review of epidemiological evidence. Front. Microbiol. 2012, 3, 322. [Google Scholar] [CrossRef]
- Hermine, O.; Ramos, J.C.; Tobinai, K. A Review of New Findings in Adult T-cell Leukemia-Lymphoma: A Focus on Current and Emerging Treatment Strategies. Adv. Ther. 2018, 35, 135–152. [Google Scholar] [CrossRef]
- Shimoyama, M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984–87). Br. J. Haematol. 1991, 79, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Kirk, P.D.; Huvet, M.; Melamed, A.; Maertens, G.N.; Bangham, C.R. Retroviruses integrate into a shared, non-palindromic DNA motif. Nat. Microbiol. 2016, 2, 16212. [Google Scholar] [CrossRef]
- Schnell, A.P.; Kohrt, S.; Thoma-Kress, A.K. Latency Reversing Agents: Kick and Kill of HTLV-1? Int. J. Mol. Sci. 2021, 22, 5545. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, K.; Rowan, A.G.; Tanaka, Y.; Taylor, G.P.; Bangham, C.R. CADM1/TSLC1 Identifies HTLV-1-Infected Cells and Determines Their Susceptibility to CTL-Mediated Lysis. PLoS Pathog. 2016, 12, e1005560. [Google Scholar] [CrossRef] [PubMed]
- Furuta, R.; Yasunaga, J.I.; Miura, M.; Sugata, K.; Saito, A.; Akari, H.; Ueno, T.; Takenouchi, N.; Fujisawa, J.I.; Koh, K.R.; et al. Human T-cell leukemia virus type 1 infects multiple lineage hematopoietic cells in vivo. PLoS Pathog. 2017, 13, e1006722. [Google Scholar] [CrossRef]
- Suehiro, Y.; Hasegawa, A.; Iino, T.; Sasada, A.; Watanabe, N.; Matsuoka, M.; Takamori, A.; Tanosaki, R.; Utsunomiya, A.; Choi, I.; et al. Clinical outcomes of a novel therapeutic vaccine with Tax peptide-pulsed dendritic cells for adult T cell leukaemia/lymphoma in a pilot study. Br. J. Haematol. 2015, 169, 356–367. [Google Scholar] [CrossRef]
- Harashima, N.; Kurihara, K.; Utsunomiya, A.; Tanosaki, R.; Hanabuchi, S.; Masuda, M.; Ohashi, T.; Fukui, F.; Hasegawa, A.; Masuda, T.; et al. Graft-versus-Tax response in adult T-cell leukemia patients after hematopoietic stem cell transplantation. Cancer Res. 2004, 64, 391–399. [Google Scholar] [CrossRef]
- Harashima, N.; Tanosaki, R.; Shimizu, Y.; Kurihara, K.; Masuda, T.; Okamura, J.; Kannagi, M. Identification of two new HLA-A*1101-restricted tax epitopes recognized by cytotoxic T lymphocytes in an adult T-cell leukemia patient after hematopoietic stem cell transplantation. J. Virol. 2005, 79, 10088–10092. [Google Scholar] [CrossRef] [PubMed]
- Kannagi, M.; Shida, H.; Igarashi, H.; Kuruma, K.; Murai, H.; Aono, Y.; Maruyama, I.; Osame, M.; Hattori, T.; Inoko, H.; et al. Target epitope in the Tax protein of human T-cell leukemia virus type I recognized by class I major histocompatibility complex-restricted cytotoxic T cells. J. Virol. 1992, 66, 2928–2933. [Google Scholar] [CrossRef] [PubMed]
- Kannagi, M.; Hasegawa, A.; Nagano, Y.; Iino, T.; Okamura, J.; Suehiro, Y. Maintenance of long remission in adult T-cell leukemia by Tax-targeted vaccine: A hope for disease-preventive therapy. Cancer Sci. 2019, 110, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Tanaka, Y.; Nakasone, H.; Ishihara, Y.; Kako, S.; Kobayashi, S.; Tanaka, Y.; Ohmori, T.; Uchimaru, K.; Okamoto, S.; et al. Development of a Unique T Cell Receptor Gene-Transferred Tax-Redirected T Cell Immunotherapy for Adult T Cell Leukemia. Biol. Blood Marrow Transpl. 2020, 26, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Revaud, D.; Bejanariu, A.; Loussaief, L.; Sarry, E.; Zemmar, A.; Deplaine, G.; Coman, T.; Rossignol, J.; Hermine, O.; Bauche, C. Development of an Anti-HTLV-1 Vaccine for the Treatment of Adult T-Cell Leukemia/Lymphoma. Blood 2015, 126, 4010. [Google Scholar] [CrossRef]
- Ishizawa, M.; Ganbaatar, U.; Hasegawa, A.; Takatsuka, N.; Kondo, N.; Yoneda, T.; Katagiri, K.; Masuda, T.; Utsunomiya, A.; Kannagi, M. Short-term cultured autologous peripheral blood mononuclear cells as a potential immunogen to activate Tax-specific CTL response in adult T-cell leukemia patients. Cancer Sci. 2021, 112, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Jo, T.; Kubota-Koketsu, R.; Kaneko, Y.; Sakai, T.; Noguchi, K.; Irie, S.; Matsuo, M.; Taguchi, J.; Abe, K.; Shigematsu, K. Live attenuated VZV vaccination induces antitumor immunity in ATLL patients. Cancer Immunol. Immunother. 2023, 72, 929–944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xu, M. EBV-induced T-cell responses in EBV-specific and nonspecific cancers. Front. Immunol. 2023, 14, 1250946. [Google Scholar] [CrossRef] [PubMed]
- Murray, R.J.; Kurilla, M.G.; Brooks, J.M.; Thomas, W.A.; Rowe, M.; Kieff, E.; Rickinson, A.B. Identification of target antigens for the human cytotoxic T cell response to Epstein-Barr virus (EBV): Implications for the immune control of EBV-positive malignancies. J. Exp. Med. 1992, 176, 157–168. [Google Scholar] [CrossRef]
- Steven, N.M.; Leese, A.M.; Annels, N.E.; Lee, S.P.; Rickinson, A.B. Epitope focusing in the primary cytotoxic T cell response to Epstein-Barr virus and its relationship to T cell memory. J. Exp. Med. 1996, 184, 1801–1813. [Google Scholar] [CrossRef]
- von Witzleben, A.; Wang, C.; Laban, S.; Savelyeva, N.; Ottensmeier, C.H. HNSCC: Tumour Antigens and Their Targeting by Immunotherapy. Cells 2020, 9, 2103. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, E.B.; Ladanyi, M.; Emanuel, D.; Mackinnon, S.; Boulad, F.; Carabasi, M.H.; Castro-Malaspina, H.; Childs, B.H.; Gillio, A.P.; Small, T.N.; et al. Infusions of donor leukocytes to treat Epstein-Barr virus-associated lymphoproliferative disorders after allogeneic bone marrow transplantation. N. Engl. J. Med. 1994, 330, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Bollard, C.M.; Gottschalk, S.; Torrano, V.; Diouf, O.; Ku, S.; Hazrat, Y.; Carrum, G.; Ramos, C.; Fayad, L.; Shpall, E.J.; et al. Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein-Barr virus latent membrane proteins. J. Clin. Oncol. 2014, 32, 798–808. [Google Scholar] [CrossRef]
- Chia, W.K.; Teo, M.; Wang, W.W.; Lee, B.; Ang, S.F.; Tai, W.M.; Chee, C.L.; Ng, J.; Kan, R.; Lim, W.T.; et al. Adoptive T-cell transfer and chemotherapy in the first-line treatment of metastatic and/or locally recurrent nasopharyngeal carcinoma. Mol. Ther. 2014, 22, 132–139. [Google Scholar] [CrossRef]
- Cho, S.G.; Kim, N.; Sohn, H.J.; Lee, S.K.; Oh, S.T.; Lee, H.J.; Cho, H.I.; Yim, H.W.; Jung, S.E.; Park, G.; et al. Long-term Outcome of Extranodal NK/T Cell Lymphoma Patients Treated with Postremission Therapy Using EBV LMP1 and LMP2a-specific CTLs. Mol. Ther. 2015, 23, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, L.P.; Rouce, R.; Gottschalk, S.; Torrano, V.; Carrum, G.; Wu, M.F.; Hoq, F.; Grilley, B.; Marcogliese, A.M.; Hanley, P.J.; et al. EBV/LMP-specific T cells maintain remissions of T- and B-cell EBV lymphomas after allogeneic bone marrow transplantation. Blood 2018, 132, 2351–2361. [Google Scholar] [CrossRef] [PubMed]
- Comoli, P.; Pedrazzoli, P.; Maccario, R.; Basso, S.; Carminati, O.; Labirio, M.; Schiavo, R.; Secondino, S.; Frasson, C.; Perotti, C.; et al. Cell therapy of stage IV nasopharyngeal carcinoma with autologous Epstein-Barr virus-targeted cytotoxic T lymphocytes. J. Clin. Oncol. 2005, 23, 8942–8949. [Google Scholar] [CrossRef] [PubMed]
- Fae, D.A.; Martorelli, D.; Mastorci, K.; Muraro, E.; Dal Col, J.; Franchin, G.; Barzan, L.; Comaro, E.; Vaccher, E.; Rosato, A.; et al. Broadening Specificity and Enhancing Cytotoxicity of Adoptive T Cells for Nasopharyngeal Carcinoma Immunotherapy. Cancer Immunol. Res. 2016, 4, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.I.; Kim, U.H.; Shin, A.R.; Won, J.N.; Lee, H.J.; Sohn, H.J.; Kim, T.G. A novel Epstein-Barr virus-latent membrane protein-1-specific T-cell receptor for TCR gene therapy. Br. J. Cancer 2018, 118, 534–545. [Google Scholar] [CrossRef]
- Orentas, R.J.; Roskopf, S.J.; Nolan, G.P.; Nishimura, M.I. Retroviral transduction of a T cell receptor specific for an Epstein-Barr virus-encoded peptide. Clin. Immunol. 2001, 98, 220–228. [Google Scholar] [CrossRef]
- Kobayashi, E.; Mizukoshi, E.; Kishi, H.; Ozawa, T.; Hamana, H.; Nagai, T.; Nakagawa, H.; Jin, A.; Kaneko, S.; Muraguchi, A. A new cloning and expression system yields and validates TCRs from blood lymphocytes of patients with cancer within 10 days. Nat. Med. 2013, 19, 1542–1546. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Shao, Q.; Sun, H.; Mu, X.; Gao, Y.; Jiang, R.; Hou, J.; Yao, K.; Chen, Y.; Sun, B. Evaluation of Epstein-Barr virus latent membrane protein 2 specific T-cell receptors driven by T-cell specific promoters using lentiviral vector. Clin. Dev. Immunol. 2011, 2011, 716926. [Google Scholar] [CrossRef] [PubMed]
- Schaft, N.; Lankiewicz, B.; Drexhage, J.; Berrevoets, C.; Moss, D.J.; Levitsky, V.; Bonneville, M.; Lee, S.P.; McMichael, A.J.; Gratama, J.W.; et al. T cell re-targeting to EBV antigens following TCR gene transfer: CD28-containing receptors mediate enhanced antigen-specific IFNgamma production. Int. Immunol. 2006, 18, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Lo, W.F.; Lee, T.H.; Ren, Y.; Hwang, S.L.; Cheng, Y.F.; Chen, C.L.; Chang, Y.S.; Lee, S.P.; Rickinson, A.B.; et al. Immunization with Epstein-Barr Virus (EBV) peptide-pulsed dendritic cells induces functional CD8+ T-cell immunity and may lead to tumor regression in patients with EBV-positive nasopharyngeal carcinoma. Cancer Res. 2002, 62, 6952–6958. [Google Scholar] [PubMed]
- Henle, W.; Henle, G.; Niederman, J.C.; Klemola, E.; Haltia, K. Antibodies to early antigens induced by Epstein-Barr virus in infectious mononucleosis. J. Infect. Dis. 1971, 124, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Dastmalchi, F.; Karachi, A.; Mitchell, D. The role of CMV in glioblastoma and implications for immunotherapeutic strategies. Oncoimmunology 2019, 8, e1514921. [Google Scholar] [CrossRef] [PubMed]
- Dziurzynski, K.; Chang, S.M.; Heimberger, A.B.; Kalejta, R.F.; McGregor Dallas, S.R.; Smit, M.; Soroceanu, L.; Cobbs, C.S.; Hcmv; Gliomas, S. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol. 2012, 14, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Wakefield, A.; Pignata, A.; Ghazi, A.; Ashoori, A.; Hegde, M.; Landi, D.; Gray, T.; Scheurer, M.E.; Chintagumpala, M.; Adesina, A.; et al. Is CMV a target in pediatric glioblastoma? Expression of CMV proteins, pp65 and IE1-72 and CMV nucleic acids in a cohort of pediatric glioblastoma patients. J. Neurooncol. 2015, 125, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Lucas, K.G.; Bao, L.; Bruggeman, R.; Dunham, K.; Specht, C. The detection of CMV pp65 and IE1 in glioblastoma multiforme. J. Neurooncol. 2011, 103, 231–238. [Google Scholar] [CrossRef]
- Bahador, M.; Gras Navarro, A.; Rahman, M.A.; Dominguez-Valentin, M.; Sarowar, S.; Ulvestad, E.; Njolstad, G.; Lie, S.A.; Kristoffersen, E.K.; Bratland, E.; et al. Increased infiltration and tolerised antigen-specific CD8+ T(EM) cells in tumor but not peripheral blood have no impact on survival of HCMV+ glioblastoma patients. Oncoimmunology 2017, 6, e1336272. [Google Scholar] [CrossRef]
- Kim, J.W.; Kane, J.R.; Panek, W.K.; Young, J.S.; Rashidi, A.; Yu, D.; Kanojia, D.; Hasan, T.; Miska, J.; Gomez-Lim, M.A.; et al. A Dendritic Cell-Targeted Adenoviral Vector Facilitates Adaptive Immune Response Against Human Glioma Antigen (CMV-IE) and Prolongs Survival in a Human Glioma Tumor Model. Neurotherapeutics 2018, 15, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Gehrcken, L.; Sauerer, T.; Schaft, N.; Dorrie, J. T-Cell Responses in Merkel Cell Carcinoma: Implications for Improved Immune Checkpoint Blockade and Other Therapeutic Options. Int. J. Mol. Sci. 2021, 22, 8679. [Google Scholar] [CrossRef] [PubMed]
- Houben, R.; Celikdemir, B.; Kervarrec, T.; Schrama, D. Merkel Cell Polyomavirus: Infection, Genome, Transcripts and Its Role in Development of Merkel Cell Carcinoma. Cancers 2023, 15, 444. [Google Scholar] [CrossRef] [PubMed]
- Schrama, D.; Sarosi, E.M.; Adam, C.; Ritter, C.; Kaemmerer, U.; Klopocki, E.; Konig, E.M.; Utikal, J.; Becker, J.C.; Houben, R. Characterization of six Merkel cell polyomavirus-positive Merkel cell carcinoma cell lines: Integration pattern suggest that large T antigen truncating events occur before or during integration. Int. J. Cancer 2019, 145, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Iyer, J.G.; Afanasiev, O.K.; McClurkan, C.; Paulson, K.; Nagase, K.; Jing, L.; Marshak, J.O.; Dong, L.; Carter, J.; Lai, I.; et al. Merkel cell polyomavirus-specific CD8+ and CD4+ T-cell responses identified in Merkel cell carcinomas and blood. Clin. Cancer Res. 2011, 17, 6671–6680. [Google Scholar] [CrossRef] [PubMed]
- Lyngaa, R.; Pedersen, N.W.; Schrama, D.; Thrue, C.A.; Ibrani, D.; Met, O.; Straten, P.T.; Nghiem, P.; Becker, J.C.; Hadrup, S.R. T-cell responses to oncogenic merkel cell polyomavirus proteins distinguish patients with merkel cell carcinoma from healthy donors. Clin. Cancer Res. 2014, 20, 1768–1778. [Google Scholar] [CrossRef] [PubMed]
- Gerer, K.F.; Erdmann, M.; Hadrup, S.R.; Lyngaa, R.; Martin, L.M.; Voll, R.E.; Schuler-Thurner, B.; Schuler, G.; Schaft, N.; Hoyer, S.; et al. Preclinical evaluation of NF-kappaB-triggered dendritic cells expressing the viral oncogenic driver of Merkel cell carcinoma for therapeutic vaccination. Ther. Adv. Med. Oncol. 2017, 9, 451–464. [Google Scholar] [CrossRef]
- Samimi, M.; Benlalam, H.; Aumond, P.; Gaboriaud, P.; Fradin, D.; Kervarrec, T.; Florenceau, L.; Vignard, V.; Blom, A.; Touze, A.; et al. Viral and tumor antigen-specific CD8 T-cell responses in Merkel cell carcinoma. Cell Immunol. 2019, 344, 103961. [Google Scholar] [CrossRef] [PubMed]
- Hansen, U.K.; Lyngaa, R.; Ibrani, D.; Church, C.; Verhaegen, M.; Dlugosz, A.A.; Becker, J.C.; Straten, P.T.; Nghiem, P.; Hadrup, S.R. Extended T-Cell Epitope Landscape in Merkel Cell Polyomavirus Large T and Small T Oncoproteins Identified Uniquely in Patients with Cancer. J. Investig. Dermatol. 2022, 142, 239–243.e13. [Google Scholar] [CrossRef]
- Schulz, T.F.; Freise, A.; Stein, S.C. Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen: More than a key mediator of viral persistence. Curr. Opin. Virol. 2023, 61, 101336. [Google Scholar] [CrossRef]
- Toptan, T.; Fonseca, L.; Kwun, H.J.; Chang, Y.; Moore, P.S. Complex alternative cytoplasmic protein isoforms of the Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 generated through noncanonical translation initiation. J. Virol. 2013, 87, 2744–2755. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, G.; Chan, B.; Samarina, N.; Abere, B.; Weidner-Glunde, M.; Buch, A.; Pich, A.; Brinkmann, M.M.; Schulz, T.F. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl. Acad. Sci. USA 2016, 113, E1034–E1043. [Google Scholar] [CrossRef]
- Davis, D.A.; Naiman, N.E.; Wang, V.; Shrestha, P.; Haque, M.; Hu, D.; Anagho, H.A.; Carey, R.F.; Davidoff, K.S.; Yarchoan, R. Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses. PLoS Pathog. 2015, 11, e1005064. [Google Scholar] [CrossRef] [PubMed]
- Nalwoga, A.; Roshan, R.; Moore, K.; Marshall, V.; Miley, W.; Labo, N.; Nakibuule, M.; Cose, S.; Rochford, R.; Newton, R.; et al. Kaposi’s sarcoma-associated herpesvirus T cell responses in HIV seronegative individuals from rural Uganda. Nat. Commun. 2021, 12, 7323. [Google Scholar] [CrossRef]
- Brander, C.; Suscovich, T.; Lee, Y.; Nguyen, P.T.; O’Connor, P.; Seebach, J.; Jones, N.G.; van Gorder, M.; Walker, B.D.; Scadden, D.T. Impaired CTL recognition of cells latently infected with Kaposi’s sarcoma-associated herpes virus. J. Immunol. 2000, 165, 2077–2083. [Google Scholar] [CrossRef]
- Sabbah, S.; Jagne, Y.J.; Zuo, J.; de Silva, T.; Ahasan, M.M.; Brander, C.; Rowland-Jones, S.; Flanagan, K.L.; Hislop, A.D. T-cell immunity to Kaposi sarcoma-associated herpesvirus: Recognition of primary effusion lymphoma by LANA-specific CD4+ T cells. Blood 2012, 119, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- Roshan, R.; Labo, N.; Trivett, M.; Miley, W.; Marshall, V.; Coren, L.; Cornejo Castro, E.M.; Perez, H.; Holdridge, B.; Davis, E.; et al. T-cell responses to KSHV infection: A systematic approach. Oncotarget 2017, 8, 109402–109416. [Google Scholar] [CrossRef] [PubMed]
- Broussard, G.; Damania, B. KSHV: Immune Modulation and Immunotherapy. Front. Immunol. 2019, 10, 3084. [Google Scholar] [CrossRef]
- Munz, C. The Role of Dendritic Cells in Immune Control and Vaccination against-Herpesviruses. Viruses 2019, 11, 1125. [Google Scholar] [CrossRef]
- de Martel, C.; Maucort-Boulch, D.; Plummer, M.; Franceschi, S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology 2015, 62, 1190–1200. [Google Scholar] [CrossRef]
- Tumen, D.; Heumann, P.; Gulow, K.; Demirci, C.N.; Cosma, L.S.; Muller, M.; Kandulski, A. Pathogenesis and Current Treatment Strategies of Hepatocellular Carcinoma. Biomedicines 2022, 10, 3202. [Google Scholar] [CrossRef]
- Yang, J.D.; Roberts, L.R. Hepatocellular carcinoma: A global view. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 448–458. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Amaddeo, G.; Cao, Q.; Ladeiro, Y.; Imbeaud, S.; Nault, J.C.; Jaoui, D.; Gaston Mathe, Y.; Laurent, C.; Laurent, A.; Bioulac-Sage, P.; et al. Integration of tumour and viral genomic characterizations in HBV-related hepatocellular carcinomas. Gut 2015, 64, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Jayasuryan, N.; Kumar, R. A truncated mutant (residues 58-140) of the hepatitis B virus X protein retains transactivation function. Proc. Natl. Acad. Sci. USA 1996, 93, 5647–5652. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Budzinska, M.A.; Shackel, N.A.; Urban, S. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses 2017, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.T.; Yang, N.; Lee Krishnamoorthy, T.; Oei, V.; Chua, A.; Zhao, X.; Tan, H.S.; Chia, A.; Le Bert, N.; Low, D.; et al. Use of Expression Profiles of HBV-DNA Integrated Into Genomes of Hepatocellular Carcinoma Cells to Select T Cells for Immunotherapy. Gastroenterology 2019, 156, 1862–1876.e9. [Google Scholar] [CrossRef]
- de Beijer, M.T.A.; Jansen, D.; Dou, Y.; van Esch, W.J.E.; Mok, J.Y.; Maas, M.J.P.; Brasser, G.; de Man, R.A.; Woltman, A.M.; Buschow, S.I. Discovery and Selection of Hepatitis B Virus-Derived T Cell Epitopes for Global Immunotherapy Based on Viral Indispensability, Conservation, and HLA-Binding Strength. J. Virol. 2020, 94, e01663-19. [Google Scholar] [CrossRef] [PubMed]
- de Beijer, M.T.A.; Bezstarosti, K.; Luijten, R.; Doff, W.A.S.; Boor, P.P.C.; Pieterman, R.F.A.; Bouzid, R.; Biesta, P.J.; Ijzermans, J.N.M.; Doukas, M.; et al. Immunopeptidome of hepatocytes isolated from patients with HBV infection and hepatocellular carcinoma. JHEP Rep. 2022, 4, 100576. [Google Scholar] [CrossRef]
- Tagliamonte, M.; Petrizzo, A.; Mauriello, A.; Tornesello, M.L.; Buonaguro, F.M.; Buonaguro, L. Potentiating cancer vaccine efficacy in liver cancer. Oncoimmunology 2018, 7, e1488564. [Google Scholar] [CrossRef]
- Loffler, M.W.; Gori, S.; Izzo, F.; Mayer-Mokler, A.; Ascierto, P.A.; Konigsrainer, A.; Ma, Y.T.; Sangro, B.; Francque, S.; Vonghia, L.; et al. Phase I/II Multicenter Trial of a Novel Therapeutic Cancer Vaccine, HepaVac-101, for Hepatocellular Carcinoma. Clin. Cancer Res. 2022, 28, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Honda, T. Roles of Human Endogenous Retroviruses and Endogenous Virus-Like Elements in Cancer Development and Innate Immunity. Biomolecules 2023, 13, 1706. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Cherkasova, E.; Malinzak, E.; Rao, S.; Takahashi, Y.; Senchenko, V.N.; Kudryavtseva, A.V.; Nickerson, M.L.; Merino, M.; Hong, J.A.; Schrump, D.S.; et al. Inactivation of the von Hippel-Lindau tumor suppressor leads to selective expression of a human endogenous retrovirus in kidney cancer. Oncogene 2011, 30, 4697–4706. [Google Scholar] [CrossRef]
- Zapatka, M.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sultmann, H.; Moch, H.; Pathogens, P.; et al. The landscape of viral associations in human cancers. Nat. Genet. 2020, 52, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Cuffel, C.; Rivals, J.P.; Zaugg, Y.; Salvi, S.; Seelentag, W.; Speiser, D.E.; Lienard, D.; Monnier, P.; Romero, P.; Bron, L.; et al. Pattern and clinical significance of cancer-testis gene expression in head and neck squamous cell carcinoma. Int. J. Cancer 2011, 128, 2625–2634. [Google Scholar] [CrossRef]
- Cherkasova, E.; Scrivani, C.; Doh, S.; Weisman, Q.; Takahashi, Y.; Harashima, N.; Yokoyama, H.; Srinivasan, R.; Linehan, W.M.; Lerman, M.I.; et al. Detection of an Immunogenic HERV-E Envelope with Selective Expression in Clear Cell Kidney Cancer. Cancer Res. 2016, 76, 2177–2185. [Google Scholar] [CrossRef]
- Rycaj, K.; Plummer, J.B.; Yin, B.; Li, M.; Garza, J.; Radvanyi, L.; Ramondetta, L.M.; Lin, K.; Johanning, G.L.; Tang, D.G.; et al. Cytotoxicity of human endogenous retrovirus K-specific T cells toward autologous ovarian cancer cells. Clin. Cancer Res. 2015, 21, 471–483. [Google Scholar] [CrossRef]
| Mechanism of Formation | Treatment Options Based on Neo-Antigens | Cancers | Reference |
|---|---|---|---|
| nsSNV * | Personalized treatment: passenger mutations, e.g., mRNA-4157/V940 vaccine | Cutaneous melanoma with LN metastases | [41] |
| Off-the-shelf treatment: driver mutations, e.g., ACT HLA-C*08:02 KRASG12D-specific TCR data | Metastatic pancreatic cancer, Colorectal cancer with lung metastases | [47,49] | |
| Indels | Personalized treatment | ccRCC and other | [26,57] |
| Off-the-shelf treatment: MMR deficiency | MSI-H associated cancers: colorectal, endometrial, stomach cancer, etc. | [61,62,63] | |
| Gene fusions | Personalized treatment | CML, synovial sarcoma | [72,73,74,75] |
| Mechanism of Formation | Treatments Based on Neo-Antigens | Cancers | Reference | |
|---|---|---|---|---|
| Noncoding DNA | Off-the-shelf treatment, e.g., hERVs *, pseudogenes, and lncRNA | ccRCC, NSCLC, breast cancer, and MMR-proficient colorectal cancer | [78,81,82,84] | |
| Splicing | Cis-acting mutations | Personalized treatment: splice site mutations | NSCLC | [94] |
| Trans-acting mutations | Off-the-shelf treatment: splice factor mutations, e.g., SF3B1 mutation | Breast cancer, ovarian serous carcinoma, and hematological malignancies | [91,113] | |
| Pharmacologic splicing inhibitors | Off-the-shelf treatment, e.g., Indisulam | Colon and lung cancer (mouse model) | [115] | |
| Translation | Initiation | Off-the-shelf treatment: near-cognate start codon | RCC | [130] |
| Elongation | Off-the-shelf treatment: sloppiness codon reassignment | Melanoma, skin, breast, ovarian, lung, colorectal cancer, hepatocellular, head and neck squamous-cell carcinoma, and lung cancer | [120,122,133,134] | |
| Termination | Rarely observed, clinical consequence unclear | [135] | ||
| PTMs | Phosphorylation | Off-the-shelf treatment: phosphopeptides | Melanoma and ovarian carcinoma | [146,147] |
| Glycosylation | Off-the-shelf treatment: glycosylated mucins | Breast cancer and pancreatic cancer | [7,153] | |
| Off-the-shelf treatment: GlcNAc | AML, ALL, and CLL | [162] | ||
| Virus | Mechanism of Integration/Role in Carcinogenesis | Viral Neo-Antigens |
|---|---|---|
| HPV * | Integration causes genomic instability such as rearrangements, translocations, amplifications, and ploidy changes | E6, E7 |
| HTLV-1 | Integration at nonpalindromic DNA motif; during latency state, only viral protein expression by mitotic and clonally expanding infected cells | Tax, HTLV-1 basic leucine zipper, p121, p30II |
| EBV | Switch from latency into lytic replication triggers malignant transformation | EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, EBNA-LP, LMP1, LMP2A, LMP2B, BZLF1, BRLF1 |
| CMV | Role of CMV in carcinogenesis not yet understood | pp65, IE1 |
| MCPyV | Integration at random sites, e.g., via viral fragmentation during replication; mutation in viral LT before or during viral integration | LT |
| KSHV | Isoforms via internal translation initiation, premature termination, internal frameshifting, or proteolytic cleavage | LANA, gB, K8.1, K12 |
| HBV and HCV | Integrates via double-strand DNA breaks which causes mutations in the viral genome causing truncations or cellular fusion proteins Integration of viral genes into intron region of CCNA2 leads to transcription of noncoding region | HBX, CCNA2 (endogen) |
| hERV | Endogenous integration; role in carcinogenesis not yet understood | ERV1, HERV-K-MEL, HERV-E |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emilius, L.; Bremm, F.; Binder, A.K.; Schaft, N.; Dörrie, J. Tumor Antigens beyond the Human Exome. Int. J. Mol. Sci. 2024, 25, 4673. https://doi.org/10.3390/ijms25094673
Emilius L, Bremm F, Binder AK, Schaft N, Dörrie J. Tumor Antigens beyond the Human Exome. International Journal of Molecular Sciences. 2024; 25(9):4673. https://doi.org/10.3390/ijms25094673
Chicago/Turabian StyleEmilius, Lisabeth, Franziska Bremm, Amanda Katharina Binder, Niels Schaft, and Jan Dörrie. 2024. "Tumor Antigens beyond the Human Exome" International Journal of Molecular Sciences 25, no. 9: 4673. https://doi.org/10.3390/ijms25094673
APA StyleEmilius, L., Bremm, F., Binder, A. K., Schaft, N., & Dörrie, J. (2024). Tumor Antigens beyond the Human Exome. International Journal of Molecular Sciences, 25(9), 4673. https://doi.org/10.3390/ijms25094673







