Comparative Analysis of Olive-Derived Phenolic Compounds’ Pro-Melanogenesis Effects on B16F10 Cells and Epidermal Human Melanocytes
Abstract
1. Introduction
2. Results
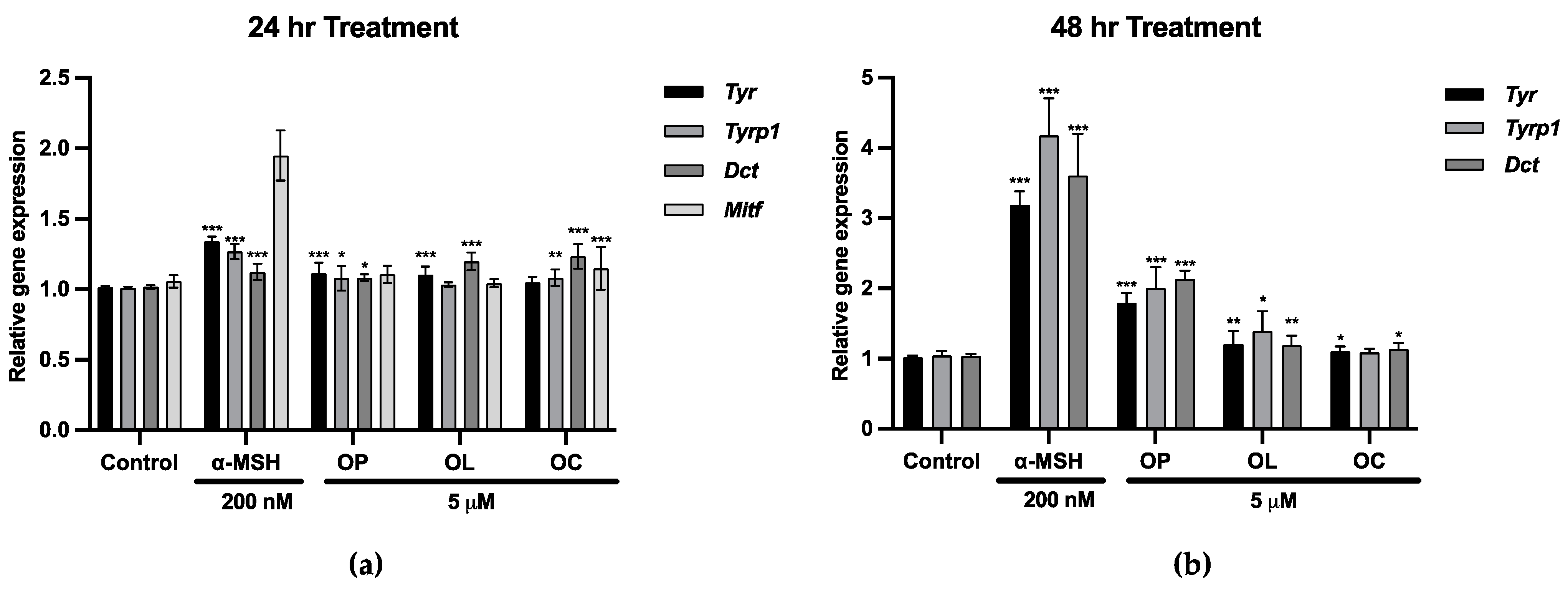
2.1. Upregulation of Mitf and Melanogenic Enzymes in B16F10 Cells
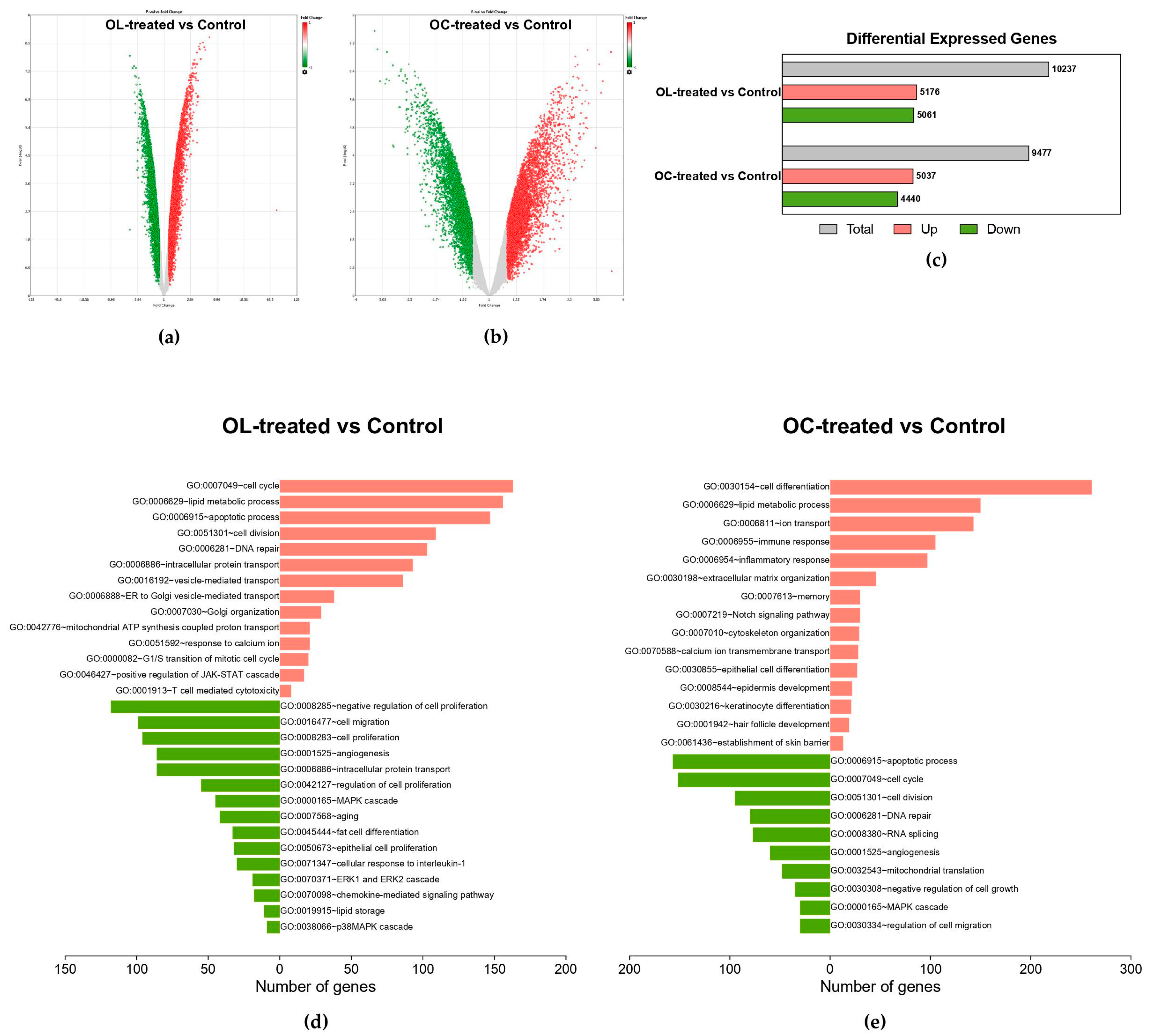
2.2. Impact of OL and OC on Global Gene Expression in B16F10 Cells
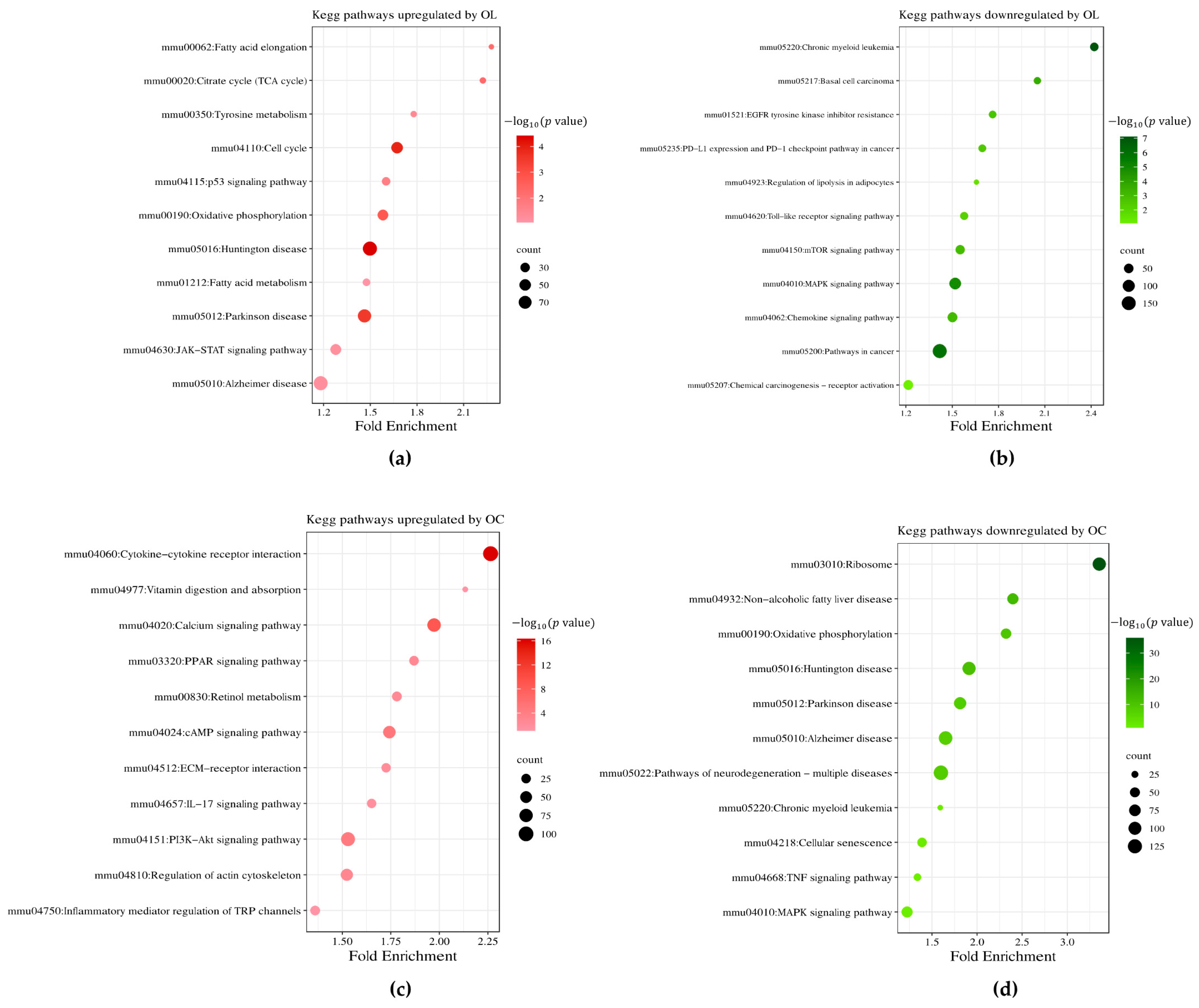
2.3. Hierarchical Clustering of DEGs of Enriched Functions in OL- and OC-Treated B16F10 Cells
2.4. Validation of Melanogenesis and Keratinocyte Differentiation-Associated Genes Regulated by OL and OC
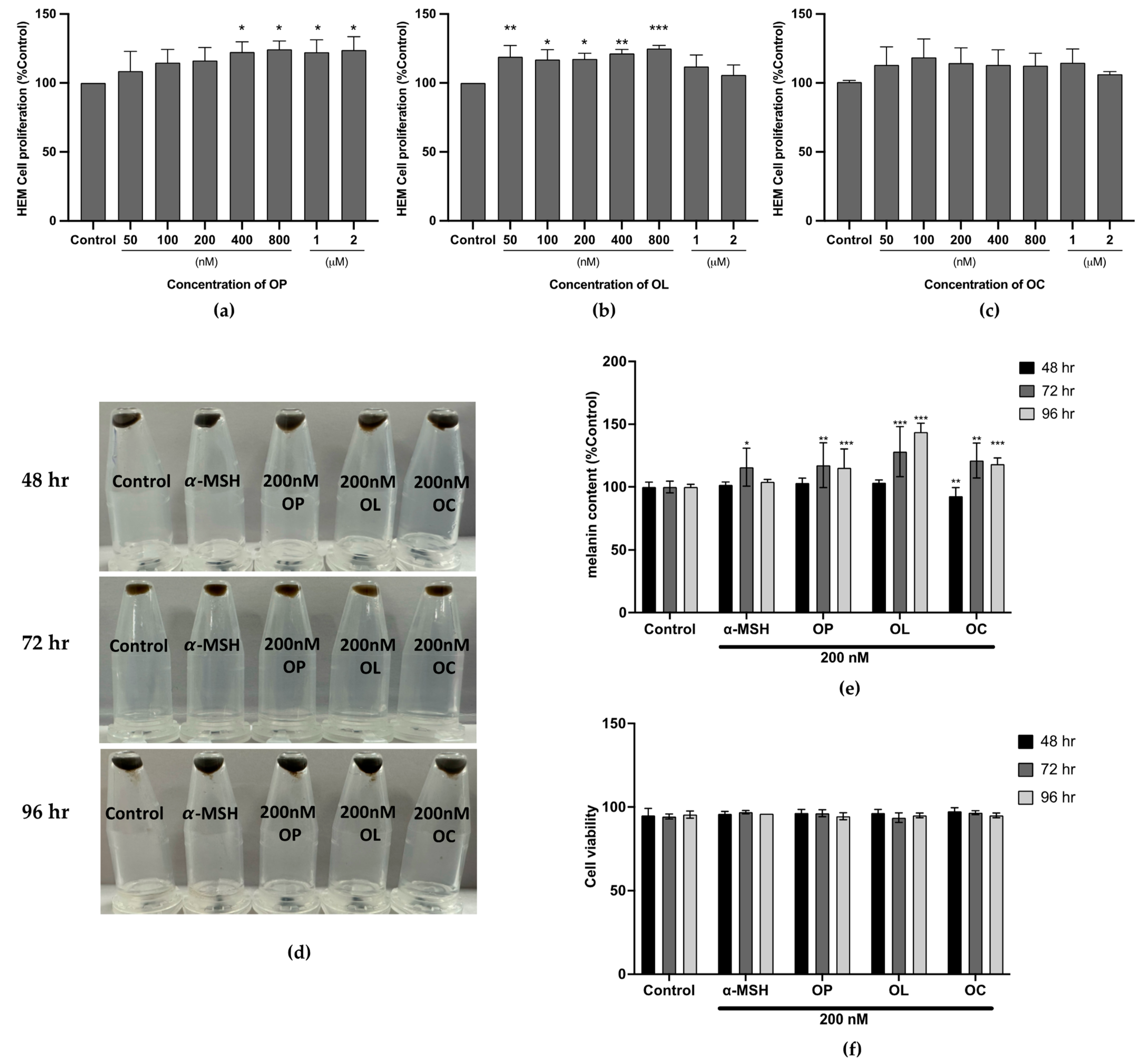
2.5. Effect of the Olive-Derived Phenolic Compounds on Melanin Production in Human Epidermal Melanocyte (HEM)
2.6. Expression of Transcription Factor MITF and Melanogenic Enzymes Were Upregulated in HEM Cells
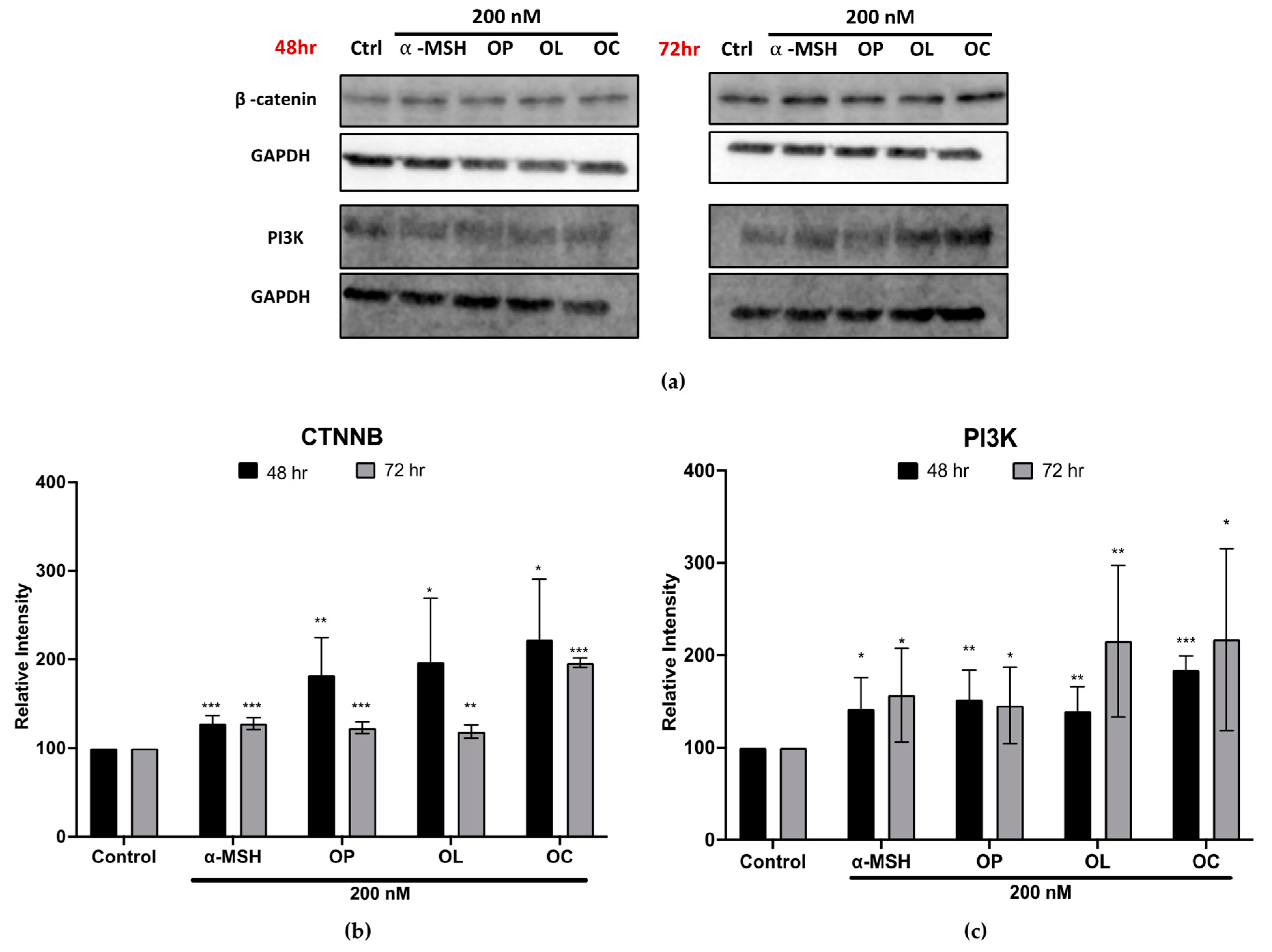
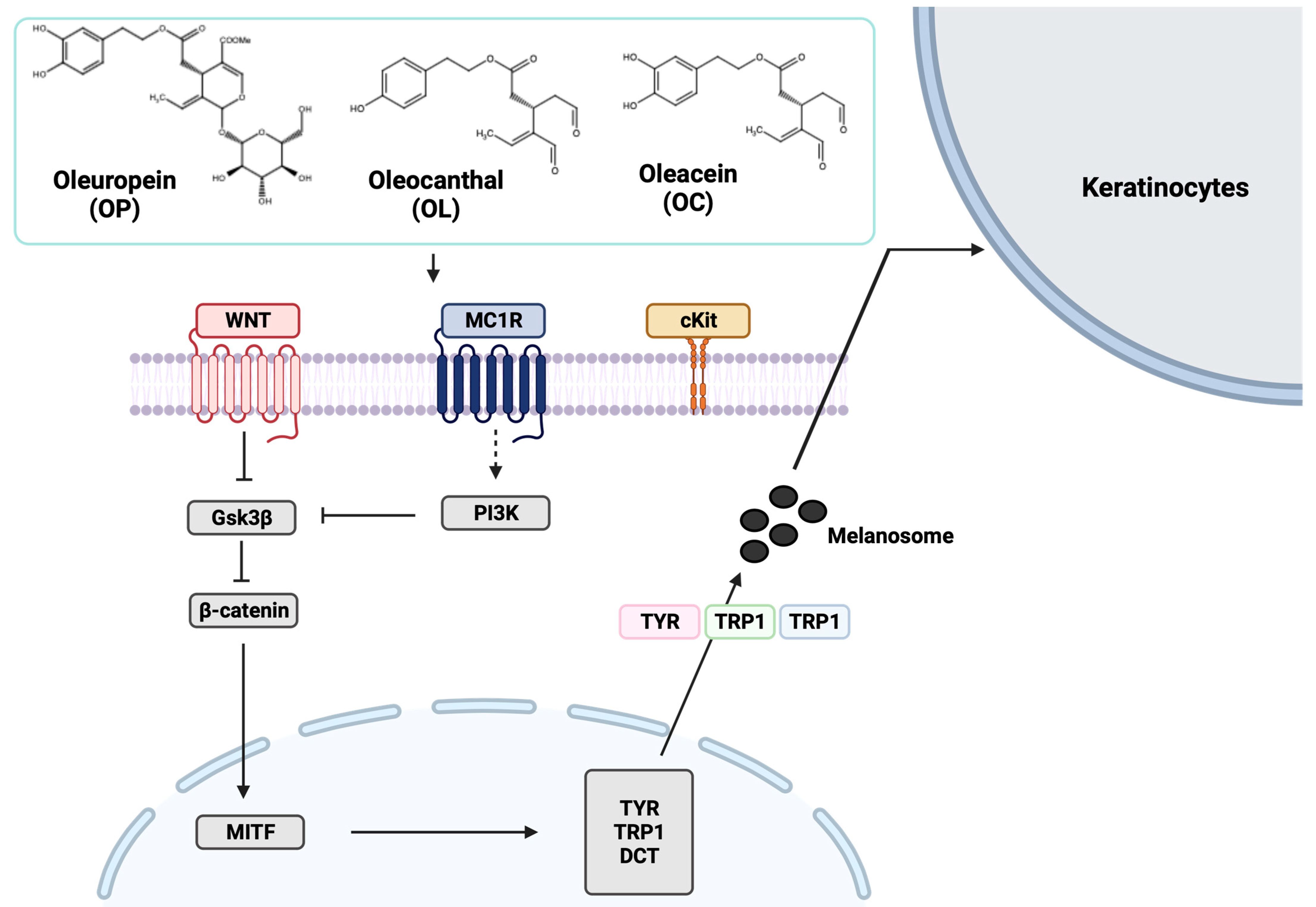
2.7. Elucidation of Molecular Mechanisms through Validation of Downstream Markers
2.8. Molecular Interaction of MC1R with Olive-Derived Compounds
3. Discussion
4. Materials and Methods
4.1. Sample Reagent Preparation
4.2. Cells and Cell Culture
4.3. Cell Proliferation Assay
4.4. RNA Extraction
4.5. DNA Microarray Analysis
4.6. Quantitative Real-Time PCR Analysis
4.7. Melanin Quantification
4.8. Western Blot
4.9. In Silico Molecular Docking
4.10. Surface Plasmon Resonance (SPR) Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dąbrowska, A.; Spano, F.; Derler, S.; Adlhart, C.; Spencer, N.D.; Rossi, R.M. The relationship between skin function, barrier properties, and body-dependent factors. Ski. Res. Technol. 2018, 24, 165–174. [Google Scholar] [CrossRef]
- Baroni, A.; Buommino, E.; De Gregorio, V.; Ruocco, E.; Ruocco, V.; Wolf, R. Structure and function of the epidermis related to barrier properties. Clin. Dermatol. 2012, 30, 257–262. [Google Scholar] [CrossRef]
- Costin, G.-E.; Hearing, V.J. Human skin pigmentation: Melanocytes modulate skin color in response to stress. FASEB J. 2007, 21, 976–994. [Google Scholar] [CrossRef]
- Mayer, T.C. The migratory pathway of neural crest cells into the skin of mouse embryos. Dev. Biol. 1973, 34, 39–46. [Google Scholar] [CrossRef]
- Sulem, P.; Gudbjartsson, D.F.; Stacey, S.N.; Helgason, A.; Rafnar, T.; Magnusson, K.P.; Manolescu, A.; Karason, A.; Palsson, A.; Thorleifsson, G. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007, 39, 1443–1452. [Google Scholar] [CrossRef]
- Westerhof, W. The discovery of the human melanocyte. Pigment Cell Res. 2006, 19, 183–193. [Google Scholar] [CrossRef]
- Eiberger, W.; Volkmer, B.; Amouroux, R.; Dhérin, C.; Radicella, J.P.; Epe, B. Oxidative stress impairs the repair of oxidative DNA base modifications in human skin fibroblasts and melanoma cells. DNA Repair 2008, 7, 912–921. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, K.B.; Son, Y.H.; Shin, J.; Lee, J.H.; Kim, H.J.; Hong, A.Y.; Bae, H.W.; Kwon, M.A.; Lee, W.J.; et al. Transglutaminase 2 mediates UV-induced skin inflammation by enhancing inflammatory cytokine production. Cell Death Dis. 2017, 8, e3148. [Google Scholar] [CrossRef]
- D’Orazio, J.; Jarrett, S.; Amaro-Ortiz, A.; Scott, T. UV radiation and the skin. Int. J. Mol. Sci. 2013, 14, 12222–12248. [Google Scholar] [CrossRef]
- Park, H.Y.; Kosmadaki, M.; Yaar, M.; Gilchrest, B.A. Cellular mechanisms regulating human melanogenesis. Cell. Mol. Life Sci. 2009, 66, 1493–1506. [Google Scholar] [CrossRef]
- Sarkar, S.; Gaddameedhi, S. Solar ultraviolet-induced DNA damage response: Melanocytes story in transformation to environmental melanomagenesis. Environ. Mol. Mutagen. 2020, 61, 736–751. [Google Scholar] [CrossRef]
- Moreiras, H.; Pereira, F.J.C.; Neto, M.V.; Bento-Lopes, L.; Festas, T.C.; Seabra, M.C.; Barral, D.C. The exocyst is required for melanin exocytosis from melanocytes and transfer to keratinocytes. Pigment Cell Melanoma Res. 2020, 33, 366–371. [Google Scholar] [CrossRef]
- Tarafder, A.K.; Bolasco, G.; Correia, M.S.; Pereira, F.J.C.; Iannone, L.; Hume, A.N.; Kirkpatrick, N.; Picardo, M.; Torrisi, M.R.; Rodrigues, I.P.; et al. Rab11b Mediates Melanin Transfer between Donor Melanocytes and Acceptor Keratinocytes via Coupled Exo/Endocytosis. J. Investig. Dermatol. 2014, 134, 1056–1066. [Google Scholar] [CrossRef]
- Fukuda, M. Rab GTPases: Key players in melanosome biogenesis, transport, and transfer. Pigment Cell Melanoma Res. 2021, 34, 222–235. [Google Scholar] [CrossRef]
- Wu, X.S.; Masedunskas, A.; Weigert, R.; Copeland, N.G.; Jenkins, N.A.; Hammer, J.A. Melanoregulin regulates a shedding mechanism that drives melanosome transfer from melanocytes to keratinocytes. Proc. Natl. Acad. Sci. USA 2012, 109, E2101–E2109. [Google Scholar] [CrossRef]
- Le, L.; Sirés-Campos, J.; Raposo, G.; Delevoye, C.; Marks, M.S. Melanosome biogenesis in the pigmentation of mammalian skin. Integr. Comp. Biol. 2021, 61, 1517–1545. [Google Scholar] [CrossRef]
- Hossain, M.R.; Kimura-Sashikawa, M.; Komine, M. Genetic Abnormalities, Melanosomal Transfer, and Degradation inside Keratinocytes Affect Skin Pigmentation. In Keratinocyte Biology—Structure and Function in the Epidermis; IntechOpen: London, UK, 2022. [Google Scholar]
- Hadley, M.E.; Quevedo, W.C. Vertebrate epidermal melanin unit. Nature 1966, 209, 1334–1335. [Google Scholar] [CrossRef]
- D’Mello, S.A.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Manickam, M.; Jung, S.-H. Recent development of signaling pathways inhibitors of melanogenesis. Cell. Signal. 2017, 40, 99–115. [Google Scholar] [CrossRef]
- Herraiz, C.; Martínez-Vicente, I.; Maresca, V. The α-melanocyte-stimulating hormone/melanocortin-1 receptor interaction: A driver of pleiotropic effects beyond pigmentation. Pigment Cell Melanoma Res. 2021, 34, 748–761. [Google Scholar] [CrossRef]
- Abdel-Malek, Z.A.; Ruwe, A.; Kavanagh-Starner, R.; Kadekaro, A.L.; Swope, V.; Haskell-Luevano, C.; Koikov, L.; Knittel, J.J. α-MSH tripeptide analogs activate the melanocortin 1 receptor and reduce UV-induced DNA damage in human melanocytes. Pigment Cell Melanoma Res. 2009, 22, 635–644. [Google Scholar] [CrossRef]
- Herraiz, C.; Garcia-Borron, J.C.; Jiménez-Cervantes, C.; Olivares, C. MC1R signaling. Intracellular partners and pathophysiological implications. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2017, 1863, 2448–2461. [Google Scholar] [CrossRef]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef]
- Yun, C.Y.; Roh, E.; Kim, S.H.; Han, J.; Lee, J.; Jung, D.E.; Kim, G.H.; Jung, S.H.; Cho, W.J.; Han, S.B.; et al. Stem Cell Factor-Inducible MITF-M Expression in Therapeutics for Acquired Skin Hyperpigmentation. Theranostics 2020, 10, 340–352. [Google Scholar] [CrossRef]
- Uto, T.; Ohta, T.; Katayama, K.; Shoyama, Y. Silibinin promotes melanogenesis through the PKA and p38 MAPK signaling pathways in melanoma cells. Biomed. Res. 2022, 43, 31–39. [Google Scholar] [CrossRef]
- Shin, S.; Ko, J.; Kim, M.; Song, N.; Park, K. Morin induces melanogenesis via activation of MAPK signaling pathways in B16F10 mouse melanoma cells. Molecules 2021, 26, 2150. [Google Scholar] [CrossRef]
- Choi, H.; Yoon, J.-H.; Youn, K.; Jun, M. Decursin prevents melanogenesis by suppressing MITF expression through the regulation of PKA/CREB, MAPKs, and PI3K/Akt/GSK-3β cascades. Biomed. Pharmacother. 2022, 147, 112651. [Google Scholar] [CrossRef]
- Yamada, T.; Hasegawa, S.; Inoue, Y.; Date, Y.; Yamamoto, N.; Mizutani, H.; Nakata, S.; Matsunaga, K.; Akamatsu, H. Wnt/β-catenin and kit signaling sequentially regulate melanocyte stem cell differentiation in UVB-induced epidermal pigmentation. J. Investig. Dermatol. 2013, 133, 2753–2762. [Google Scholar] [CrossRef]
- Bellei, B.; Pitisci, A.; Catricalà, C.; Larue, L.; Picardo, M. Wnt/β-catenin signaling is stimulated by α-melanocyte-stimulating hormone in melanoma and melanocyte cells: Implication in cell differentiation. Pigment Cell Melanoma Res. 2011, 24, 309–325. [Google Scholar] [CrossRef]
- Lin, X.; Meng, X.; Lin, J. The possible role of Wnt/β-catenin signalling in vitiligo treatment. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 2208–2221. [Google Scholar] [CrossRef]
- Guo, H.; Xing, Y.; Liu, Y.; Luo, Y.; Deng, F.; Yang, T.; Yang, K.; Li, Y. Wnt/β-catenin signaling pathway activates melanocyte stem cells in vitro and in vivo. J. Dermatol. Sci. 2016, 83, 45–51. [Google Scholar] [CrossRef]
- Kim, J.-H.; Hong, A.-r.; Kim, Y.-H.; Yoo, H.; Kang, S.-W.; Chang, S.E.; Song, Y. JNK suppresses melanogenesis by interfering with CREB-regulated transcription coactivator 3-dependent MITF expression. Theranostics 2020, 10, 4017. [Google Scholar] [CrossRef]
- Vachtenheim, J.; Borovanský, J. “Transcription physiology” of pigment formation in melanocytes: Central role of MITF. Exp. Dermatol. 2010, 19, 617–627. [Google Scholar] [CrossRef]
- Li, P.-H.; Liu, L.-H.; Chang, C.-C.; Gao, R.; Leung, C.-H.; Ma, D.-L.; David Wang, H.-M. Silencing stem cell factor gene in fibroblasts to regulate paracrine factor productions and enhance c-Kit expression in melanocytes on melanogenesis. Int. J. Mol. Sci. 2018, 19, 1475. [Google Scholar] [CrossRef]
- Brenner, M.; Hearing, V.J. The protective role of melanin against UV damage in human skin. Photochem. Photobiol. 2008, 84, 539–549. [Google Scholar] [CrossRef]
- Solano, F. Photoprotection and Skin Pigmentation: Melanin-Related Molecules and Some Other New Agents Obtained from Natural Sources. Molecules 2020, 25, 1537. [Google Scholar] [CrossRef]
- Cripps, D.J. Natural and artificial photoprotection. J. Investig. Dermatol. 1981, 77, 154–157. [Google Scholar] [CrossRef][Green Version]
- Plensdorf, S.; Livieratos, M.; Dada, N. Pigmentation Disorders: Diagnosis and Management. Am. Fam. Physician 2017, 96, 797–804. [Google Scholar]
- Rao, M.; Young, K.; Jackson-Cowan, L.; Kourosh, A.; Theodosakis, N. Post-Inflammatory Hypopigmentation: Review of the Etiology, Clinical Manifestations, and Treatment Options. J. Clin. Med. 2023, 12, 1243. [Google Scholar] [CrossRef]
- Boskou, D. Olive fruit, table olives, and olive oil bioactive constituents. In Olive and Olive Oil Bioactive Constituents; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1–30. [Google Scholar]
- Wahrburg, U.; Kratz, M.; Cullen, P. Mediterranean diet, olive oil and health. Eur. J. Lipid Sci. Technol. 2002, 104, 698–705. [Google Scholar] [CrossRef]
- Mentella, M.C.; Scaldaferri, F.; Ricci, C.; Gasbarrini, A.; Miggiano, G.A.D. Cancer and Mediterranean diet: A review. Nutrients 2019, 11, 2059. [Google Scholar] [CrossRef] [PubMed]
- Talhaoui, N.; Taamalli, A.; Gómez-Caravaca, A.M.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Phenolic compounds in olive leaves: Analytical determination, biotic and abiotic influence, and health benefits. Food Res. Int. 2015, 77, 92–108. [Google Scholar] [CrossRef]
- Sofi, F.; Cesari, F.; Abbate, R.; Gensini, G.F.; Casini, A. Adherence to Mediterranean diet and health status: Meta-analysis. BMJ 2008, 337, a1344. [Google Scholar] [CrossRef] [PubMed]
- Fabiani, R.; Sepporta, M.V.; Mazza, T.; Rosignoli, P.; Fuccelli, R.; De Bartolomeo, A.; Crescimanno, M.; Taticchi, A.; Esposto, S.; Servili, M. Influence of cultivar and concentration of selected phenolic constituents on the in vitro chemiopreventive potential of olive oil extracts. J. Agric. Food Chem. 2011, 59, 8167–8174. [Google Scholar] [CrossRef] [PubMed]
- Ahmad-Qasem, M.H.; Canovas, J.; Barrajon-Catalan, E.; Carreres, J.E.; Micol, V.; Garcia-Perez, J.V. Influence of olive leaf processing on the bioaccessibility of bioactive polyphenols. J. Agric. Food Chem. 2014, 62, 6190–6198. [Google Scholar] [CrossRef] [PubMed]
- Castejón, M.L.; Montoya, T.; Alarcón-de-la-Lastra, C.; Sánchez-Hidalgo, M. Potential Protective Role Exerted by Secoiridoids from Olea europaea L. in Cancer, Cardiovascular, Neurodegenerative, Aging-Related, and Immunoinflammatory Diseases. Antioxidants 2020, 9, 149. [Google Scholar] [CrossRef] [PubMed]
- Miho, H.; Díez, C.; Mena-Bravo, A.; de Medina, V.S.; Moral, J.; Melliou, E.; Magiatis, P.; Rallo, L.; Barranco, D.; Priego-Capote, F. Cultivar influence on variability in olive oil phenolic profiles determined through an extensive germplasm survey. Food Chem. 2018, 266, 192–199. [Google Scholar] [CrossRef]
- Sarikaki, G.; Christoforidou, N.; Gaboriaud-Kolar, N.; Smith III, A.B.; Kostakis, I.K.; Skaltsounis, A.-L. Biomimetic synthesis of oleocanthal, oleacein, and their analogues starting from oleuropein, a major compound of olive leaves. J. Nat. Prod. 2020, 83, 1735–1739. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, Y.; Fujitani, T.; Uchiage, E.; Isoda, H.; Tominaga, K.-i. Solid acid-catalyzed one-step synthesis of oleacein from oleuropein. Sci. Rep. 2023, 13, 8275. [Google Scholar] [CrossRef]
- Melguizo-Rodríguez, L.; González-Acedo, A.; Illescas-Montes, R.; García-Recio, E.; Ramos-Torrecillas, J.; Costela-Ruiz, V.J.; García-Martínez, O. Biological effects of the olive tree and its derivatives on the skin. Food Funct. 2022, 13, 11410–11424. [Google Scholar] [CrossRef]
- Rodrigues, F.; Pimentel, F.B.; Oliveira, M.B.P. Olive by-products: Challenge application in cosmetic industry. Ind. Crop. Prod. 2015, 70, 116–124. [Google Scholar] [CrossRef]
- Dauber, C.; Parente, E.; Zucca, M.P.; Gámbaro, A.; Vieitez, I. Olea europea and By-Products: Extraction Methods and Cosmetic Applications. Cosmetics 2023, 10, 112. [Google Scholar] [CrossRef]
- Goenka, S.; Simon, S.R. A novel pro-melanogenic effect of standardized dry olive leaf extract on primary human melanocytes from lightly pigmented and moderately pigmented skin. Pharmaceuticals 2021, 14, 252. [Google Scholar] [CrossRef] [PubMed]
- Omar, S.H. Oleuropein in olive and its pharmacological effects. Sci. Pharm. 2010, 78, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Perugini, P.; Vettor, M.; Rona, C.; Troisi, L.; Villanova, L.; Genta, I.; Conti, B.; Pavanetto, F. Efficacy of oleuropein against UVB irradiation: Preliminary evaluation. Int. J. Cosmet. Sci. 2008, 30, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Serre, C.; Busuttil, V.; Botto, J.M. Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. Int. J. Cosmet. Sci. 2018, 40, 328–347. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, G.; Li, G. Targeting MAPK pathway in melanoma therapy. Cancer Metastasis Rev. 2013, 32, 567–584. [Google Scholar] [CrossRef]
- Wu, B.; Shen, R.N.; Wang, W.X.; Broxmeyer, H.E.; Lu, L. Antitumor effect of interleukin 7 in combination with local hyperthermia in mice bearing B16a melanoma cells. Stem Cells 1993, 11, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Kollmann, K.; Briand, C.; Bellutti, F.; Schicher, N.; Blunder, S.; Zojer, M.; Hoeller, C. The interplay of CDK4 and CDK6 in melanoma. Oncotarget 2019, 10, 1346. [Google Scholar] [CrossRef]
- Zheng, J.; Yun, W.; Park, J.; Kang, P.J.; Lee, G.; Song, G.; Kim, I.Y.; You, S. Long-term expansion of directly reprogrammed keratinocyte-like cells and in vitro reconstitution of human skin. J. Biomed. Sci. 2020, 27, 56. [Google Scholar] [CrossRef]
- Arena, C.; Carpi, S.; Fogli, S.; Bertini, S.; Gado, F.; Saba, A.; Saccomanni, G.; Nieri, P.; Manera, C.; Macchia, M. Anti-proliferative activity of oleocanthal in human malignant cutaneous melanoma cells. In Book of Abstracts; Divisione di Chimica Farmaceutica della Società Chimica Italiana: Bologna, Italy, 2015. [Google Scholar]
- Gu, Y.; Wang, J.; Peng, L. (-)-Oleocanthal exerts anti-melanoma activities and inhibits STAT3 signaling pathway. Oncol. Rep. 2017, 37, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Carpi, S.; Polini, B.; Manera, C.; Digiacomo, M.; Salsano, J.E.; Macchia, M.; Scoditti, E.; Nieri, P. miRNA modulation and antitumor activity by the extra-virgin olive oil polyphenol oleacein in human melanoma cells. Front. Pharmacol. 2020, 11, 574317. [Google Scholar] [CrossRef]
- Polini, B.; Digiacomo, M.; Carpi, S.; Bertini, S.; Gado, F.; Saccomanni, G.; Macchia, M.; Nieri, P.; Manera, C.; Fogli, S. Oleocanthal and oleacein contribute to the in vitro therapeutic potential of extra virgin oil-derived extracts in non-melanoma skin cancer. Toxicol. Vitr. 2018, 52, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zeng, H.; Huang, J.; Lei, L.; Tong, X.; Li, S.; Zhou, Y.; Guo, H.; Khan, M.; Luo, L. Epigenetic regulation of melanogenesis. Ageing Res. Rev. 2021, 69, 101349. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, H.; Burg, M.B.; Ferraris, J.D. Inhibitory phosphorylation of GSK-3β by AKT, PKA, and PI3K contributes to high NaCl-induced activation of the transcription factor NFAT5 (TonEBP/OREBP). Am. J. Physiol. Ren. Physiol. 2013, 304, F908–F917. [Google Scholar] [CrossRef] [PubMed]
- Proksch, E.; Brandner, J.M.; Jensen, J.M. The skin: An indispensable barrier. Exp. Dermatol. 2008, 17, 1063–1072. [Google Scholar] [CrossRef]
- Upadhyay, P.R.; Ho, T.; Abdel-Malek, Z.A. Participation of keratinocyte- and fibroblast-derived factors in melanocyte homeostasis, the response to UV, and pigmentary disorders. Pigment Cell Melanoma Res. 2021, 34, 762–776. [Google Scholar] [CrossRef]
- Cichorek, M.; Wachulska, M.; Stasiewicz, A.; Tymińska, A. Skin melanocytes: Biology and development. Adv. Dermatol. Allergol. Postępy Dermatol. Alergol. 2013, 30, 30–41. [Google Scholar] [CrossRef]
- Hou, L.; Panthier, J.-J.; Arnheiter, H. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: Interactions between KIT and MITF. Development 2000, 127, 5379–5389. [Google Scholar] [CrossRef]
- Passeron, T.; Valencia, J.C.; Bertolotto, C.; Hoashi, T.; Le Pape, E.; Takahashi, K.; Ballotti, R.; Hearing, V.J. SOX9 is a key player in ultraviolet B-induced melanocyte differentiation and pigmentation. Proc. Natl. Acad. Sci. USA 2007, 104, 13984–13989. [Google Scholar] [CrossRef]
- Hirobe, T. How are proliferation and differentiation of melanocytes regulated? Pigment Cell Melanoma Res. 2011, 24, 462–478. [Google Scholar] [CrossRef]
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Saldana-Caboverde, A.; Kos, L. Roles of endothelin signaling in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010, 23, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Grichnik, J.M.; Burch, J.A.; Burchette, J.; Shea, C.R. The SCF/KIT pathway plays a critical role in the control of normal human melanocyte homeostasis. J. Investig. Dermatol. 1998, 111, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Hyun, C.-G. Mechanistic insights into the ameliorating effect of melanogenesis of psoralen derivatives in B16F10 melanoma cells. Molecules 2022, 27, 2613. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-J.; Lyu, J.-L.; Kuo, Y.-H.; Chiu, C.-Y.; Wen, K.-C.; Chiang, H.-M. The anti-melanogenesis effect of 3,4-dihydroxybenzalacetone through downregulation of melanosome maturation and transportation in B16F10 and human epidermal melanocytes. Int. J. Mol. Sci. 2021, 22, 2823. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.C.; Hyun, C.-G. Inhibitory effects of pinostilbene hydrate on melanogenesis in B16F10 melanoma cells via ERK and p38 signaling pathways. Int. J. Mol. Sci. 2020, 21, 4732. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Kim, J.Y.; Heo, J.; Cho, D.-H.; Kim, H.-S.; An, I.-S.; An, S.; Bae, S. The inhibition of melanogenesis via the PKA and ERK signaling pathways by Chlamydomonas reinhardtii extract in B16F10 melanoma cells and artificial human skin equivalents. J. Microbiol. Biotechnol. 2018, 28, 2121–2132. [Google Scholar] [CrossRef] [PubMed]
- Yardman-Frank, J.M.; Fisher, D.E. Skin pigmentation and its control: From ultraviolet radiation to stem cells. Exp. Dermatol. 2021, 30, 560–571. [Google Scholar] [CrossRef]
- Slominski, R.M.; Sarna, T.; Płonka, P.M.; Raman, C.; Brożyna, A.A.; Slominski, A.T. Melanoma, melanin, and melanogenesis: The Yin and Yang relationship. Front. Oncol. 2022, 12, 842496. [Google Scholar] [CrossRef]
- Premi, S. Role of melanin chemiexcitation in melanoma progression and drug resistance. Front. Oncol. 2020, 10, 1305. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, Z.; Luo, T.; Shi, H. Targeting the PI3K/AKT/mTOR and RAF/MEK/ERK pathways for cancer therapy. Mol. Biomed. 2022, 3, 47. [Google Scholar] [CrossRef] [PubMed]
- Nymark Aasen, S.; Parajuli, H.; Hoang, T.; Feng, Z.; Stokke, K.; Wang, J.; Roy, K.; Bjerkvig, R.; Knappskog, S.; Thorsen, F. Effective treatment of metastatic melanoma by combining MAPK and PI3K signaling pathway inhibitors. Int. J. Mol. Sci. 2019, 20, 4235. [Google Scholar] [CrossRef] [PubMed]
- Yajima, I.; Kumasaka, M.Y.; Thang, N.D.; Goto, Y.; Takeda, K.; Yamanoshita, O.; Iida, M.; Ohgami, N.; Tamura, H.; Kawamoto, Y. RAS/RAF/MEK/ERK and PI3K/PTEN/AKT signaling in malignant melanoma progression and therapy. Dermatol. Res. Pract. 2012, 2012, 354191. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, Y.; Zhou, C.; Mei, W.; Zeng, C. PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we making headway? Front. Oncol. 2022, 12, 819128. [Google Scholar] [CrossRef] [PubMed]
- Wiedlocha, A.; Haugsten, E.M.; Zakrzewska, M. Roles of the FGF-FGFR signaling system in cancer development and inflammation. Cells 2021, 10, 2231. [Google Scholar] [CrossRef] [PubMed]
- Presta, M.; Chiodelli, P.; Giacomini, A.; Rusnati, M.; Ronca, R. Fibroblast growth factors (FGFs) in cancer: FGF traps as a new therapeutic approach. Pharmacol. Ther. 2017, 179, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, S.; Patinen, T.; Deen, A.J.; Pitkänen, S.; Härkönen, J.; Kansanen, E.; Küblbeck, J.; Levonen, A.-L. The KEAP1-NRF2 pathway: Targets for therapy and role in cancer. Redox Biol. 2023, 63, 102726. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef]
- Gibbs, S.; Silva Pinto, A.N.; Murli, S.; Huber, M.; Hohl, D.; Ponec, M. Epidermal growth factor and keratinocyte growth factor differentially regulate epidermal migration, growth, and differentiation. Wound Repair Regen. 2000, 8, 192–203. [Google Scholar] [CrossRef]
- Eckhart, L.; Declercq, W.; Ban, J.; Rendl, M.; Lengauer, B.; Mayer, C.; Lippens, S.; Vandenabeele, P.; Tschachler, E. Terminal differentiation of human keratinocytes and stratum corneum formation is associated with caspase-14 activation. J. Investig. Dermatol. 2000, 115, 1148–1151. [Google Scholar] [CrossRef] [PubMed]
- Natsuga, K. Epidermal barriers. Cold Spring Harb. Perspect. Med. 2014, 4, a018218. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xiao Liu, Z.; Oh, J.; Shin, K.; Kim, R.; Jiang, M.; Park, N.; Kang, M. Grainyhead-like 2 (GRHL2) inhibits keratinocyte differentiation through epigenetic mechanism. Cell Death Dis. 2012, 3, e450. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Villareal, M.O.; Kume, S.; Neffati, M.; Isoda, H. Upregulation of Mitf by phenolic compounds-rich cymbopogon schoenanthus treatment promotes melanogenesis in b16 melanoma cells and human epidermal melanocytes. BioMed Res. Int. 2017, 2017, 8303671. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chen, H.; Wang, Y.Y.; Wang, X.Q.; Chen, H.Y.; Zhang, M.; Tang, Y.; Zhang, B. Network pharmacological mechanisms of Vernonia anthelmintica (L.) in the treatment of vitiligo: Isorhamnetin induction of melanogenesis via up-regulation of melanin-biosynthetic genes. BMC Syst. Biol. 2017, 11, 103. [Google Scholar] [CrossRef]
- Abdel-Malek, Z.A.; Swope, V.B. Epidermal melanocytes: Regulation of their survival, proliferation, and function in human skin. In Melanoma Development: Molecular Biology, Genetics and Clinical Application; Springer: Berlin/Heidelbreg, Germany, 2011; pp. 7–33. [Google Scholar]




| Kinetics Model | Target | Analyte | () | ka (1/Ms) | kd (1/s) | KD (M) | Rmax (RU) |
|---|---|---|---|---|---|---|---|
| 1:1 binding | MC1R | OP | - | - | - | - | - |
| 1:1 binding | MC1R | OL | 3.21 | 12.05 | 0.002320 | 39.73 | |
| 1:1 binding | MC1R | OC | 17.1 | 0.001252 | 7.676 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, J.; Bejaoui, M.; Tominaga, K.; Isoda, H. Comparative Analysis of Olive-Derived Phenolic Compounds’ Pro-Melanogenesis Effects on B16F10 Cells and Epidermal Human Melanocytes. Int. J. Mol. Sci. 2024, 25, 4479. https://doi.org/10.3390/ijms25084479
Cho J, Bejaoui M, Tominaga K, Isoda H. Comparative Analysis of Olive-Derived Phenolic Compounds’ Pro-Melanogenesis Effects on B16F10 Cells and Epidermal Human Melanocytes. International Journal of Molecular Sciences. 2024; 25(8):4479. https://doi.org/10.3390/ijms25084479
Chicago/Turabian StyleCho, Juhee, Meriem Bejaoui, Kenichi Tominaga, and Hiroko Isoda. 2024. "Comparative Analysis of Olive-Derived Phenolic Compounds’ Pro-Melanogenesis Effects on B16F10 Cells and Epidermal Human Melanocytes" International Journal of Molecular Sciences 25, no. 8: 4479. https://doi.org/10.3390/ijms25084479
APA StyleCho, J., Bejaoui, M., Tominaga, K., & Isoda, H. (2024). Comparative Analysis of Olive-Derived Phenolic Compounds’ Pro-Melanogenesis Effects on B16F10 Cells and Epidermal Human Melanocytes. International Journal of Molecular Sciences, 25(8), 4479. https://doi.org/10.3390/ijms25084479







