Studying the Effects and Competitive Mechanisms of YOYO-1 on the Binding Characteristics of DOX and DNA Molecules Based on Surface-Enhanced Raman Spectroscopy and Molecular Docking Techniques
Abstract
1. Introduction
2. Results and Discussion
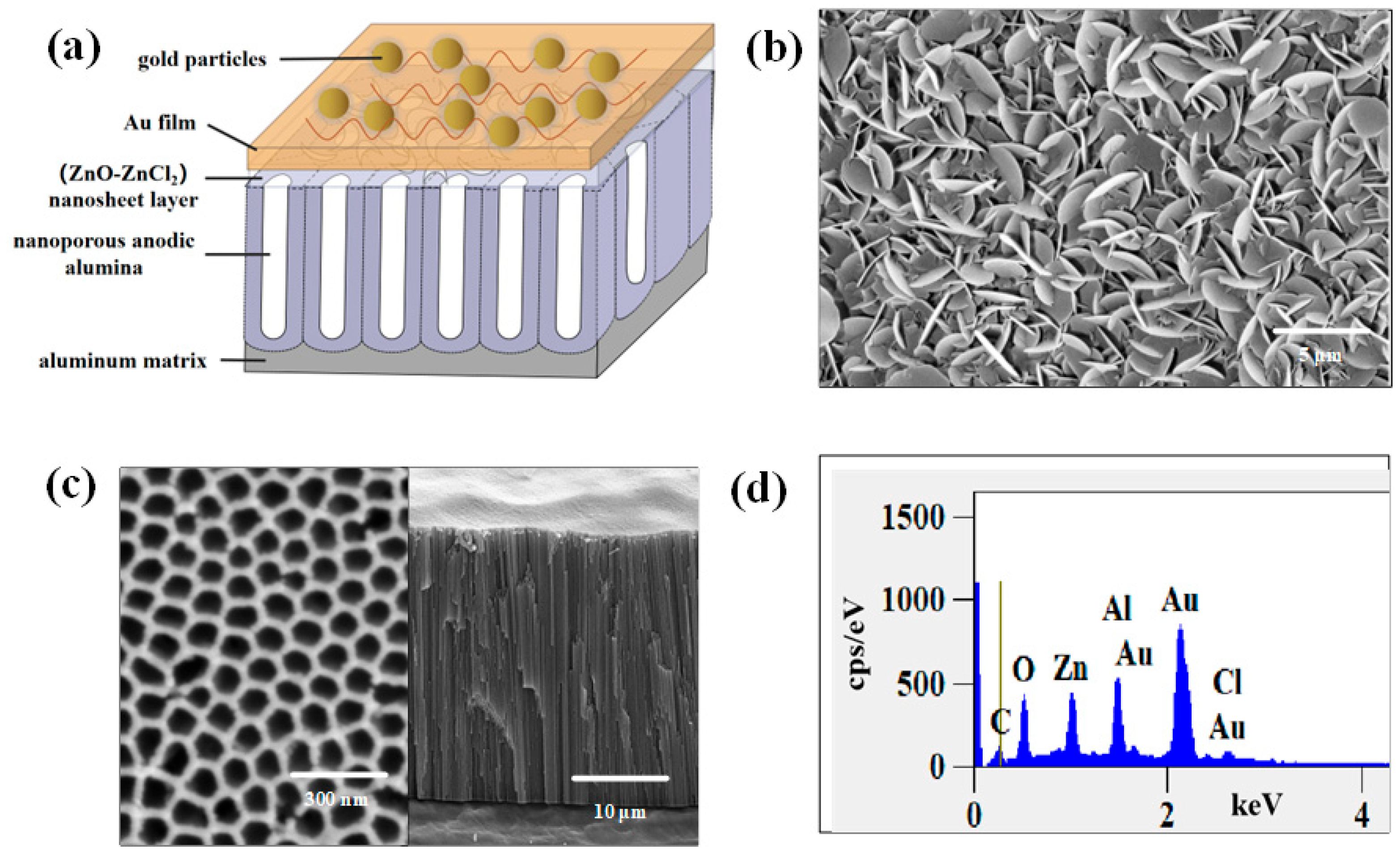
2.1. NpAA/(ZnO-ZnCl2)/AuNPs Active SERS Solid Substrate
2.1.1. Structural Characteristics
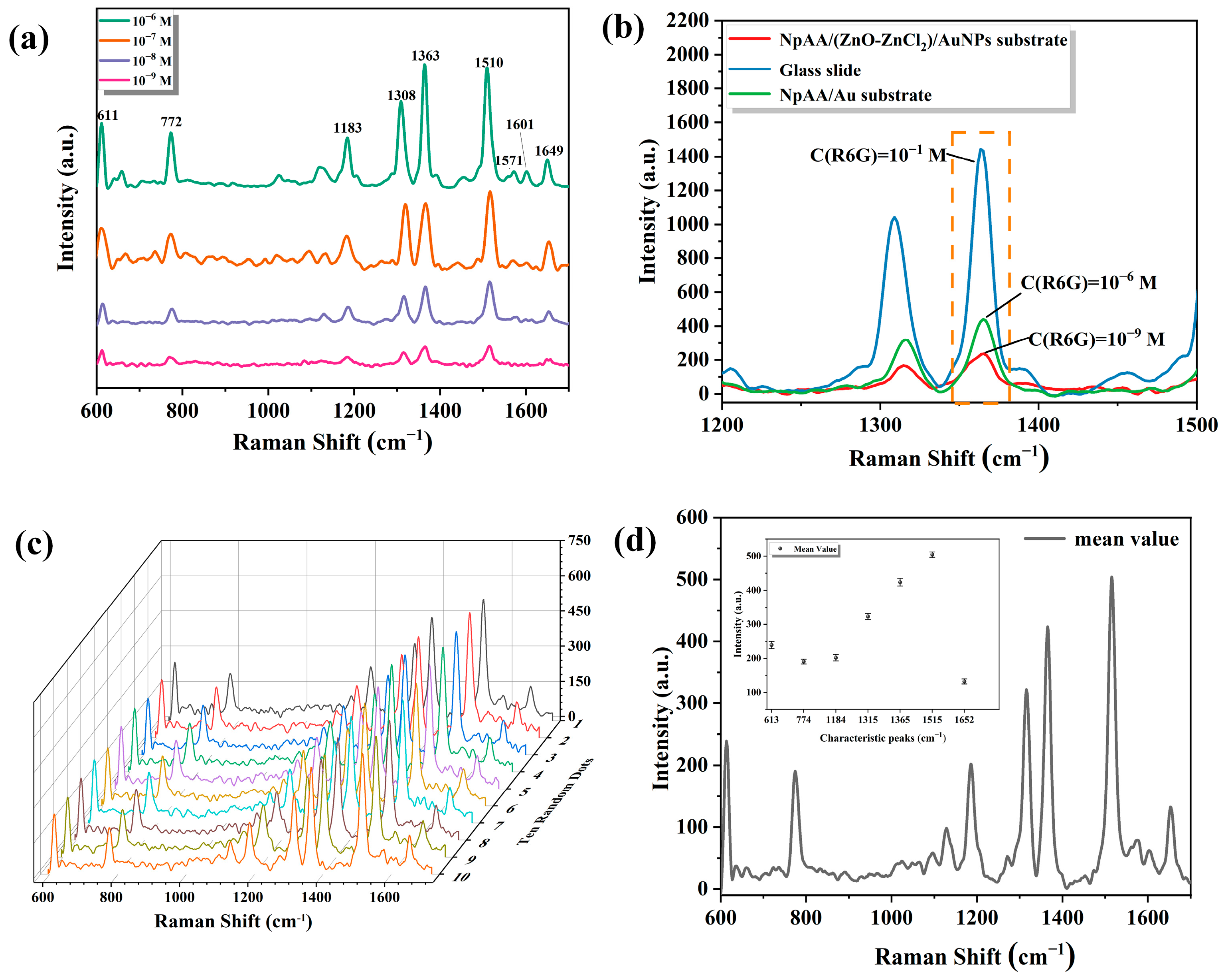
2.1.2. SERS Active Performance
2.2. SERS Spectra
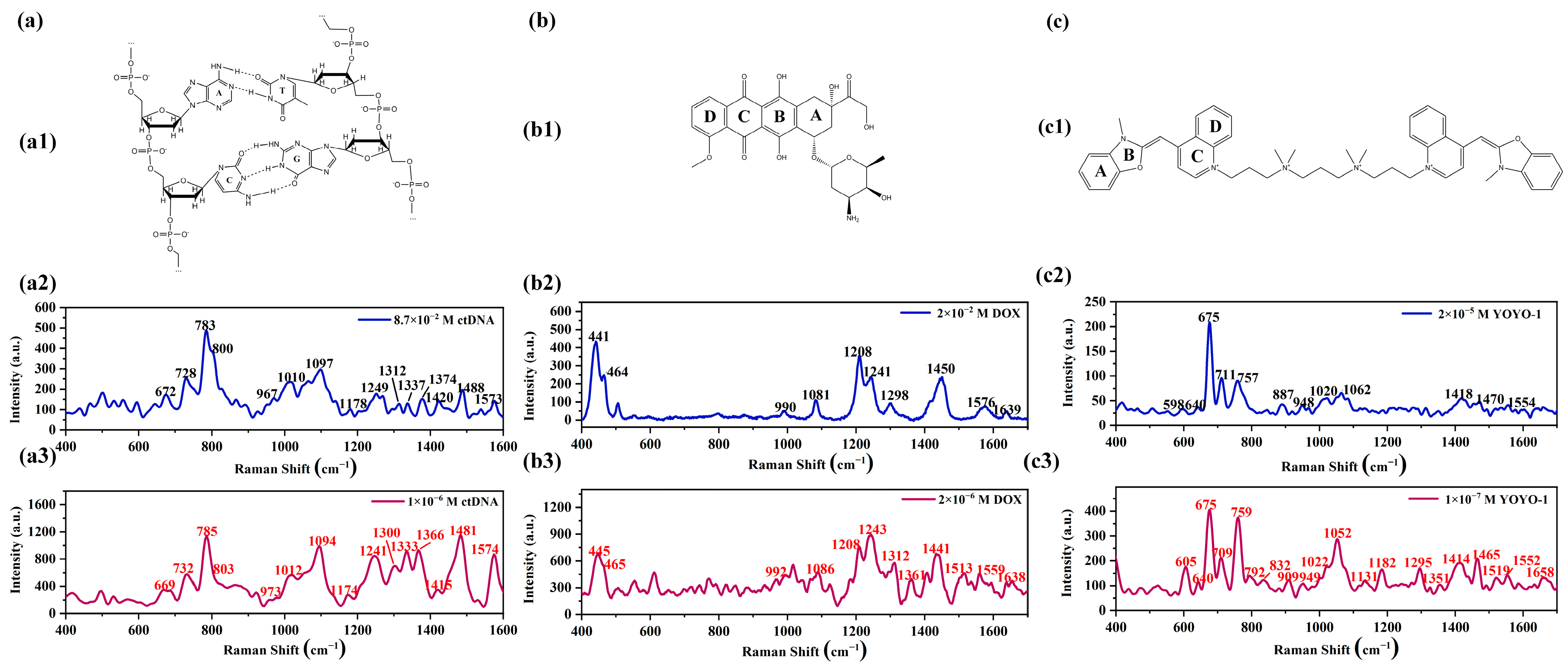
2.2.1. Raman Spectra and SERS Spectra of ctDNA, DOX, and YOYO-1
2.2.2. SERS Spectrum of YOYO-1 and DOX Mixture
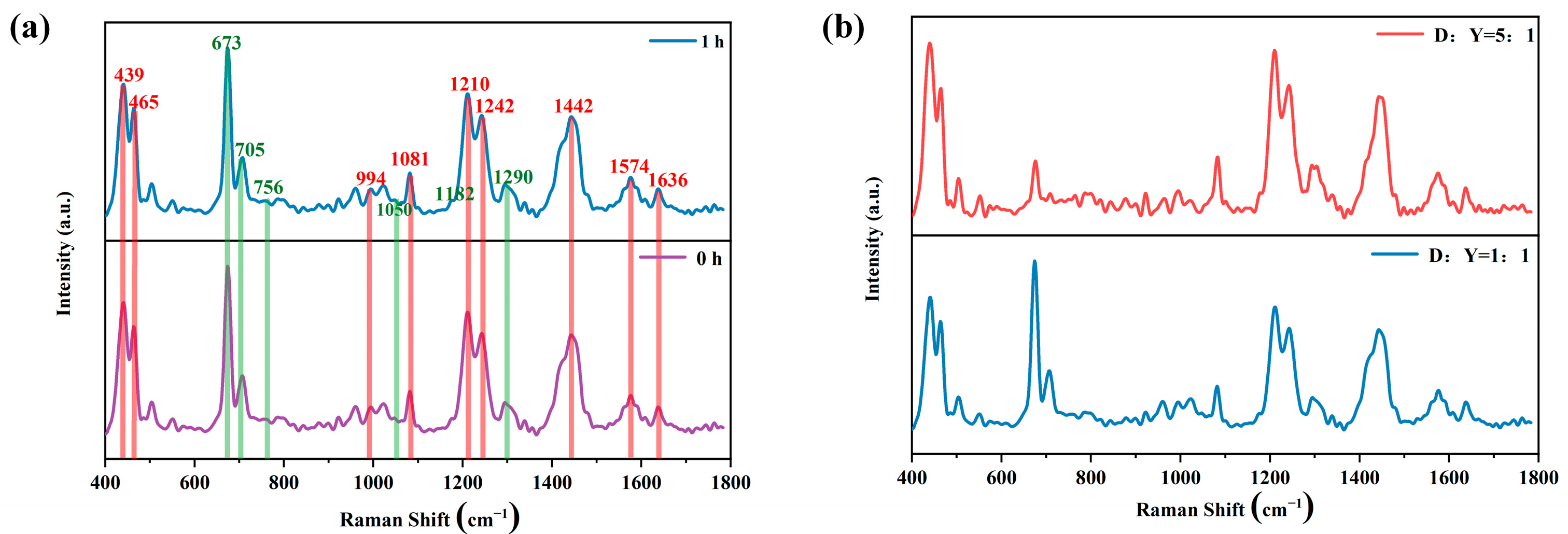
2.2.3. SERS Spectra of ctDNA-DOX and ctDNA-DOX + YOYO-1
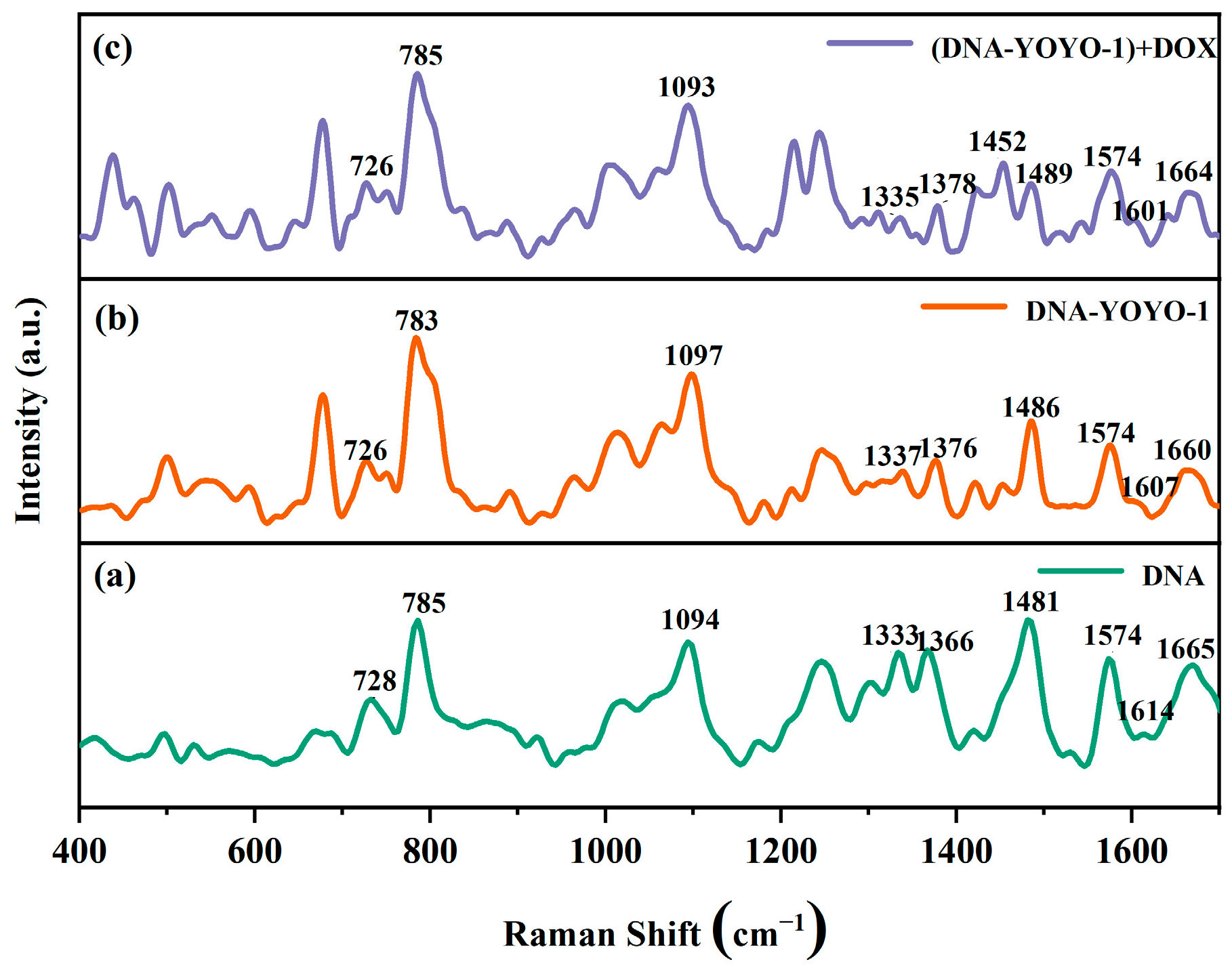
2.2.4. SERS Spectra of ctDNA-YOYO-1 and ctDNA-YOYO-1 + DOX
2.3. Fluorescence Experiment
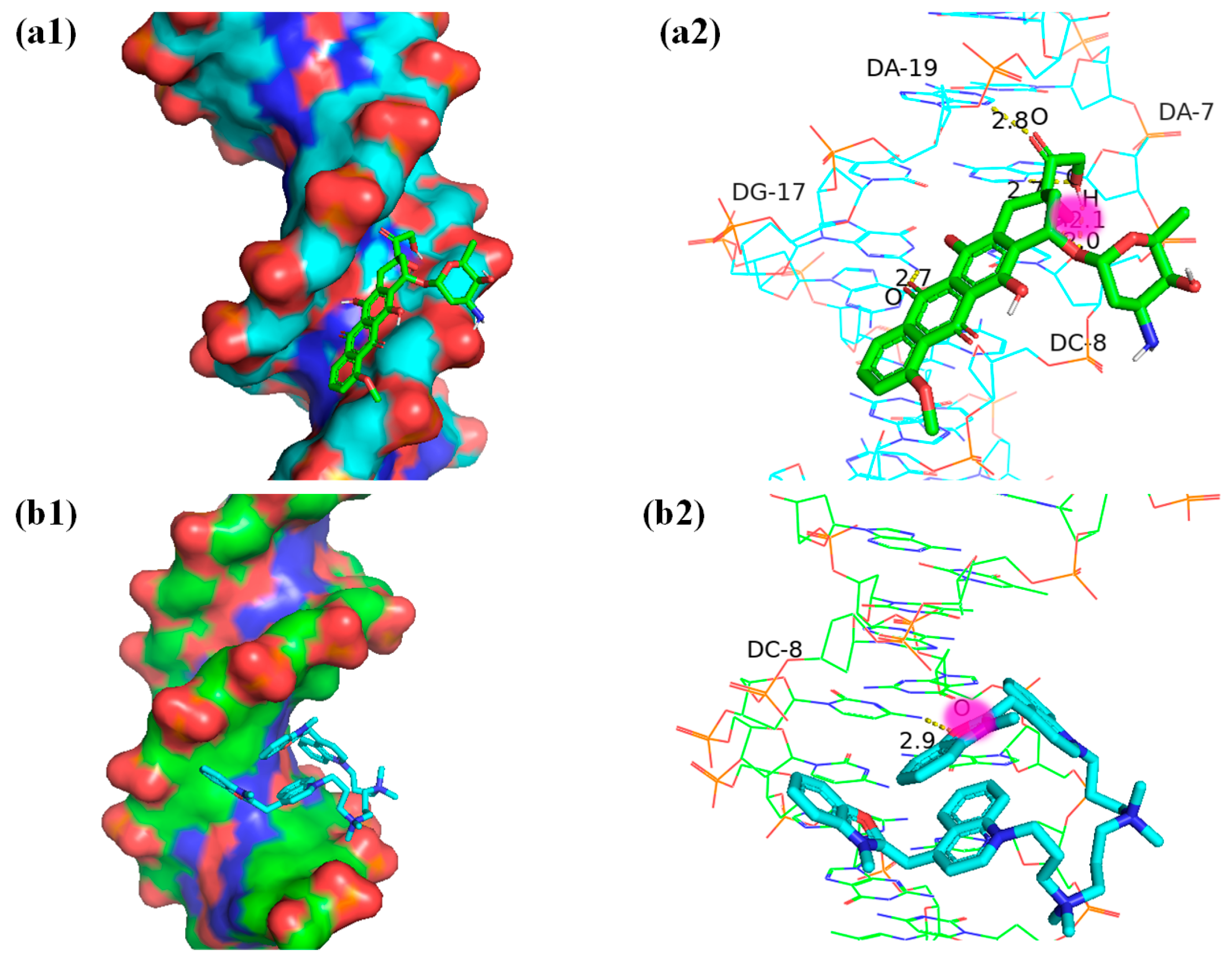
2.4. Molecular Docking
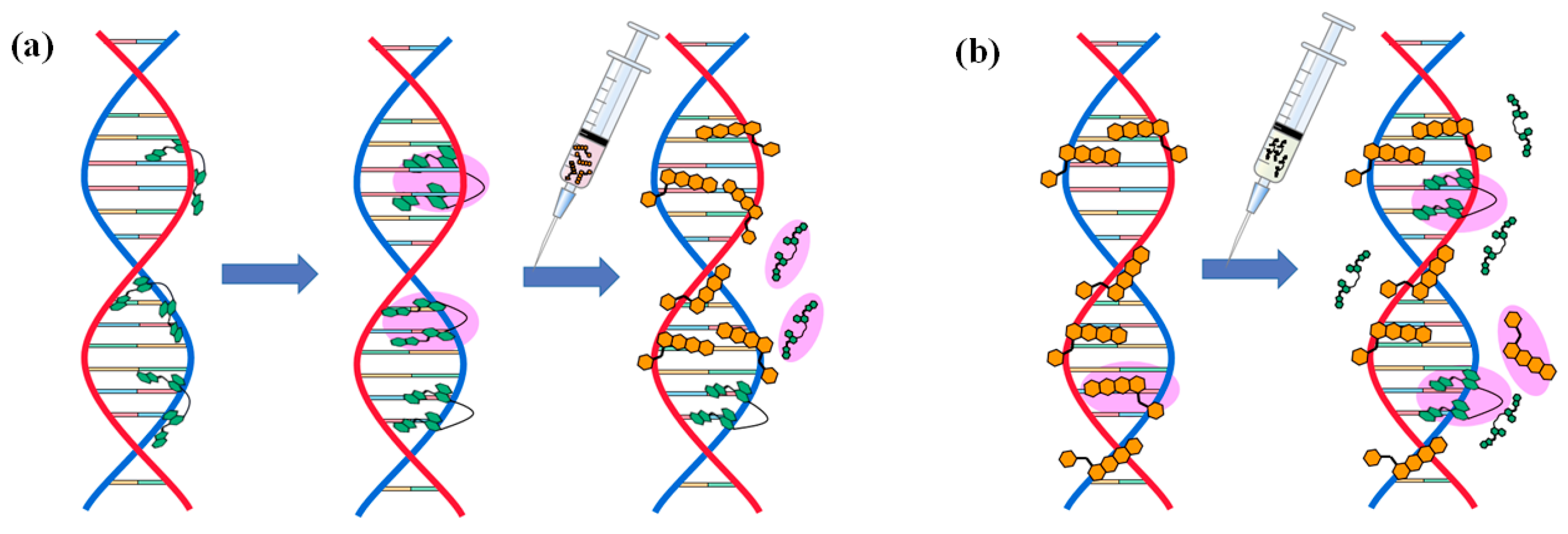
2.5. Mechanism of Action
3. Materials and Methods
3.1. Materials
3.2. NpAA/(ZnO-ZnCl2)/AuNPs Substrate
3.3. Methods
- Surface-enhanced Raman spectrometer
- Scanning Electron Microscope (SEM)
- Inverted Fluorescence Microscope
- Molecular Docking Software
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Yan, Q.; Fan, C.; Mo, Y.; Wang, Y.; Li, X.; Liao, Q.; Guo, C.; Li, G.; Zeng, Z. Overview and countermeasures of cancer burden in China. Sci. China Life Sci. 2023, 66, 2515–2526. [Google Scholar] [CrossRef] [PubMed]
- Weingart, S.N.; Zhang, L.; Sweeney, M.; Hassett, M. Chemotherapy medication errors. Lancet Oncol. 2018, 19, e191–e199. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yin, F.; Song, L.; Mao, X.; Li, F.; Fan, C.; Zuo, X.; Xia, Q. Nucleic acid tests for clinical translation. Chem. Rev. 2021, 121, 10469–10558. [Google Scholar] [CrossRef] [PubMed]
- Pugazhendhi, A.; Edison, T.N.J.I.; Velmurugan, B.K.; Jacob, J.A.; Karuppusamy, I. Toxicity of Doxorubicin (Dox) to different experimental organ systems. Life Sci. 2018, 200, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Rivankar, S. An overview of doxorubicin formulations in cancer therapy. J. Cancer Res. Ther. 2014, 10, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Wiles, M.E.; Bell, C.; Landfair, D.; Lynam, E.; Bendele, R. Anthracycline efficacy in vitro: Cytotoxicity of liposomal/nonliposomal daunorubicin and doxorubicin for multiple tumor cell types. Drug Deliv. 1997, 4, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Kciuk, M.; Gielecińska, A.; Mujwar, S.; Kołat, D.; Kałuzińska-Kołat, Ż.; Celik, I.; Kontek, R. Doxorubicin—An Agent with Multiple Mechanisms of Anticancer Activity. Cells 2023, 12, 659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Poniewierski, A.; Sozański, K.; Zhou, Y.; Brzozowska-Elliott, A.; Holyst, R. Fluorescence correlation spectroscopy for multiple-site equilibrium binding: A case of doxorubicin–DNA interaction. Phys. Chem. Chem. Phys. 2019, 21, 1572–1577. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gou, X.; Pu, X. The mechanism of doxorubicin hydrochloride interaction with DNA was investigated by spectroscopic and isothermal titration calorimetry. Spectrosc. Spectr. Anal 2018, 38, 540–545. [Google Scholar]
- Pérez-Arnaiz, C.; Busto, N.; Leal, J.M.; García, B. New insights into the mechanism of the DNA/doxorubicin interaction. J. Phys. Chem. B 2014, 118, 1288–1295. [Google Scholar] [CrossRef]
- Pei, Y.; Liu, Y.; Xie, C.; Zhang, X.; You, H. Detecting the formation kinetics of doxorubicin-DNA interstrand cross-link at the single-molecule level and clinically relevant concentrations of doxorubicin. Anal. Chem. 2020, 92, 4504–4511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhu, J.; Sun, D.; Li, J.; Yao, L.; Meng, S.; Li, Y.; Dang, Y.; Wang, K. The Mechanism of Dynamic Interaction between Doxorubicin and Calf Thymus DNA at the Single-Molecule Level Based on Confocal Raman Spectroscopy. Micromachines 2022, 13, 940. [Google Scholar] [CrossRef] [PubMed]
- Sheibani, M.; Azizi, Y.; Shayan, M.; Nezamoleslami, S.; Eslami, F.; Farjoo, M.H.; Dehpour, A.R. Doxorubicin-induced cardiotoxicity: An overview on pre-clinical therapeutic approaches. Cardiovasc. Toxicol. 2022, 22, 292–310. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.U.; Sarwar, T.; Husain, M.A.; Ishqi, H.M.; Tabish, M. Studying non-covalent drug–DNA interactions. Arch. Biochem. Biophys. 2015, 576, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.; Chiorcea-Paquim, A.; Ribeiro, S.; Melo, A.; Vivan, M.; Oliveira-Brett, A. In situ electrochemical and AFM study of thalidomide–DNA interaction. Bioelectrochemistry 2009, 76, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.T.; Silva, G.; Pungartnik, C.; Brendel, M. Study of DNA–emodin interaction by FTIR and UV–vis spectroscopy. J. Photochem. Photobiol. B Biol. 2012, 111, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Wang, K.; Zhou, Y.; Sui, C.; Wang, L.; Bai, R.; Yang, Z. Label-free detecting of the compaction and decompaction of ctDNA molecules induced by surfactants with SERS based on a nanoPAA-ZnCl2-AuLs solid substrate. ACS Omega 2020, 5, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Ilkhani, H.; Hughes, T.; Li, J.; Zhong, C.J.; Hepel, M. Nanostructured SERS-electrochemical biosensors for testing of anticancer drug interactions with DNA. Biosens. Bioelectron. 2016, 80, 257–264. [Google Scholar] [CrossRef]
- Yao, L.; Li, Y.; Zuo, Z.; Gong, Z.; Zhu, J.; Feng, X.; Sun, D.; Wang, K. Studying the Interaction between Bendamustine and DNA Molecule with SERS Based on AuNPs/ZnCl2/NpAA Solid-State Substrate. Int. J. Mol. Sci. 2023, 24, 13517. [Google Scholar] [CrossRef]
- Kang, Y.; An, S.; Min, D.; Lee, J.Y. Single-molecule fluorescence imaging techniques reveal molecular mechanisms underlying deoxyribonucleic acid damage repair. Front. Bioeng. Biotechnol. 2022, 10, 973314. [Google Scholar] [CrossRef]
- Ali, A.; Bhattacharya, S. DNA binders in clinical trials and chemotherapy. Bioorg. Med. Chem. 2014, 22, 4506–4521. [Google Scholar] [CrossRef] [PubMed]
- Murade, C.U.; Subramaniam, V.; Otto, C.; Bennink, M.L. Interaction of oxazole yellow dyes with DNA studied with hybrid optical tweezers and fluorescence microscopy. Biophys. J. 2009, 97, 835–843. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Persil, O.; Hud, N.V. Harnessing DNA intercalation. Trends Biotechnol. 2007, 25, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.; Dryden, D.T. The kinetics of YOYO-1 intercalation into single molecules of double-stranded DNA. Biochem. Biophys. Res. Commun. 2010, 403, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Perkins, T.T.; Quake, S.R.; Smith, D.E.; Chu, S. Relaxation of a single DNA molecule observed by optical microscopy. Science 1994, 264, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Qi, Y.; Mestre, H.; Canavesi, C.; Marola, O.J.; Cogliati, A.; Nedergaard, M.; Libby, R.T.; Rolland, J.P. Gabor domain optical coherence microscopy combined with laser scanning confocal fluorescence microscopy. Biomed. Opt. Express 2019, 10, 6242–6257. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Widengren, J.; Lee, J.-C. Fluorescent probes for STED optical nanoscopy. Nanomaterials 2021, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R.; Denny, W.A. Genotoxicity of non-covalent interactions: DNA intercalators. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2007, 623, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Duan, J.; Wang, Y.; Zhang, Y.; Guo, Y.; Guo, H.; Kang, X. Nanopore single-molecule analysis of DNA–doxorubicin interactions. Anal. Chem. 2015, 87, 338–342. [Google Scholar] [CrossRef]
- Lee, C.; Kim, Y.-J.; Kim, K.S.; Lee, J.Y.; Kim, D.-N. Modulating the chemo-mechanical response of structured DNA assemblies through binding molecules. Nucleic Acids Res. 2021, 49, 12591–12599. [Google Scholar] [CrossRef]
- Müller, V.; Dvirnas, A.; Andersson, J.; Singh, V.; Kk, S.; Johansson, P.; Ebenstein, Y.; Ambjörnsson, T.; Westerlund, F. Enzyme-free optical DNA mapping of the human genome using competitive binding. Nucleic Acids Res. 2019, 47, e89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zeng, T.; Tan, X.; Wu, W.; Tang, Y.; Zhang, H. A facile surface-enhanced Raman scattering (SERS) detection of rhodamine 6G and crystal violet using Au nanoparticle substrates. Appl. Surf. Sci. 2015, 347, 569–573. [Google Scholar] [CrossRef]
- Wang, Z.; Li, S.; Wang, J.; Shao, Y.; Mei, L. A recyclable graphene/Ag/TiO2 SERS substrate with high stability and reproducibility for detection of dye molecules. New J. Chem. 2022, 46, 18787–18795. [Google Scholar] [CrossRef]
- Pilot, R.; Bozio, R. Validation of SERS enhancement factor measurements. J. Raman Spectrosc. 2018, 49, 462–471. [Google Scholar] [CrossRef]
- Ghanashyam, C.; Sinha, R.K.; Bankapur, A. Surface-Plasmon-Polaritons for Reversible Assembly of Gold Nanoparticles, In Situ Nanogap Tuning, and SERS. Small Methods 2023, 8, 2301086. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, R.; Jangir, D.K.; Agarwal, S.; Ray, B.; Singh, P.; Srivastava, A. Interaction studies of anticancer drug lomustine with calf thymus DNA using surface enhanced Raman spectroscopy. Mapan 2013, 28, 273–277. [Google Scholar] [CrossRef]
- Farhane, Z.; Bonnier, F.; Maher, M.A.; Bryant, J.; Casey, A.; Byrne, H.J. Differentiating responses of lung cancer cell lines to Doxorubicin exposure: In vitro Raman micro spectroscopy, oxidative stress and bcl-2 protein expression. J. Biophotonics 2017, 10, 151–165. [Google Scholar] [CrossRef]
- Beljebbar, A.; Sockalingum, G.; Angiboust, J.; Manfait, M. Comparative FT SERS, resonance Raman and SERRS studies of doxorubicin and its complex with DNA. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 1995, 51, 2083–2090. [Google Scholar] [CrossRef]
- Selvarajan, A. Raman Spectrum of Dimethyl Sulfoxide (DMSO) and the Influence of Solvents; Proceedings of the Indian Academy of Sciences—Section A; Springer: Berlin/Heidelberg, Germany, 1966; pp. 44–50. [Google Scholar]
- Martens, W.N.; Frost, R.L.; Kristof, J.; Theo Kloprogge, J. Raman spectroscopy of dimethyl sulphoxide and deuterated dimethyl sulphoxide at 298 and 77 K. J. Raman Spectrosc. 2002, 33, 84–91. [Google Scholar] [CrossRef]
- Griffiths, P.R. The handbook of infrared and Raman characteristic frequencies of organic molecules. Vib. Spectrosc. 1992, 4, 121. [Google Scholar] [CrossRef]
- Mary, Y.S.; Jojo, P.; Panicker, C.Y.; Van Alsenoy, C.; Ataei, S.; Yildiz, I. Quantum mechanical and spectroscopic (FT-IR, FT-Raman, 1H NMR and UV) investigations of 2-(phenoxymethyl) benzimidazole. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 125, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Mary, Y.S.; Raju, K.; Yildiz, I.; Temiz-Arpaci, O.; Nogueira, H.I.; Granadeiro, C.M.; Van Alsenoy, C. FT-IR, FT-Raman, SERS and computational study of 5-ethylsulphonyl-2-(o-chlorobenzyl) benzoxazole. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 96, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Caprara, D.; Ripanti, F.; Capocefalo, A.; Ceccarini, M.; Petrillo, C.; Postorino, P. Exploiting SERS sensitivity to monitor DNA aggregation properties. Int. J. Biol. Macromol. 2021, 170, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sierra, M.; Quiñones, E. Assays for the determination of the activity of DNA nucleases based on the fluorometric properties of the YOYO dye. Arch. Biochem. Biophys. 2015, 570, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Usman, A.; Ahmad, M. Binding of Bisphenol-F, a bisphenol analogue, to calf thymus DNA by multi-spectroscopic and molecular docking studies. Chemosphere 2017, 181, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Ponkarpagam, S.; Mahalakshmi, G.; Vennila, K.; Elango, K.P. Multi-spectroscopic, voltammetric and molecular docking studies on binding of anti-diabetic drug rosigiltazone with DNA. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 234, 118268. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shuang, S.; Dong, C. Study on the phosphorescence characterizations of palmatine chloride on the solid substrate and its interaction with ctDNA. Talanta 2009, 77, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lv, J.; Zhang, G.; Wang, G.; Liu, Q. Interaction of an anthracycline disaccharide with ctDNA: Investigation by spectroscopic technique and modeling studies. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 75, 1511–1515. [Google Scholar] [CrossRef] [PubMed]
- Sui, C.; Wang, K.; Wang, S.; Ren, J.; Bai, X.; Bai, J. SERS activity with tenfold detection limit optimization on a type of nanoporous AAO-based complex multilayer substrate. Nanoscale 2016, 8, 5920–5927. [Google Scholar] [CrossRef]
- Gong, Z.; Dang, Y.; Zhu, J.; Zheng, J.; Zhang, C.; Zhao, W.; Wang, K. Reflection Interference Spectroscopy Technology Monitoring the Synthesis of ZnCl2-ZnO Nanosheets on Nanoporous Anodic Alumina Substrate in Real Time. Photonics 2023, 10, 552. [Google Scholar] [CrossRef]
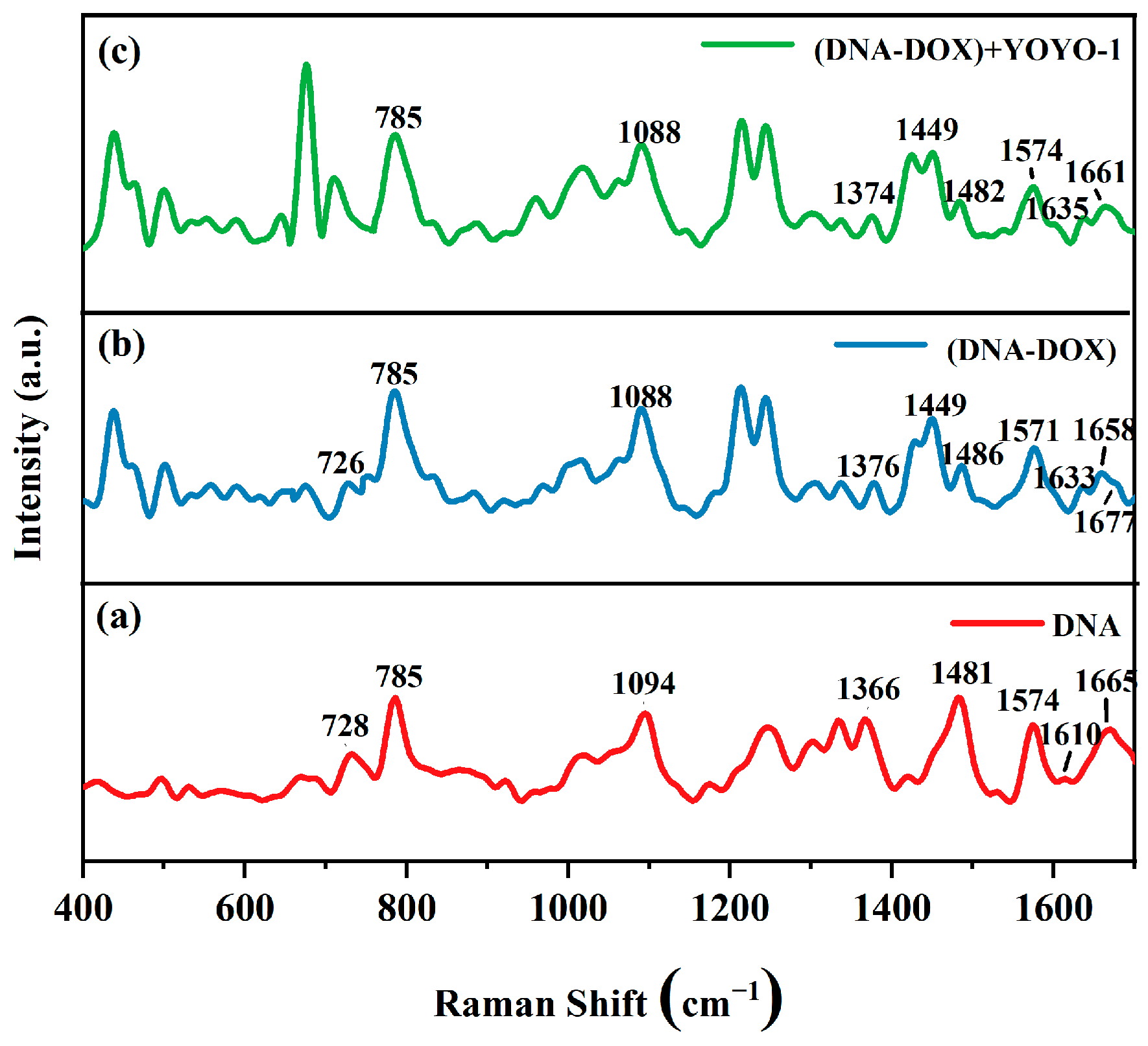

| ctDNA | ctDNA-DOX | ctDNA-DOX + YOYO-1 | Assignment |
|---|---|---|---|
| 728 | 726 | / | A |
| 785 | 785 | 785 | C |
| 1094 | 1088 | 1088 | O=P=O |
| 1366 | 1376 | 1374 | T |
| / | 1449 | 1449 | Deoxyribose |
| 1481 | 1486 | 1482 | G |
| 1574 | 1571 | 1574 | A |
| 1610 | 1633 | 1635 | A |
| 1665 | 1658 | 1661 | C |
| ctDNA | (ctDNA-YOYO-1) | (ctDNA-YOYO-1) + DOX | Assignment |
|---|---|---|---|
| 728 | 726 | 726 | A |
| 785 | 783 | 785 | C |
| 1094 | 1097 | 1093 | O=P=O |
| 1366 | 1376 | 1378 | T |
| 1481 | 1486 | 1489 | G |
| 1574 | 1574 | 1574 | A |
| 1614 | 1607 | 1601 | A |
| 1665 | 1660 | 1664 | C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Li, Z.; Yun, P.; Sun, D.; Niu, Y.; Yao, B.; Wang, K. Studying the Effects and Competitive Mechanisms of YOYO-1 on the Binding Characteristics of DOX and DNA Molecules Based on Surface-Enhanced Raman Spectroscopy and Molecular Docking Techniques. Int. J. Mol. Sci. 2024, 25, 3804. https://doi.org/10.3390/ijms25073804
Li Y, Li Z, Yun P, Sun D, Niu Y, Yao B, Wang K. Studying the Effects and Competitive Mechanisms of YOYO-1 on the Binding Characteristics of DOX and DNA Molecules Based on Surface-Enhanced Raman Spectroscopy and Molecular Docking Techniques. International Journal of Molecular Sciences. 2024; 25(7):3804. https://doi.org/10.3390/ijms25073804
Chicago/Turabian StyleLi, Yanjie, Zhiwei Li, Penglun Yun, Dan Sun, Yong Niu, Baoli Yao, and Kaige Wang. 2024. "Studying the Effects and Competitive Mechanisms of YOYO-1 on the Binding Characteristics of DOX and DNA Molecules Based on Surface-Enhanced Raman Spectroscopy and Molecular Docking Techniques" International Journal of Molecular Sciences 25, no. 7: 3804. https://doi.org/10.3390/ijms25073804
APA StyleLi, Y., Li, Z., Yun, P., Sun, D., Niu, Y., Yao, B., & Wang, K. (2024). Studying the Effects and Competitive Mechanisms of YOYO-1 on the Binding Characteristics of DOX and DNA Molecules Based on Surface-Enhanced Raman Spectroscopy and Molecular Docking Techniques. International Journal of Molecular Sciences, 25(7), 3804. https://doi.org/10.3390/ijms25073804







