Abstract
The gut–brain axis is increasingly understood to play a role in neuropsychiatric disorders. The probiotic bacterium Lactobacillus (L.) reuteri and products of tryptophan degradation, specifically the neuroactive kynurenine pathway (KP) metabolite kynurenic acid (KYNA), have received special attention in this context. We, therefore, assessed relevant features of KP metabolism, namely, the cellular uptake of the pivotal metabolite kynurenine and its conversion to its primary products KYNA, 3-hydroxykynurenine and anthranilic acid in L. reuteri by incubating the bacteria in Hank’s Balanced Salt solution in vitro. Kynurenine readily entered the bacterial cells and was preferentially converted to KYNA, which was promptly released into the extracellular milieu. De novo production of KYNA increased linearly with increasing concentrations of kynurenine (up to 1 mM) and bacteria (107 to 109 CFU/mL) and with incubation time (1–3 h). KYNA neosynthesis was blocked by two selective inhibitors of mammalian kynurenine aminotransferase II (PF-048559989 and BFF-122). In contrast to mammals, however, kynurenine uptake was not influenced by other substrates of the mammalian large neutral amino acid transporter, and KYNA production was not affected by the presumed competitive enzyme substrates (glutamine and α-aminoadipate). Taken together, these results reveal substantive qualitative differences between bacterial and mammalian KP metabolism.
1. Introduction
The gut–brain axis has recently emerged as a major topic of interest in neurobiology, especially with regard to the pathophysiology of neuropsychiatric disorders. Mounting evidence supports the hypothesis that gut microbiota regulate brain development and behavior [1,2,3,4,5] and that dysbiosis contributes to cognitive deficits and other behavioral abnormalities [6,7,8] and may be critically involved in a variety of psychiatric diseases [9,10,11,12].
Studies in germ-free and antibiotic-treated rodents suggest that the essential amino acid tryptophan plays a special role in this context [13]. In the mammalian gut, most dietary tryptophan is either absorbed in the small intestine or locally converted to biologically active metabolites that then enter the circulation [14]. About 5% of tryptophan is metabolized by specific gut bacteria, mostly to indole derivatives [15,16,17], and, notably, elevated circulating tryptophan levels are associated with a decrease in gut microbiota [13,18].
In mammals, 95% of dietary tryptophan is degraded via the kynurenine pathway (KP), which is named after the pivotal metabolite L-kynurenine (L-KYN). Conversion of tryptophan to L-KYN is catalyzed by indoleamine 2,3-dioxygenases (IDO 1 and IDO 2), which are present in the intestine, brain and immune cells and activated by inflammatory stimuli and tryptophan 2,3-dioxygenase (TDO), which is mostly expressed in the liver. Subsequently, L-KYN is metabolized to the free radical generator 3-hydroxykynurenine (3-HK) by kynurenine 3-monooxygenase or to anthranilic acid (ANA) by kynureninase (Figure 1). Of special significance for brain function and dysfunction [19,20,21,22,23], L-KYN is also converted by kynurenine aminotransferases (KATs) to kynurenic acid (KYNA), an antagonist of N-methyl-D-aspartate (NMDA; [24,25], and α7 nicotinic acetylcholine (α7nACh; [26] receptors and an agonist at an orphan G-protein-coupled receptor (GPR35; [27]. Interestingly, KYNA can also regulate the immune system through its agonistic effects on the aryl hydrocarbon receptor (AhR; [28] and is an effective scavenger of reactive oxygen species [29,30].
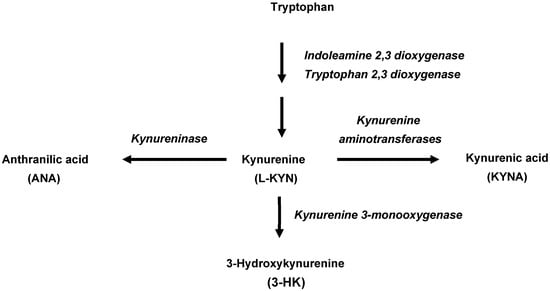
Figure 1.
Initial enzymatic processes involved in the kynurenine pathway of tryptophan metabolism in mammals.
Although the formation of KP metabolites by intestinal microbiota impacts host function [31], and in spite of the fact that experimental manipulation of the gut microbiota affects KP metabolism in the host [6,32,33,34,35,36], only relatively little is known about the ability of individual intestinal bacteria to express functional KP enzymes and to produce KYNA, 3-HK and ANA [37,38,39,40].
Lactobacillus (L.) reuteri, a well-studied probiotic bacterium that colonizes the gastrointestinal tract of both humans and animals, can influence the composition of the gut microbiota [41] and has been shown to exert a remarkable range of beneficial effects in both humans and relevant animal models. Notably, L. reuteri administration affects neurodevelopment and prevents social deficits and depressive-like behaviors in experimental animals [41,42,43,44,45,46,47]. Interestingly, though the underlying mechanisms have not been identified so far, L. reuteri is able to normalize the impaired plasma levels of KP metabolites caused by chronic stress and to improve associated behavioral abnormalities [35].
The present study was designed to examine the intricacies of KP metabolism in this translationally relevant bacterial species in greater depth. To this end, we investigated the ability of L. reuteri to accumulate exogenous L-KYN and to produce KYNA, 3-HK and ANA under a variety of experimental conditions in vitro.
2. Results
2.1. L-KYN Uptake into Live L. reuteri
To characterize KP metabolism in live L. reuteri, we first tested the ability of L-KYN to enter the cells. Incubation resulted in increased levels of intracellular L-KYN, starting at 5 min and slowly rising until 180 min, indicating that L-KYN rapidly enters and accumulates in the bacterial cells (Figure 2). Under standard conditions (1 h incubation at 37 °C), significantly less L-KYN was recovered from heat-inactivated bacteria (9.8 ± 1.3 vs. 3.1 ± 1.0 fmoles, n = 3; p < 0.05; Student’s t-test).
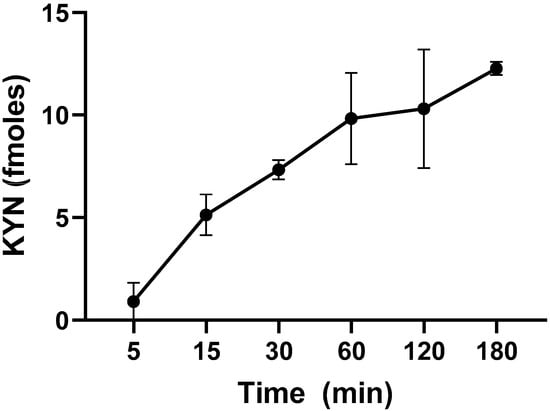
Figure 2.
Time-dependence of L-KYN uptake into live L. reuteri (109 CFU/mL). Bacteria were incubated in the presence of 100 µM L-KYN and 0.2 µCi 3H-L-KYN (see text for experimental details). Data are the mean ± SEM of 3 experiments.
We next tested selected amino acids that are known to compete with L-KYN as substrates of the large neutral amino acid transporter (LAT) in mammals [48,49] for their ability to affect L-KYN uptake into bacterial cells (Table 1). Unexpectedly, none of the tested compounds affected L-KYN uptake at a concentration of 5 mM (p > 0.05; one-way ANOVA, followed by Bonferroni’s post hoc test). Five mM BCH, an inhibitor of L-type amino acid transporters including the LAT [50], also failed to affect L-KYN uptake (p > 0.05; one-way ANOVA, followed by Bonferroni’s post hoc test), indicating substantive qualitative differences between L-KYN uptake into bacterial and mammalian cells.

Table 1.
Effects of selected compounds (final concentration: 5 mM) on L-KYN uptake into live L. reuteri.
2.2. Basal Levels and De Novo Production of KYNA, 3-HK and ANA
Next, we examined the presence of endogenous levels of KP metabolites in live L. reuteri and their de novo production after incubation with L-KYN for 3 h. Under these conditions, the basal levels of KYNA (2.4 ± 1.7 fmoles/µL, n = 4) and ANA (1.9 ± 0.7 fmoles/µL, n = 3) were clearly measurable, but basal 3-HK levels were below our limit of detection (<0.5 fmoles/µL).
Incubation with 100 µM L-KYN induced a significant increase in the levels of KYNA (p < 0.0001; Student’s t-test) and raised the levels of 3-HK above the detection limit (2.8 ± 0.6 fmoles/µL); however, it did not significantly elevate the levels of ANA (p > 0.05; Student’s t-test), demonstrating that L-KYN in L. reuteri is preferentially converted to KYNA (Figure 3).
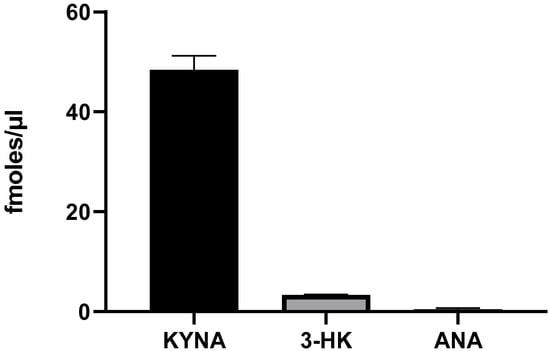
Figure 3.
De novo production of KYNA, 3-HK and ANA by live L. reuteri (108 CFU/mL). Bacteria were incubated for 3 h with 100 µM L-KYN, and metabolites were recovered from the extracellular milieu. See text for experimental details. Data are the mean ± SEM of 3–4 experiments.
In view of these results, subsequent experiments focused exclusively on evaluating the de novo synthesis of KYNA and its regulation.
2.3. Optimization of De Novo synthesis of KYNA from L-KYN by Live L. reuteri
To determine the optimal conditions for the neosynthesis of KYNA production in live L. reuteri, we tested different L-KYN and bacterial concentrations as well as various incubation times (Figure 4). A dose-dependent elevation in KYNA levels was observed with increasing L-KYN concentrations (ranging from 10 to 1000 µM; Figure 4A) and bacterial density (from to 107 to 109 CFU/mL; Figure 4B). Increasing the incubation time from 1 to 3 h linearly raised KYNA formation (Figure 4C), and 99.6% of total KYNA produced from L-KYN was released into the extracellular milieu (Figure 4D). Longer incubation times (tested up to 24 h) failed to result in further increases. Based on these results, all subsequent experiments were conducted using 108 CFU/mL bacteria, which were incubated for 3 h with 100 µM L-KYN.
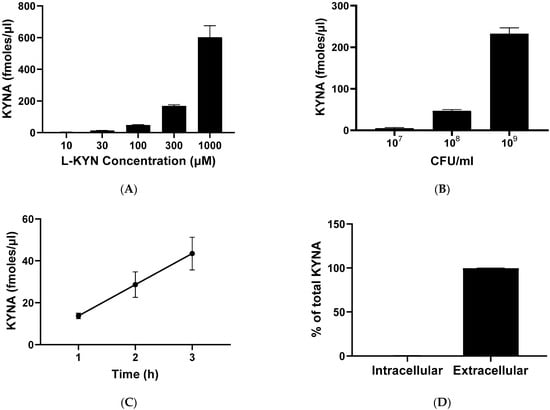
Figure 4.
De novo synthesis of KYNA from L-KYN in live L. reuteri. (A–C): concentration of KYNA recovered from the extracellular milieu (see text for experimental details). (A) KYNA production in bacteria (108 CFU/mL) incubated for 3 h with increasing concentrations of L-KYN. (B) KYNA formation after incubation of different concentrations of L. reuteri with 100 µM L-KYN for 3 h. (C) Effect of incubation time on KYNA neosynthesis using 100 µM L-KYN and 108 CFU/mL L. reuteri. (D) Percentage of KYNA detected in the intracellular vs. the extracellular compartment following a 3 h incubation of L. reuteri (108 CFU/mL) with 100 µM L-KYN. In all cases, data are the mean ± SEM of 3–4 experiments. See text for experimental details.
In light of the ability of bacteria—in contrast to mammalian cells—to synthesize D-amino acids [51,52], we also investigated the production of KYNA from D-KYN. Incubation of L. reuteri with 100 µM D-KYN under standard experimental conditions (108 CFU/mL bacteria, 3 h) caused a significant elevation in KYNA over endogenous levels (p < 0.01; Student’s t-test); however, the increase in extracellular KYNA was only ~30% of the effect resulting from incubation with L-KYN (18.1 ± 3.5 vs. 65.1 ± 3.5 fmoles/µL, n = 3; p < 0.05; Student’s t-test).
Notably, no de novo formation of KYNA was detected when L. reuteri was incubated with 1 mM of L-tryptophan under standard conditions.
2.4. Pharmacological Regulation of KYNA Production in Live L. reuteri
To examine the role of KAT enzymes (KAT I, KAT II, KAT III and KAT IV; [53,54] in KYNA formation in live L. reuteri, we tested the effect of endogenous amino acids, which are known to compete with L-KYN as respective substrates of the mammalian enzymes (glutamine for KAT I and KAT III, α-aminoadipate for KAT II and aspartate for KAT IV), as well as the non-specific aminotransferase inhibitor AOAA and the synthetic KAT II inhibitors BFF-122 [55] and PF-04859989 [56] (Table 2). Whereas 1 mM AOAA essentially totally prevented the de novo production of KYNA from L-KYN (99.7%, p < 0.0001) and 10 mM aspartate induced a small reduction (~25%, p < 0.01), neither glutamine nor α-aminoadipate (both at 10 mM) had an effect (p > 0.05). Interestingly, both BFF-122 (1 mM) and PF-04859989 (100 µM) significantly inhibited the bacterial neosynthesis of KYNA (p < 0.0001). Subsequent dose-dependency experiments showed higher efficacy of PF-04859989 compared to BFF-122 (>90% and <50% inhibition, respectively, at 100 µM; Figure 5).

Table 2.
Effects of selected endogenous and exogenous KAT inhibitors on the de novo synthesis of KYNA from L-KYN in live L. reuteri.
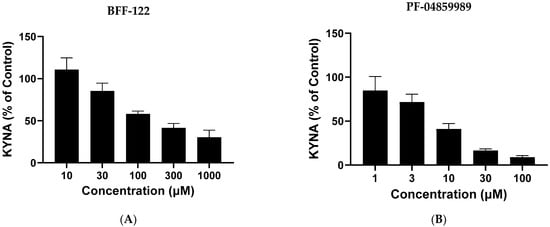
Figure 5.
Dose-dependent inhibition of KYNA neosynthesis by BFF-122 (A) and PF-04859989 (B). L. reuteri (108 CFU/mL) was incubated for 3 h with 100 µM L-kynurenine. Control values: 48.4 ± 2.8 fmoles/µL. See text for experimental details. Data are the mean ± SEM of 3 experiments.
We also studied the pro-cognitive and antioxidant compound N-acetylcysteine (NAC), which inhibits mammalian KAT II both in vitro and in vivo [57]. NAC showed only weak activity, with less than 40% inhibition of KYNA neosynthesis at 10 mM (n = 6; p < 0.05; Student’s t-test). To examine, more generally, the possible involvement of oxidative processes in the de novo formation of KYNA from L-KYN in live L. reuteri, we then tested the antioxidant ascorbic acid (500 µM) under the same experimental conditions. Ascorbic acid had no significant effect on KYNA production (n = 3; p > 0.05; Student’s t-test).
Finally, we examined the ability of several 2-oxoacids, which are established co-substrates of KATs and readily stimulate KYNA synthesis in mammalian tissues [58]. When added to the incubation buffer at a final concentration of 5 mM, only some of the compounds significantly enhanced L-KYN conversion to KYNA in the live bacteria, with α-ketoisovalerate having the greatest effect of the 2-oxoacids tested (increase to ~190% of control values; p < 0.0001). In contrast, the inclusion of oxaloacetate in the incubation medium unexpectedly caused a reduction in KYNA neosynthesis (p < 0.05) (Table 3).

Table 3.
Effects of various oxoacids (5 mM) on the neosynthesis of KYNA from L-KYN in live L. reuteri.
2.5. KYNA Production in Homogenized Bacteria
To examine the de novo formation of KYNA from L-KYN in lysed cells, sonicated bacterial tissue was incubated under conditions that are optimal for the mammalian KAT II enzyme [59]. L-KYN was readily converted to KYNA and, as in live bacteria, 10 mM glutamine caused only a small, non-significant reduction in KYNA neosynthesis from L-KYN in the homogenate (cf. Table 2 and Table 4). However, the addition of α-aminoadipate or aspartate (both at 10 mM) had stronger effects in the homogenized bacteria, with reductions in KYNA formation of ~20% and ~70%, respectively (p < 0.05 and p < 0.0001). Notably, in contrast to their quantitatively different effects in live bacteria, the two synthetic KAT II inhibitors BFF-122 and PF-04859989 (both at 1 mM) had similar potency in bacterial homogenates (~95% and ~88% inhibition, respectively) (both p < 0.0001) (cf. Table 2 and Table 4).

Table 4.
Effects of selected KAT substrates and inhibitors on KYNA synthesis from L-KYN in homogenized bacteria.
3. Discussion
The present study was designed to evaluate the role of the probiotic bacterium L. reuteri in regulating KP metabolism and, in particular, its ability to synthesize KYNA, 3-HK and ANA from KYN. We demonstrated that L-KYN rapidly entered the bacterial cells, but its uptake was not affected by other substrates of mammalian LAT. Although we were able to detect endogenous levels of both KYNA and ANA in live bacteria, our findings revealed that L-KYN is preferentially metabolized to KYNA, which is then promptly released into the extracellular milieu.
Interestingly, we did not observe de novo formation of KYNA from tryptophan under our experimental conditions. Although not examined in the present study, L. reuteri, like several other intestinal bacteria, may, therefore, preferentially convert tryptophan to a number of biologically active indoles instead of forming KP metabolites [14]. On the other hand, L. reuteri could generate KYNA from D-KYN, though with lower efficiency than from L-enantiomer. This finding clearly deserves further scrutiny and may be of (patho)physiological relevance in view of the fact that bacteria, unlike eukaryotes, have the ability to synthesize D-amino acids [51,60]. Also of interest in this context, D-KYN is a better precursor of KYNA in L. reuteri than in rodents or humans [61,62].
In mammals, L-KYN enters cells through LATs, which are able to transport both branched (valine, leucine, isoleucine) and aromatic (tryptophan, phenylalanine) amino acids, all of which compete for entrance into the cells [48,49]. Although L-KYN was shown here to rapidly enter the bacterial cells, its uptake was not inhibited by tryptophan or other presumably competing amino acids. Similarly, the LAT inhibitor BCH, which effectively interferes with L-KYN uptake in both rat brain slices in vitro and mouse brain in vivo [50], did not affect L-KYN uptake into L. reuteri, indicating that the bacterial transporter differs substantively from the mammalian LAT.
Although not studied systematically so far, attempts to examine enzymatic L-KYN degradation in individual bacteria have revealed remarkable qualitative strain differences. For example, both Cytophaga hutchinsonii and Pseudomonas fluorescens express kynurenine 3-monooxygenase, the enzyme responsible for the synthesis of 3-HK [63,64], Pseudomonas fluorescens also contains kynureninase to produce ANA [65,66], and Pseudomonas aeruginosa is capable of enzymatically producing all three primary L-KYN metabolites, i.e., KYNA, 3-HK and ANA [37] (cf. Figure 1). In the present study, we observed that L. reuteri preferentially converts L-KYN to KYNA and appears to contain very little kynureninase and kynurenine 3-monooxygenase. These results also suggest that the low endogenous levels of 3-HK and ANA in L. reuteri may be related to non-enzymatic degradative processes [67] and, in the case of ANA, may involve alternative synthetic routes [68].
Although the presence of KAT in bacteria has been known for decades [69], the examination of bacterial KYNA neosynthesis has attracted only limited interest so far [37,38,70]. The dominant conversion of L-KYN to KYNA seen in L. reuteri prompted us to study this mechanism in greater detail. The fact that ascorbic acid did not significantly reduce the bacterial formation of KYNA suggested that KYNA synthesis from L-KYN in L. reuteri was not caused by the non-enzymatic oxidation of L-KYN [71,72] but was mainly enzymatic in nature. Although confirmed in principle by the very effective blockade of KYNA formation by AOAA, the mechanism of bacterial KYNA production showed major qualitative differences from mammalian cells, however [59]. Thus, glutamine, a highly competitive substrate of mammalian KAT I (= glutamine aminotransferase), was unable to inhibit the de novo KYNA production in both live and homogenized bacteria, even at a high concentration (10 mM). Similarly, 10 mM α-aminoadipate, a competing substrate of mammalian KAT II (=α-aminoadipate aminotransferase), did not interfere with KYNA formation from L-KYN in live L. reuteri and caused only a small decrease (~20%) in homogenized bacteria. Interestingly, though, two synthetic inhibitors of mammalian KAT II (BFF-122 and PF-04859989; [55,56] substantially reduced KYNA production. These results, as well as the modest but significant effects caused by the addition of aspartate, a relatively weak inhibitor of mammalian KAT II [73], suggest that bacterial enzyme(s) bearing some similarity to mammalian KAT II or KAT IV [53] account for the neosynthesis of KYNA from L-KYN in L. reuteri. The distinct nature of this enzymatic process, which is further supported by the fact that several 2-oxoacids known to serve as amino-acceptors of mammalian KATs [74] failed to show the expected effects in the present study, clearly requires further clarification. Notably, future experiments should consider the growth rate and gene activity of L. reuteri, which were not taken into account in the present study.
The present results raise the possibility that KYNA plays a role in the remarkable beneficial effects of L. reuteri administration in both humans and rodents. Thus, through its antioxidant properties and/or by targeting several receptors with critical roles in both physiology as well as pathology, an elevation in KYNA levels may have considerable functional consequences in both the periphery and brain (cf. Introduction). For example, increased formation and release of KYNA by L. reuteri may regulate the enteric nervous system [75], affect the growth and viability of probiotic bacteria in the digestive system [76], alleviate various gastrointestinal pathologies [77] and have anti-inflammatory effects by inhibiting Th17 cell differentiation and the increase in TNFα in monocytes and leukocytes [20,78]. Notably, though the mechanistic link clearly needs to be investigated further, KYNA generated by L. reuteri—alone or in combination with other probiotics—may also participate in the attenuation of depressive-like symptoms associated with chronic stress [41] and in the reduction of obesity-related behavioral impairments and related microglial activation [42,43,46].
4. Materials and Methods
4.1. Materials
L. reuteri (F 275T = ATCC 23272T = DSM 20016T = JCM 1112T = LMG 9213T = LMG 13557T) bacteria were obtained from the American Type Culture Collection (Manassas, VA, USA). 3H-L-kynurenine (3H-KYN) (16 Ci/mmol) was purchased from Amersham (Buckinghamshire, UK). Aminooxyacetic acid (AOAA), aspartate, α-aminoadipate, glutamine, pyruvate, α-ketoglutarate, α-ketoisocaproate, α-ketoisovalerate, oxaloacetate, 2-amino-2-norbornanecarboxylic acid (BCH) and PF-04859989 were obtained from Sigma-Aldrich (St. Louis, MO, USA). L-Kynurenine sulfate (L-KYN) was acquired from Sai Advantium (Hyderabad, India) and D-kynurenine sulfate (D-KYN) from Shanghai Hanhong Chemical Co. (Shanghai, China). BFF-122 [(S)-(-)-9-(4-aminopiperazine-1-yl)-8-fluoro-3-methyl-6-oxo-2,3,5,6-tetrahydro-4H-1-oxa-3a-azaphenalene-5-carboxylic acid] was kindly provided by Dr. Y. Kajii (Mitsubishi-Tanabe Pharma Corp., Yokohama, Japan). All other chemicals used were purchased from commercial suppliers and were of the highest available purity.
4.2. Preparation of the Bacteria
L. reuteri bacteria were grown on deMan, Rogosa and Sharpe (MRS) agar (Hardy Diagnostics, Santa Maria, CA, USA) for 24 h at 37 °C. The bacteria were then transferred to an MRS broth (Hardy Diagnostics) and quantified by serial dilution plating and counting colonies. Bacteria were diluted to 1010 or 109 CFU/mL for storage, aliquoted and frozen at −80 °C. For all assays, bacterial aliquots were thawed at room temperature, centrifuged (6000× g, 2 min), rinsed with cold sterile Hank’s Balanced Salt Solution (HBSS; containing 1.0 g/L glucose, 0.011 g/L phenol red and 0.35 g/L sodium bicarbonate, pH 7.1–7.5; H9269; Sigma-Aldrich) and then resuspended in HBSS and diluted to the concentrations used in the respective assays.
The viability of the bacteria under standard experimental conditions (108 CFU/mL live bacteria in HBSS, 100 µM KYN, 3 h incubation) was evaluated by serial dilution plating and counting colonies.
4.3. L-KYN Uptake by Live Bacteria
3H-KYN was purified by high-performance liquid chromatography (HPLC) prior to its use in uptake experiments [79]. L-KYN uptake was assessed by incubating 109 CFU/mL live L. reuteri in HBSS at 37 °C for 5 to 180 min in the presence of 0.2 µCi of 3H-KYN (20 µL), 100 µM non-radioactive L-KYN (10 µL) and water (replaced by dissolved test compounds when indicated), in a total volume of 200 µL. Blank values were routinely obtained by incubating the bacteria on ice. Inactivated bacteria (cells heated for 2 min at 100 °C before incubation) were examined in some experiments. Incubations were performed using a Roto-Therm H2020 Fixed Speed Incubated Tube Rotator (Benchmark Scientific, Edison, NJ, USA) at 24 rpm.
Following incubation, the assay mixture was immediately centrifuged (6000× g, 2 min), and the supernatant was discarded. The remaining pellet was placed on ice, and residual radioactivity was eliminated by adding 500 µL of cold HBSS, gentle mixing and further centrifugation (6000× g, 2 min). After removal of the supernatant, the pellet was suspended in 200 µL HBSS and transferred to vials containing 10 mL scintillation fluid. Radioactivity was measured by liquid scintillation spectrometry (Packard Tri-Carb 2200CA LCA, Perkin Elmer, Boston, MA, USA).
4.4. KYNA, 3-HK and ANA Production by Live Bacteria
De novo synthesis of KYNA, 3-HK and ANA from L-KYN by live bacteria was assessed by incubation at 37 °C in a total volume of 200 µL [160 µL of bacteria in HBSS, 20 µL of L-KYN and 20 µL of water (replaced by KAT inhibitors or 2-oxoacids in some experiments)] using a Roto-Therm H2020 Fixed Speed Incubated Tube Rotator at 24 rpm. Incubation times (1, 2 or 3 h), bacterial density (107–109 CFU/mL) and L-KYN concentrations (10–1000 µM) varied during method development. The reaction was terminated by the addition of 40 µL of 50% trichloroacetic acid (TCA). Samples were then centrifuged (15,800× g, 10 min), and the supernatant was removed and analyzed as detailed below. Blanks were obtained by incubating bacteria in the absence of L-KYN.
To measure intracellular KYNA, the pellet containing the bacterial cells was resuspended in 200 µL of water and sonicated. After centrifugation (15,800× g, 5 min), KYNA was determined in the supernatant.
4.5. KYNA Synthesis in Lysed L. reuteri Cells
The neosynthesis of KYNA was also examined in homogenized L. reuteri cells. To this end, frozen bacteria were thawed and centrifuged (6000× g, 10 min). The MRS broth was replaced with 0.5 M Tris-acetate buffer (pH 8.0) to obtain a final bacterial concentration of 108 CFU/mL, and the solution was sonicated to lyse the bacterial cells. Assays were performed in a total volume of 200 µL [80 µL of sonicated 108 CFU/mL bacteria, 100 µL of assay cocktail (100 µM L-KYN, 80 µM pyridoxal-5′-phosphate and 1 mM pyruvate in 150 mM Tris-acetate buffer, pH 7.4) and 20 µL of water (replaced by KAT inhibitors in some experiments)]. Blanks were obtained by adding the non-specific aminotransferase inhibitor AOAA (final concentration: 1 mM) to the solution. After incubation for 3 h at 37 °C, the reaction was terminated by the addition of 20 µL of cold 50% trichloroacetic acid (TCA) and 1 mL 0.1 N HCl. Samples were kept on ice and centrifuged (15,800× g, 10 min), and KYNA was analyzed in the supernatant.
4.6. KYNA, 3-HK and ANA Measurement
KYNA quantification was performed by high-performance liquid chromatography (HPLC) with fluorescent detection. In total, 20 µL of the supernatant was injected onto a BDS Hypersil C18 column (100 mm × 4.6 mm, particle size 3 µm; Thermo Fisher Scientific, Waltham, MA, USA) with a mobile phase consisting of 3% acetonitrile, 250 mM sodium acetate and 50 mM zinc acetate (pH 6.2) at a flow rate of 1 mL/min. Fluorescence detection was performed using a Perkin Elmer Series 200a instrument (Perkin Elmer, Shelton, CT, USA) (excitation: 344 nm, emission: 398 nm). The retention time was approximately 8 min.
3-HK quantification was performed by HPLC with electrochemical detection. To this end, 20 µL of the supernatant was injected onto a C18 reverse phase column (HR-80; 80 mm × 4.6 mm; particle size 3 µm; Thermo Fisher Scientific) with a mobile phase consisting of 1.5% acetonitrile, 0.9% trimethyl amine, 0.59% phosphoric acid, 0.27 mM EDTA and 8.9 mM sodium heptane sulfonic acid at a flow rate of 0.5 mL/min. 3-HK was detected electrochemically using an HTEC 500 detector (Eicom, San Diego, CA, USA) and had a retention time of approximately 11 min.
For ANA detection, 20 μL of the supernatant was applied to a 5 μm C18 reverse phase column (Adsorbosil; 150 mm × 4.6 mm; Dr. Maisch GmbH, Ammerbuch, Germany) using a mobile phase containing 100 mM sodium acetate (pH 5.8) and 1% acetonitrile at a flow rate of 1.0 mL/min. ANA was detected fluorimetrically in the eluate (excitation: 340 nm; emission: 410 nm; 2475 fluorescence detector; Waters, Milford, MA, USA). The retention time was approximately 6 min [80].
4.7. Statistical Analysis
All data are expressed as the mean ± SEM. Statistical analyses were performed using Graphpad Prism 9 (San Diego, CA, USA). Student’s t-test or one-way ANOVA followed by Bonferroni’s post hoc test was used to determine significance in selected experiments. A p value < 0.05 was considered significant.
5. Conclusions
In summary, the present in vitro study not only provided further evidence of the well-established ability of gut microbiota to metabolize the major tryptophan metabolite L-KYN but revealed that L. reuteri, in contrast to other bacteria, preferentially synthesizes KYNA, which is then promptly released into the extracellular milieu. Notably, an assessment of the underlying biochemical mechanisms revealed substantive qualitative differences from mammalian KP metabolism. As the administration of L. reuteri has remarkably beneficial effects in both animals and humans and since KYNA is increasingly understood to play substantive roles in mammalian biology, our results may have significant translational implications. Future studies should, therefore, be designed to manipulate KYNA production in L. reuteri and selected other bacteria by pharmacological and genetic means and to examine the physiological and pathological consequences of such bacterial modifications in the host in vivo.
Author Contributions
Conceptualization: R.S.; Methodology: K.V.S. and F.M.N.; Validation: F.M.N.; Formal analysis, K.V.S. and F.M.N.; Data curation: A.F. and F.M.N.; Writing (original draft): F.M.N.; Writing (writing and editing): R.S., K.V.S. and F.M.N.; Supervision: K.V.S.; Funding acquisition: R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by NIMH grant P50 MH103222 (Silvio O. Conte Center for Translational Mental Health Research).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
3-HK: 3-Hydroxykynurenine; ANA: Anthranilic acid; AOAA: Aminooxy acetic acid; KP: Kynurenine pathway; KYN: Kynurenine; KYNA: Kynurenic acid; LAT: Large neutral amino acid transporter; α7nACh: α7 Nicotinic acetylcholine; NMDA: N-methyl-D-aspartate
References
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Diaz Heijtz, R.; Wang, S.; Anuar, F.; Qian, Y.; Bjorkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Morais, L.H.; Schreiber, H.L.t.; Mazmanian, S.K. The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.B.; Keating, D.J.; Young, R.L.; Wong, M.L.; Licinio, J.; Wesselingh, S. From gut dysbiosis to altered brain function and mental illness: Mechanisms and pathways. Mol. Psychiatry 2016, 21, 738–748. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Backhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Desbonnet, L.; Clarke, G.; Traplin, A.; O’Sullivan, O.; Crispie, F.; Moloney, R.D.; Cotter, P.D.; Dinan, T.G.; Cryan, J.F. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav. Immun. 2015, 48, 165–173. [Google Scholar] [CrossRef]
- Hoban, A.E.; Moloney, R.D.; Golubeva, A.V.; McVey Neufeld, K.A.; O’Sullivan, O.; Patterson, E.; Stanton, C.; Dinan, T.G.; Clarke, G.; Cryan, J.F. Behavioural and neurochemical consequences of chronic gut microbiota depletion during adulthood in the rat. Neuroscience 2016, 339, 463–477. [Google Scholar] [CrossRef]
- Raber, J.; Sharpton, T.J. Gastrointestinal dysfunction in neurological and neurodegenerative disorders. Semin. Neurol. 2023, 43, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Bastiaanssen, T.F.S.; Cowan, C.S.M.; Claesson, M.J.; Dinan, T.G.; Cryan, J.F. Making sense of ... the microbiome in psychiatry. Int. J. Neuropsychopharmacol. 2019, 22, 37–52. [Google Scholar] [CrossRef]
- Connell, E.; Le Gall, G.; Pontifex, M.G.; Sami, S.; Cryan, J.F.; Clarke, G.; Muller, M.; Vauzour, D. Microbial-derived metabolites as a risk factor of age-related cognitive decline and dementia. Mol. Neurodegener. 2022, 17, 43. [Google Scholar] [CrossRef]
- Socala, K.; Doboszewska, U.; Szopa, A.; Serefko, A.; Wlodarczyk, M.; Zielinska, A.; Poleszak, E.; Fichna, J.; Wlaz, P. The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol. Res. 2021, 172, 105840. [Google Scholar] [CrossRef]
- Szeligowski, T.; Yun, A.L.; Lennox, B.R.; Burnet, P.W.J. The Gut microbiome and schizophrenia: The current state of the field and clinical applications. Front. Psychiatry 2020, 11, 156. [Google Scholar] [CrossRef]
- Gao, K.; Mu, C.L.; Farzi, A.; Zhu, W.Y. Tryptophan metabolism: A link between the gut microbiota and brain. Adv. Nutr. 2020, 11, 709–723. [Google Scholar] [CrossRef]
- Hyland, N.P.; Cavanaugh, C.R.; Hornby, P.J. Emerging effects of tryptophan pathway metabolites and intestinal microbiota on metabolism and intestinal function. Amino Acids 2022, 54, 57–70. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Ranhotra, H.S. Discrete interplay of gut microbiota L-tryptophan metabolites in host biology and disease. Mol. Cell. Biochem. 2023. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Bosi, A.; Banfi, D.; Bistoletti, M.; Giaroni, C.; Baj, A. Tryptophan metabolites along the microbiota-gut-brain axis: An interkingdom communication system influencing the gut in health and disease. Int. J. Tryptophan Res. 2020, 13, 1178646920928984. [Google Scholar] [CrossRef]
- Erhardt, S.; Olsson, S.K.; Engberg, G. Pharmacological manipulation of kynurenic acid: Potential in the treatment of psychiatric disorders. CNS Drugs 2009, 23, 91–101. [Google Scholar] [CrossRef]
- Moroni, F.; Cozzi, A.; Sili, M.; Mannaioni, G. Kynurenic acid: A metabolite with multiple actions and multiple targets in brain and periphery. J. Neural Transm. 2012, 119, 133–139. [Google Scholar] [CrossRef]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.Q. Kynurenines in the mammalian brain: When physiology meets pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef]
- Stone, T.W.; Forrest, C.M.; Darlington, L.G. Kynurenine pathway inhibition as a therapeutic strategy for neuroprotection. FEBS J. 2012, 279, 1386–1397. [Google Scholar] [CrossRef] [PubMed]
- Szalardy, L.; Zadori, D.; Toldi, J.; Fulop, F.; Klivenyi, P.; Vecsei, L. Manipulating kynurenic acid levels in the brain-on the edge between neuroprotection and cognitive dysfunction. Curr. Top. Med. Chem. 2012, 12, 1797–1806. [Google Scholar] [CrossRef] [PubMed]
- Birch, P.J.; Grossman, C.J.; Hayes, A.G. Kynurenic acid antagonises responses to NMDA via an action at the strychnine-insensitive glycine receptor. Eur. J. Pharmacol. 1988, 154, 85–87. [Google Scholar] [CrossRef] [PubMed]
- Perkins, M.N.; Stone, T.W. An iontophoretic investigation of the actions of convulsant kynurenines and their interaction with the endogenous excitant quinolinic acid. Brain Res. 1982, 247, 184–187. [Google Scholar] [CrossRef]
- Hilmas, C.; Pereira, E.F.; Alkondon, M.; Rassoulpour, A.; Schwarcz, R.; Albuquerque, E.X. The brain metabolite kynurenic acid inhibits alpha7 nicotinic receptor activity and increases non-alpha7 nicotinic receptor expression: Physiopathological implications. J. Neurosci. 2001, 21, 7463–7473. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Simonavicius, N.; Wu, X.; Swaminath, G.; Reagan, J.; Tian, H.; Ling, L. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem. 2006, 281, 22021–22028. [Google Scholar] [CrossRef] [PubMed]
- DiNatale, B.C.; Murray, I.A.; Schroeder, J.C.; Flaveny, C.A.; Lahoti, T.S.; Laurenzana, E.M.; Omiecinski, C.J.; Perdew, G.H. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol. Sci. 2010, 115, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Kubicova, L.; Hadacek, F.; Bachmann, G.; Weckwerth, W.; Chobot, V. Coordination complex formation and redox properties of kynurenic and xanthurenic acid can affect brain tissue homeodynamics. Antioxidants 2019, 8, 476. [Google Scholar] [CrossRef]
- Lugo-Huitron, R.; Blanco-Ayala, T.; Ugalde-Muniz, P.; Carrillo-Mora, P.; Pedraza-Chaverri, J.; Silva-Adaya, D.; Maldonado, P.D.; Torres, I.; Pinzon, E.; Ortiz-Islas, E.; et al. On the antioxidant properties of kynurenic acid: Free radical scavenging activity and inhibition of oxidative stress. Neurotoxicol. Teratol. 2011, 33, 538–547. [Google Scholar] [CrossRef]
- Kennedy, P.J.; Cryan, J.F.; Dinan, T.G.; Clarke, G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology 2017, 112, 399–412. [Google Scholar] [CrossRef]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef]
- Hoban, A.E.; Stilling, R.M.; Moloney, G.M.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F.; Clarke, G. Microbial regulation of microRNA expression in the amygdala and prefrontal cortex. Microbiome 2017, 5, 102. [Google Scholar] [CrossRef]
- Lin, P.; Li, D.; Shi, Y.; Li, Q.; Guo, X.; Dong, K.; Chen, Q.; Lou, X.; Li, Z.; Li, P.; et al. Dysbiosis of the gut microbiota and kynurenine (Kyn) pathway activity as potential biomarkers in patients with major depressive disorder. Nutrients 2023, 15, 1752. [Google Scholar] [CrossRef]
- Marin, I.A.; Goertz, J.E.; Ren, T.; Rich, S.S.; Onengut-Gumuscu, S.; Farber, E.; Wu, M.; Overall, C.C.; Kipnis, J.; Gaultier, A. Microbiota alteration is associated with the development of stress-induced despair behavior. Sci. Rep. 2017, 7, 43859. [Google Scholar] [CrossRef]
- Pirozzi, C.; Coretti, L.; Opallo, N.; Bove, M.; Annunziata, C.; Comella, F.; Turco, L.; Lama, A.; Trabace, L.; Meli, R.; et al. Palmitoylethanolamide counteracts high-fat diet-induced gut dysfunction by reprogramming microbiota composition and affecting tryptophan metabolism. Front. Nutr. 2023, 10, 1143004. [Google Scholar] [CrossRef]
- Bortolotti, P.; Hennart, B.; Thieffry, C.; Jausions, G.; Faure, E.; Grandjean, T.; Thepaut, M.; Dessein, R.; Allorge, D.; Guery, B.P.; et al. Tryptophan catabolism in Pseudomonas aeruginosa and potential for inter-kingdom relationship. BMC Microbiol. 2016, 16, 137. [Google Scholar] [CrossRef]
- Han, Q.; Fang, J.; Li, J. Kynurenine aminotransferase and glutamine transaminase K of Escherichia coli: Identity with aspartate aminotransferase. Biochem. J. 2001, 360, 617–623. [Google Scholar] [CrossRef]
- Jansma, J.; Chatziioannou, A.C.; Castricum, K.; van Hemert, S.; El Aidy, S. Metabolic network construction reveals probiotic-specific alterations in the metabolic activity of a synthetic small intestinal community. mSystems 2023, 8, e0033223. [Google Scholar] [CrossRef]
- Wogulis, M.; Chew, E.R.; Donohoue, P.D.; Wilson, D.K. Identification of formyl kynurenine formamidase and kynurenine aminotransferase from Saccharomyces cerevisiae using crystallographic, bioinformatic and biochemical evidence. Biochemistry 2008, 47, 1608–1621. [Google Scholar] [CrossRef]
- Xie, R.; Jiang, P.; Lin, L.; Jiang, J.; Yu, B.; Rao, J.; Liu, H.; Wei, W.; Qiao, Y. Oral treatment with Lactobacillus reuteri attenuates depressive-like behaviors and serotonin metabolism alterations induced by chronic social defeat stress. J. Psychiatr. Res. 2020, 122, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Huang, L.; Zhang, C.; Zhang, L.; Xia, X.; Zhong, Z.; Wang, B.; Wang, Y.; Man Hoi, M.P.; Ding, W.; et al. Gut commensal-derived butyrate reverses obesity-induced social deficits and anxiety-like behaviors via regulation of microglial homeostasis. Eur. J. Pharmacol. 2021, 908, 174338. [Google Scholar] [CrossRef]
- Li, C.; Su, Z.; Chen, Z.; Cao, J.; Liu, X.; Xu, F. Lactobacillus reuteri strain 8008 attenuated the aggravation of depressive-like behavior induced by CUMS in high-fat diet-fed mice through regulating the gut microbiota. Front. Pharmacol. 2023, 14, 1149185. [Google Scholar] [CrossRef]
- Montgomery, T.L.; Eckstrom, K.; Lile, K.H.; Caldwell, S.; Heney, E.R.; Lahue, K.G.; D’Alessandro, A.; Wargo, M.J.; Krementsov, D.N. Lactobacillus reuteri tryptophan metabolism promotes host susceptibility to CNS autoimmunity. Microbiome 2022, 10, 198. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of lactobacillus reuteri in human health and diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Sgritta, M.; Dooling, S.W.; Buffington, S.A.; Momin, E.N.; Francis, M.B.; Britton, R.A.; Costa-Mattioli, M. Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 2019, 101, 246–259 e246. [Google Scholar] [CrossRef]
- Sovijit, W.N.; Sovijit, W.E.; Pu, S.; Usuda, K.; Inoue, R.; Watanabe, G.; Yamaguchi, H.; Nagaoka, K. Ovarian progesterone suppresses depression and anxiety-like behaviors by increasing the Lactobacillus population of gut microbiota in ovariectomized mice. Neurosci. Res. 2021, 168, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Segawa, H.; Miyamoto, K.; Uchino, H.; Takeda, E.; Endou, H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J. Biol. Chem. 1998, 273, 23629–23632. [Google Scholar] [CrossRef]
- Segawa, H.; Fukasawa, Y.; Miyamoto, K.; Takeda, E.; Endou, H.; Kanai, Y. Identification and functional characterization of a Na+-independent neutral amino acid transporter with broad substrate selectivity. J. Biol. Chem. 1999, 274, 19745–19751. [Google Scholar] [CrossRef]
- Sekine, A.; Kuroki, Y.; Urata, T.; Mori, N.; Fukuwatari, T. Inhibition of large neutral amino acid transporters suppresses kynurenic acid production via Inhibition of kynurenine uptake in rodent brain. Neurochem. Res. 2016, 41, 2256–2266. [Google Scholar] [CrossRef]
- Lam, H.; Oh, D.C.; Cava, F.; Takacs, C.N.; Clardy, J.; de Pedro, M.A.; Waldor, M.K. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 2009, 325, 1552–1555. [Google Scholar] [CrossRef]
- Lupoli, T.J.; Tsukamoto, H.; Doud, E.H.; Wang, T.S.; Walker, S.; Kahne, D. Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc. 2011, 133, 10748–10751. [Google Scholar] [CrossRef]
- Guidetti, P.; Amori, L.; Sapko, M.T.; Okuno, E.; Schwarcz, R. Mitochondrial aspartate aminotransferase: A third kynurenate-producing enzyme in the mammalian brain. J. Neurochem. 2007, 102, 103–111. [Google Scholar] [CrossRef]
- Han, Q.; Cai, T.; Tagle, D.A.; Li, J. Structure, expression, and function of kynurenine aminotransferases in human and rodent brains. Cell. Mol. Life Sci. 2010, 67, 353–368. [Google Scholar] [CrossRef]
- Rossi, F.; Valentina, C.; Garavaglia, S.; Sathyasaikumar, K.V.; Schwarcz, R.; Kojima, S.; Okuwaki, K.; Ono, S.; Kajii, Y.; Rizzi, M. Crystal structure-based selective targeting of the pyridoxal 5’-phosphate dependent enzyme kynurenine aminotransferase II for cognitive enhancement. J. Med. Chem. 2010, 53, 5684–5689. [Google Scholar] [CrossRef]
- Kozak, R.; Campbell, B.M.; Strick, C.A.; Horner, W.; Hoffmann, W.E.; Kiss, T.; Chapin, D.S.; McGinnis, D.; Abbott, A.L.; Roberts, B.M.; et al. Reduction of brain kynurenic acid improves cognitive function. J. Neurosci. 2014, 34, 10592–10602. [Google Scholar] [CrossRef]
- Blanco-Ayala, T.; Sathyasaikumar, K.V.; Uys, J.D.; Perez-de-la-Cruz, V.; Pidugu, L.S.; Schwarcz, R. N-acetylcysteine inhibits kynurenine aminotransferase II. Neuroscience 2020, 444, 160–169. [Google Scholar] [CrossRef]
- Hodgkins, P.S.; Wu, H.Q.; Zielke, H.R.; Schwarcz, R. 2-Oxoacids regulate kynurenic acid production in the rat brain: Studies in vitro and in vivo. J. Neurochem. 1999, 72, 643–651. [Google Scholar] [CrossRef]
- Guidetti, P.; Okuno, E.; Schwarcz, R. Characterization of rat brain kynurenine aminotransferases I and II. J. Neurosci. Res. 1997, 50, 457–465. [Google Scholar] [CrossRef]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. D-amino acids trigger biofilm disassembly. Science 2010, 328, 627–629. [Google Scholar] [CrossRef]
- Perez-de la Cruz, V.; Amori, L.; Sathyasaikumar, K.V.; Wang, X.D.; Notarangelo, F.M.; Wu, H.Q.; Schwarcz, R. Enzymatic transamination of D-kynurenine generates kynurenic acid in rat and human brain. J. Neurochem. 2012, 120, 1026–1035. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Notarangelo, F.M.; Wang, J.Z.; Schwarcz, R. Kynurenic acid and 3-hydroxykynurenine production from D-kynurenine in mice. Brain Res. 2012, 1455, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Crozier-Reabe, K.R.; Phillips, R.S.; Moran, G.R. Kynurenine 3-monooxygenase from Pseudomonas fluorescens: Substrate-like inhibitors both stimulate flavin reduction and stabilize the flavin-peroxo intermediate yet result in the production of hydrogen peroxide. Biochemistry 2008, 47, 12420–12433. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.S.; Anderson, A.D.; Gentry, H.G.; Guner, O.F.; Bowen, J.P. Substrate and inhibitor specificity of kynurenine monooxygenase from Cytophaga hutchinsonii. Bioorg. Med. Chem. Lett. 2017, 27, 1705–1708. [Google Scholar] [CrossRef] [PubMed]
- Hayaishi, O.; Stanier, R.Y. The kynureninase of Pseudomonas fluorescens. J. Biol. Chem. 1952, 195, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.S. Structure, mechanism, and substrate specificity of kynureninase. Biochim. Biophys. Acta 2011, 1814, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Zhuravlev, A.V.; Zakharov, G.A.; Shchegolev, B.F.; Savvateeva-Popova, E.V. Antioxidant properties of kynurenines: Density functional theory calculations. PLoS Comput. Biol. 2016, 12, e1005213. [Google Scholar] [CrossRef]
- Romero, R.M.; Roberts, M.F.; Phillipson, J.D. Anthranilate synthase in microorganisms and plants. Phytochemistry 1995, 39, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.L.; Tsuchida, M.; Adelberg, E.A. The transamination of kynurenine. J. Biol. Chem. 1953, 203, 205–211. [Google Scholar] [CrossRef]
- Kuc, D.; Zgrajka, W.; Parada-Turska, J.; Urbanik-Sypniewska, T.; Turski, W.A. Micromolar concentration of kynurenic acid in rat small intestine. Amino Acids 2008, 35, 503–505. [Google Scholar] [CrossRef]
- Blanco Ayala, T.; Lugo Huitron, R.; Carmona Aparicio, L.; Ramirez Ortega, D.; Gonzalez Esquivel, D.; Pedraza Chaverri, J.; Perez de la Cruz, G.; Rios, C.; Schwarcz, R.; Perez de la Cruz, V. Alternative kynurenic acid synthesis routes studied in the rat cerebellum. Front. Cell Neurosci. 2015, 9, 178. [Google Scholar] [CrossRef]
- Luchowski, P.; Urbanska, E.M. SNAP and SIN-1 increase brain production of kynurenic acid. Eur. J. Pharmacol. 2007, 563, 130–133. [Google Scholar] [CrossRef]
- Kocki, T.; Luchowski, P.; Luchowska, E.; Wielosz, M.; Turski, W.A.; Urbanska, E.M. L-cysteine sulphinate, endogenous sulphur-containing amino acid, inhibits rat brain kynurenic acid production via selective interference with kynurenine aminotransferase II. Neurosci. Lett. 2003, 346, 97–100. [Google Scholar] [CrossRef]
- Hodgkins, P.S.; Schwarcz, R. Metabolic control of kynurenic acid formation in the rat brain. Dev. Neurosci. 1998, 20, 408–416. [Google Scholar] [CrossRef]
- Kaszaki, J.; Erces, D.; Varga, G.; Szabo, A.; Vecsei, L.; Boros, M. Kynurenines and intestinal neurotransmission: The role of N-methyl-D-aspartate receptors. J. Neural Transm. 2012, 119, 211–223. [Google Scholar] [CrossRef]
- Dolecka, J.; Urbanik-Sypniewska, T.; Skrzydlo-Radomanska, B.; Parada-Turska, J. Effect of kynurenic acid on the viability of probiotics in vitro. Pharmacol. Rep. 2011, 63, 548–551. [Google Scholar] [CrossRef]
- Turski, M.P.; Turska, M.; Paluszkiewicz, P.; Parada-Turska, J.; Oxenkrug, G.F. Kynurenic Acid in the digestive system-new facts, new challenges. Int. J. Tryptophan Res. 2013, 6, 47–55. [Google Scholar] [CrossRef]
- Wirthgen, E.; Hoeflich, A.; Rebl, A.; Gunther, J. Kynurenic Acid: The janus-faced role of an immunomodulatory tryptophan metabolite and its link to pathological conditions. Front. Immunol. 2017, 8, 1957. [Google Scholar] [CrossRef]
- Speciale, C.; Schwarcz, R. Uptake of kynurenine into rat brain slices. J. Neurochem. 1990, 54, 156–163. [Google Scholar] [CrossRef]
- Giorgini, F.; Huang, S.Y.; Sathyasaikumar, K.V.; Notarangelo, F.M.; Thomas, M.A.; Tararina, M.; Wu, H.Q.; Schwarcz, R.; Muchowski, P.J. Targeted deletion of kynurenine 3-monooxygenase in mice: A new tool for studying kynurenine pathway metabolism in periphery and brain. J. Biol. Chem. 2013, 288, 36554–36566. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).